Abstract
Mutations in the human FBN1 gene cause Marfan syndrome, a complex disease affecting connective tissues but with a highly variable phenotype. To identify genes that might participate in epistatic interactions with FBN1, and could therefore explain the observed phenotypic variability, we have looked for genes that are co-expressed with Fbn1 in the mouse. Microarray expression data derived from a range of primary mouse cells and cell lines were analysed using the network analysis tool BioLayout Express3D. A cluster of 205 genes, including Fbn1, were selectively expressed by mouse cell lines of different mesenchymal lineages and by mouse primary mesenchymal cells (preadipocytes, myoblasts, fibroblasts, osteoblasts). Promoter analysis of this gene set identified several candidate transcriptional regulators. Genes within this co-expressed cluster are candidate genetic modifiers for Marfan syndrome and for other connective tissue diseases.
Keywords: Fibrillin-1, Marfan syndrome, mesenchyme, connective tissue, extracellular matrix
Introduction
Marfan syndrome (OMIM 154700) is an autosomal dominant condition that affects connective tissue.1, 2, 3, 4 Individuals show overgrowth of the long bones, lack of adipose and muscle tissue, and abnormalities of the eyes and skin. The major cause of morbidity and mortality is dilatation and dissection of the ascending aorta. Marfan syndrome is usually associated with mutations in the FBN1 gene (OMIM 134797), encoding the microfibrillar protein, fibrillin-1.2, 5, 6 A bovine Marfan-like syndrome (OMIA 1204) is also due to a mutation in FBN1. Homozygous mice that lack a functional Fbn1 gene have some manifestations similar to Marfan syndrome in humans, although the heterozygous phenotype is mild.5 In addition, a transgenic mouse line carrying a mutation known to cause severe disease in humans has a dose-dependent phenotype showing aspects of Marfan syndrome.7 A homozygous lethal natural mutation involving duplication of exons 17–40 of the mouse Fbn1 gene8 causes the tight skin (Tsk) phenotype in which heterozygotes have abnormalities of skin, viscera, lungs, cartilage, bone, heart and tendons,9 with some characteristics of Marfan syndrome such as overgrowth of long bones.
The tissues primarily affected by FBN1 mutation (including bone, aorta and pulmonary artery, mitral valve, zonullar fibres of the eye, dura mater, skin and adipose) contain cells of mesenchymal origin, which synthesize connective tissue extracellular matrix (ECM), composed of fibrous proteins and glycosaminoglycans. The ECM provides strength and elasticity for these tissues. Fibrillin-1 is the major structural component of the extracellular microfibrils of the ECM10 and also seems to be involved in sequestering the growth factor TGFβ in inactive form.5, 11, 12 In adults, mesenchymal cells derive from stem cells residing in the bone marrow and mesenchymal tissues.13, 14 These stem cells retain the ability to differentiate into cells of connective tissue lineages, including adipocytes, osteoblasts, chondrocytes, smooth and skeletal muscle, endothelial cells of blood vessels and fibroblasts (reviewed in Barry and Murphy14). Differentiation of mesenchymal cells into specific cell types requires induction of a range of transcription factors14 and may also involve interaction with cells of monocyte origin.15 During organogenesis, mesenchymal cells can also undergo transition to epithelial phenotype (mesenchymal–epithelial transition), with concomitant inhibition of mesenchyme-specific genes and activation of genes required to form intercellular adhesions characteristic of epithelium. The transition between the two states is regulated by a number of cellular factors, especially TGFβ family members.16, 17 The actions of TGFβ on mesenchymal cells are mediated through transcription factors such as SNAIL and SLUG (encoded by SNAI1 and SNAI2 genes),18 and result in expression of mesenchymal genes and suppression of the epithelial marker E-cadherin.
The phenotype of Marfan syndrome is extremely variable, even among family members carrying the same mutation (see refs.4, 19, 20). Potential modifier genes for Marfan syndrome are likely to be found in the network of genes that are co-expressed in tissues affected by FBN1 mutation. Such genes would also be strong candidates for a role in diseases with related phenotypes. In this article, we identify and analyse genes that are stringently co-regulated with FBN1.
Materials and methods
Identification and annotation of an FBN1-associated cluster of genes
The analysis was performed on publicly available gene expression data (to which we contributed) generated from 44 mouse cell types and 2 mouse organs15, 21 (Supplementary Table S1) using the Affymetrix MOE430_2 GeneChip and normalized using MAS5 (Affymetrix, Santa Clara, CA, USA). The data were accessed through GEO DataSets (accession number GSE10246). Correlation networks were constructed from the data on the basis of pairwise Pearson's correlation relationships. A network graph comprising 8578 nodes (probes) and 153 418 edges was generated using BioLayout Express3D.22 The resulting graph was then clustered using the Markov Clustering algorithm at an MCL inflation value of 1.7.23 Clustering was also performed on expression data from mouse tissues.24 Because the initial data did not include chondrocytes, which are likely to be involved in the skeletal phenotype of Marfan syndrome, the expression of cluster genes was also examined in data from a published study of chondrocyte differentiation (cultured limb bud mesenchymal cells; GEO Profiles accession no. GDS1865).25 In addition, we considered data sets from developing mouse kidney (E12.5; GEO Profiles accession no. GDS1583)26 and from developing mouse gastrointestinal tract (E18.5; GEO Profiles accession no. GDS2699).27
Location and function of cluster genes
Genes in the cluster of interest were assessed for recognized homologies, cellular localization and function using publicly available databases (Ensembl, NCBI). Possible or verified involvement in disease was determined by searching the Online Mendelian Inheritance in Man (OMIM) and Online Mendelian Inheritance in Animals (OMIA) databases on the NCBI website.
Determination of functional transcription factor binding sites in promoter regions of Fbn1-associated cluster genes
The Affymetrix MOE430_2 probe set was mapped to mouse RefSeq genes and the beginning of RefSeq was taken as a predicted transcription start site. Bioinformatic analysis of motif activity and motif target predictions were performed as described previously.28 All genes represented on the microarray, which had been allocated a RefSeq (12752 in total), were classified as being either among the 205 genes of the Fbn1-associated cluster or not within the set. The proportion of genes with a z(p, m) score of greater than 1 for each transcription factor binding motif m was calculated for the two groups and a z-value for this difference was determined. This provides a measure of overrepresentation of predicted targets of the transcription factor in the mesenchymal cluster relative to other genes.
Results
Identification and annotation of Fbn1-associated genes in proliferating cells
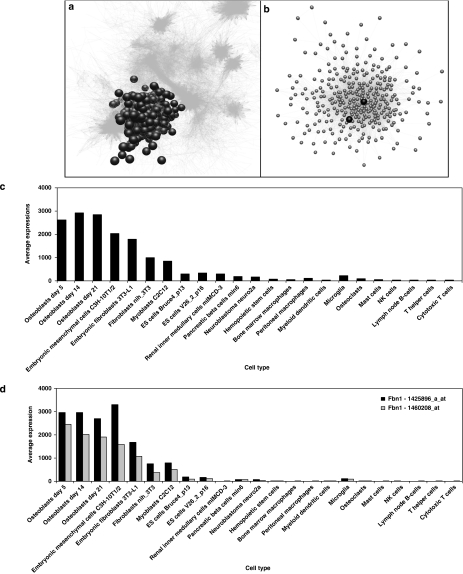
To identify genes that were strictly co-regulated with mouse Fbn1 in a cell-autonomous manner, we focused on a large data set derived from primary mouse cells, including primary calvarial osteoblasts undergoing differentiation and a range of haemopoietic cell types (see Supplementary Table S1), produced as described previously.15 BioLayout Express3D analysis of the cell line data generated 480 clusters containing at least five nodes on the basis of their connectivity within the co-expression network graph. The third largest cluster contained 304 transcripts, including two probes for Fbn1 (1425896_a_at and 1460208_at) (Figure 1a and b). In total, 205 different genes were represented by the 304 probe sets. The full list of genes represented in this cluster is available in Supplementary Material (Supplementary Table S2). This cluster was enriched for genes associated with the ECM. Fbn1 was a central gene in the cluster (Figure 1b), which was termed the Fbn1-associated cluster. The two Fbn1 probes were correlated (at r≥0.90) with 241 and 229 probes. Figure 1c shows the averaged expression in 23 cell types of the 304 probes of the cluster. Cells with a high expression of genes in this cluster included mesenchymal cell types such as preadipocytes, myoblasts, fibroblasts and osteoblasts. Fbn1 had a high expression in mesenchymal cells and minimal expression in other cell types (Figure 1d). Two other probes for Fbn1 (1438870_at and 1458593_at) did not cluster with this set of genes. This is probably because the latter two probes detected sequences with a very low expression and high variability (see expression profiles on BioGPS). Both mapped to intronic sequences (Affymetrix website) that have a low frequency of transcript initiation, indicating that these probes may detect rare variant Fbn1 transcripts that do not show clustering with the major Fbn1 probe sets. Probes for the other mouse fibrillin gene, Fbn2, which has overlapping functions with Fbn1,29 did not cluster with Fbn1 in this data set. Fbn2 showed expression only in osteoblasts and C3H 10T1/2 cells, and is therefore likely to function more specifically in bone.
Figure 1.
Characteristics of the Fbn1-associated expression cluster. (a) A three-dimensional image of the Fbn1-associated cluster (nodes shown by black spheres, edges by grey lines) within the network. Other clusters are shown by edges only. (b) A two-dimensional image of the Fbn1-associated cluster with the two Fbn1 probe sets shown as black spheres. (c) Normalized expression of genes in representative cell types, averaged across all probes in the cluster. The means of two experiments performed in triplicate are shown. (d) Expression of two Fbn1 probes, 1460208_at (black) and 1425896_a_at (grey). The means of two experiments performed in triplicate are shown for each probe. Data are available at GEO DataSets (accession number GSE10246).
Fbn1-associated cluster genes in other data sets
The cell lines assessed in the initial analysis did not include all mesenchymal cell types that would be found within tissues, nor all states of mesenchymal differentiation. To identify a subset of genes that were robustly expressed in mesenchymal tissues rather than cell lines, we clustered expression patterns across tissues in the publicly available GNF1M data set of gene expression in mouse tissues.24 Gene expression showed more diversity across these tissues and there was substantially greater noise in this data set, as evidenced by smaller clusters and overall lower correlation coefficients. Hence, a lower correlation level (at r≥0.75) was required to detect associations. A total of 119 genes clustered with Fbn1 in this analysis (Supplementary Table S3). Of these, 24 overlapped with the cluster derived from proliferating cells (indicated in Supplementary Table S3). Classic ECM genes such as Eln, Fbln2, Mfap4, Mfap5 and Fbn2 also clustered with Fbn1 in this analysis of expression in tissues.
One major mesenchyme-derived cell type excluded from the cellular data was the chondrocyte. We therefore examined a published study of the differentiation of primary chondrocytes derived from embryonic footpads.25 Results for 235 of the 304 probes were available, representing 160 different genes. In all, 81% of these genes, including Fbn1, were in the highest 25% of expression at most or all time points, extending the view that these genes are co-expressed by proliferating mesenchyme, regardless of lineage.
As noted above, mesenchyme–epithelial transition is a key event in organogenesis. The transition has been analysed separately in developing mouse kidney26 and gastrointestinal tract.27 Fbn1 expression was strongly associated with mesenchyme in these data sets. Fbn1-associated cluster genes such as Bgn (biglycan), Cald1 (caldesmon 1), Col1a2 (collagen type 1-α 2 subunit), Il6st (interleukin-6 signal transducer), Ror1 (receptor tyrosine kinase-like orphan receptor 1), Sparc (osteonectin; secreted protein, acidic, cysteine rich) and Timp 2 (tissue inhibitor of metalloproteinases 2) showed a similar pattern of expression to Fbn1 in both the data sets, whereas others were profile neighbours of Fbn1 in one or the other data set (not shown).
Cellular location and function of Fbn1-associated cluster genes
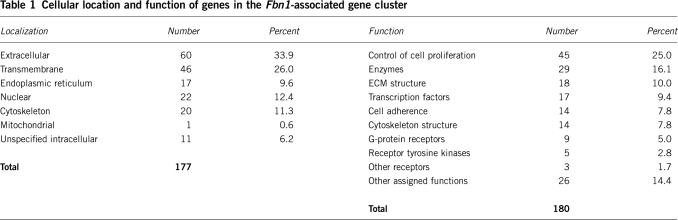
As summarized in Table 1, 171 members of the Fbn1-associated cluster could be assigned a cellular location on the basis of experimental evidence or electronic annotation. The majority were extracellular but a substantial number were involved in secretion. For example, 10% of the annotated genes encoded proteins of the endoplasmic reticulum (including trafficking proteins and molecular chaperones), indicating a surprising level of target specificity for these processing proteins. Table 1 also shows that 181 genes could be assigned a function (Table 1). The largest group (25%) was a broad category of proteins involved in regulating cell size and number. A total of 10% were involved in ECM structure. There were 17 (9.4%) genes encoding known or putative transcription factors, including some families (SLUG/SNAIL, TWIST, PRRX, NFAT, ID, SOX) known to regulate mesenchyme differentiation or function. There were nine genes for G-protein-coupled receptors and five for receptor tyrosine kinases. Most of the receptors had unknown ligands. Four genes had no informative annotation, with no similarity to known genes or assignable function or location.
Table 1. Cellular location and function of genes in the Fbn1-associated gene cluster.
| Localization | Number | Percent | Function | Number | Percent |
|---|---|---|---|---|---|
| Extracellular | 60 | 33.9 | Control of cell proliferation | 45 | 25.0 |
| Transmembrane | 46 | 26.0 | Enzymes | 29 | 16.1 |
| Endoplasmic reticulum | 17 | 9.6 | ECM structure | 18 | 10.0 |
| Nuclear | 22 | 12.4 | Transcription factors | 17 | 9.4 |
| Cytoskeleton | 20 | 11.3 | Cell adherence | 14 | 7.8 |
| Mitochondrial | 1 | 0.6 | Cytoskeleton structure | 14 | 7.8 |
| Unspecified intracellular | 11 | 6.2 | G-protein receptors | 9 | 5.0 |
| Receptor tyrosine kinases | 5 | 2.8 | |||
| Total | 177 | Other receptors | 3 | 1.7 | |
| Other assigned functions | 26 | 14.4 | |||
| Total | 180 |
The role of genes of the Fbn1-associated cluster in disease was examined by assessing entries in OMIM. Of 168 genes with an entry, 60 were associated with a phenotype in mouse (41) or human (29) (Supplementary Table S4). Of these, bones, skin, eyes and blood vessels were most frequently affected in humans, and bones, blood vessels and lung were most frequently noted for mouse. Eight mouse knockout models resulted in embryonic lethality. The results are consistent with a critical role for these genes in the development of the ECM.
Determination of common functional transcription factor binding sites in promoter regions of Fbn1-associated cluster genes
To assess the basis for their apparent co-regulation, we subjected the 205 genes of the Fbn1-associated cluster to an analysis of transcription factor binding sites.28 Table 2 lists the 15 transcription factors that had the highest positive correlations with the expression pattern of cluster genes. Comparison was also carried out between genes within the cluster and the remaining genes of the data set. Supplementary Table S5 shows the 65 transcription factor binding motifs that showed significant overrepresentation in the cluster genes. Seven transcription factor binding motifs showed a high correlation between activity and expression of the cluster genes and were consistently overrepresented in cluster gene promoters. The motifs were consensus sequences for binding proteins of the TEAD, RP58, MAZ, KLF4, IK1/IK2, BLIMP1 and CIZ families (Table 2). No Fbn1-associated cluster gene was significantly (Z>2) associated with activity of all seven of these motifs, and Fbn1 alone was associated with six. Five of the genes were significantly associated with activity of five of these motifs and fifteen were associated with four.
Table 2. Transcription factor motifs showing the highest correlations of activity with expression of Fbn1-associated cluster genes and with Fbn1.
Identification of genes highly correlated with Fbn1
When the initial clustering analysis using cell line data was repeated at a higher stringency of r≥0.95, 46 probes (31 genes) were found to be in the same cluster as Fbn1 (Supplementary Table S6). Twelve of these genes were annotated as being located in the ECM, extracellular region or extracellular space. There were five recognized transcription factors and seven receptors. Eight of these genes had no or limited annotation, including a TGFβ-induced transcript (Tgfb1i1), a steroid-sensitive coiled-coil domain protein (Ccdc80) and a transmembrane protein (Tmem45a).
As noted above, in this study Fbn1 expression showed strong association (Z>6.5) with the activity of six of the seven transcription factor motifs identified as having high activity in the cluster (for TEAD, RP58, MAZ, KLF4, BLIMP1 and CIZ family members; Table 2). Three genes (Loxl3, Nfatc4 and Atoh8) were associated with activity of five of the six motifs in common with Fbn1 and 11 were associated with four of the six motifs in common with Fbn1 (Nuak1, Col1a2, Col3a1, Gas1, Serpinh1, Cdh11, Thbs2, Tpm1, Pcdh18, Boc, Grp23). In addition, Fbn1 expression was significantly associated (Z>4) with a number of other motifs found to be overrepresented in the cluster. These included binding motifs for AP-4, MAZR, Broad Complex and SP1-gershenzon (Table 2). Five genes (Gas1, Capn6, Atoh8, Col1a2 and Snai2) were associated (Z>2) with 10–15 of the same factors as Fbn1.
Discussion
This analysis of gene expression data revealed that the mouse Fbn1 gene was in a cluster of 205 genes representing a lineage-independent expression signature for mesenchymal cells. Transcription factors binding TEAD, CIZ, RP58, KLF4, MAZ, BLIMP1 and IK1/IK2 sites are candidate regulators of this Fbn1-associated cluster. Several of these have known roles in mesenchymal cell types. MAZ (myc-associated zinc-finger protein) has been shown to regulate muscle-specific gene expression.30 RP58 has an essential role in skeletal myogenesis,31 and BLIMP1 is involved in myocyte differentiation.32 CIZ is implicated in regulation of bone mass biology.33 TEAD2 and TEAD4, although not previously implicated in mesenchyme biology, had a similar expression pattern to the genes of the Fbn1 cluster. KLF family members and IK1/IK2 are associated with transcriptional repression in haemopoietic cells, and their function may be to prevent ectopic expression of the Fbn1-associated cluster genes in non-mesenchymal cells. Fbn1 was associated with six of these seven transcription factor motifs, the only gene with this level of association. The Fbn1-associated cluster itself includes genes for a number of transcriptional regulators that are known to be involved in epithelial–mesenchyme transition, including the Snai1, Snai2, Prrx 1, Prrx2 and Twist1 genes. Our recent analysis34 detected motifs for PRRX family members in the Fbn1 proximal promoter region.
The study is limited by a number of factors. The published data were from cells of a single mouse strain, and it would be interesting to use a different strain, especially as there is considerable between-strain variability in gene expression (see Wells et al35 and mouse e-QTL data on BioGPS); we would predict that the same genes would continue to cluster on the basis of expression pattern, even though those patterns might vary with different strains. Many members of this Fbn1 cluster (including BGN, key collagen genes, SERPINH1 and the transcription factor genes SNAI2, PRRX1 and TWIST1) were also co-expressed with FBN1 in human tumours and tissues (TC Freeman and TN Doig, unpublished results), as were several minimally annotated genes such as CD248, FKBP9, LRRC17 and TGFB1I1. We did not assess all cell types that are abnormal in Marfan syndrome. For example, the main morbidity comes from dissection of the aorta, and there were no aortic cells in the study, nor were there cells from the anterior segment of the eye or from dura mater. If these cell types were included, some of the genes would drop out of the cluster and those that remained would represent tightly co-regulated genes that are powerful candidates for a role in modulating the Marfan syndrome phenotype.
The rationale behind our analysis is that genes that are co-expressed with Fbn1 are candidate modifiers of the effects of FBN1 mutation in humans and may contribute to other diseases of connective tissue with similar phenotypes. In spite of the limitations, several examples validate this rationale. For instance, a strong association of Fbn1 with the Lox and Bgn genes was noted. Biglycan protein (encoded by Bgn) has been reported to stimulate synthesis of fibrillin-1 in pressure-induced renal injury and may have a more general role in assembly of connective tissue.36 No disease has been associated with BGN mutation in humans but Bgn-deficient mice have a skeletal phenotype.37 Lysyl oxidase (Lox gene) may be important in overall assembly of elastic microfibrils (reviewed in Wagenseil and Mecham38) and may be involved in preparing tumour cells for metastasis.39 The human homologue of another of the cluster genes, SERPINH1, was recently implicated in a recessive form of osteogenesis imperfecta, a bone disease.40 Others within the cluster are not well characterized. They include novel transcription factors, G-protein-coupled receptors, nuclear receptors and receptor tyrosine kinases that are clearly potential drug targets and may be important in cell signalling during differentiation of mesenchymal cell types. A group of novel genes encoding hypothetical proteins was also present. The functions of these genes can now be inferred from their co-expression in the cluster,41 and they clearly warrant a detailed characterization in cells of mesenchymal lineage, and also consideration as modifiers of the Marfan phenotype or as candidate genes in human connective tissue diseases.
Acknowledgments
This work was supported by a grant-in-aid from the National Heart Foundation of Australia (G06B2527) and by the Biotechnology and Biological Sciences Council of the United Kingdom (Institutional Strategic Programme Grant to The Roslin Institute).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Web Resources
The URLs for data presented herein are as follows:
Affymetrix, http://www.affymetrix.com.
BioGPS, http://biogps.gnf.org.
BioLayout Express3D, http://www.biolayout.org.
Ensembl Human Genome Server, http://www.ensembl.org.
National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/.
NCBI Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/.
Online Mendelian Inheritance in Animals (OMIA), http://omia.angis.org.au/.
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/OMIM.
Supplementary Material
References
- Ramirez F, Dietz HC. Marfan syndrome: from molecular pathogenesis to clinical treatment. Curr Opin Genet Dev. 2007;17:252–258. doi: 10.1016/j.gde.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Pyeritz RE. The Marfan syndrome. Annu Rev Med. 2000;51:481–510. doi: 10.1146/annurev.med.51.1.481. [DOI] [PubMed] [Google Scholar]
- De Paepe A, Devereux RB, Dietz HC, Hennekam RC, Pyeritz RE. Revised diagnostic criteria for the Marfan syndrome. Am J Med Genet. 1996;62:417–426. doi: 10.1002/(SICI)1096-8628(19960424)62:4<417::AID-AJMG15>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Summers KM, West JA, Peterson MM, Stark D, McGill JJ, West MJ. Challenges in the diagnosis of Marfan syndrome. Med J Aust. 2006;184:627–631. doi: 10.5694/j.1326-5377.2006.tb00419.x. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Loeys B, Carta L, Ramirez F. Recent progress towards a molecular understanding of Marfan syndrome. Am J Med Genet C Semin Med Genet. 2005;139C:4–9. doi: 10.1002/ajmg.c.30068. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Arteaga-Solis E, Baldock C, et al. The molecular genetics of Marfan syndrome and related disorders. J Med Genet. 2006;43:769–787. doi: 10.1136/jmg.2005.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge DP, Biery NJ, Keene DR, et al. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest. 2004;114:172–181. doi: 10.1172/JCI20641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siracusa LD, McGrath R, Ma Q, et al. A tandem duplication within the fibrillin 1 gene is associated with the mouse tight skin mutation. Genome Res. 1996;6:300–313. doi: 10.1101/gr.6.4.300. [DOI] [PubMed] [Google Scholar]
- Green MC, Sweet HO, Bunker LE. Tight-skin, a new mutation of the mouse causing excessive growth of connective tissue and skeleton. Am J Pathol. 1976;82:493–512. [PMC free article] [PubMed] [Google Scholar]
- Jordan CD, Charbonneau NL, Sakai LY. Fibrillin microfibrils: connective tissue pathways that regulate shape and signaling. J Musculoskelet Neuronal Interact. 2006;6:366–367. [PubMed] [Google Scholar]
- Chaudhry SS, Cain SA, Morgan A, Dallas SL, Shuttleworth CA, Kielty CM. Fibrillin-1 regulates the bioavailability of TGFbeta1. J Cell Biol. 2007;176:355–367. doi: 10.1083/jcb.200608167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataatmadja M, West J, West M. Overexpression of transforming growth factor-beta is associated with increased hyaluronan content and impairment of repair in Marfan syndrome aortic aneurysm. Circulation. 2006;114:I371–I377. doi: 10.1161/CIRCULATIONAHA.105.000927. [DOI] [PubMed] [Google Scholar]
- Peng L, Jia Z, Yin X, et al. Comparative analysis of mesenchymal stem cells from bone marrow, cartilage and adipose tissue. Stem Cells Dev. 2008. [DOI] [PubMed]
- Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol. 2004;36:568–584. doi: 10.1016/j.biocel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Chang MK, Raggatt L-J, Alexander KA, et al. Osteal tissue macrophages are intercalated throughout human and mouse bone lining tissues and regulate osteoblast function in vitro and in vivo. J Immunol. 2008;181:1232–1244. doi: 10.4049/jimmunol.181.2.1232. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Nawshad A. Complexity in interpretation of embryonic epithelial–mesenchymal transition in response to transforming growth factor-signaling. Cells Tissues Organs. 2007;185:131–145. doi: 10.1159/000101314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Pimental ME, Runyan RB. Multiple transforming growth factor-beta isoforms and receptors function during epithelial–mesenchymal cell transformation in the embryonic heart. Cells Tissues Organs. 2007;185:146–156. doi: 10.1159/000101315. [DOI] [PubMed] [Google Scholar]
- Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- Summers KM, Xu D, West JA, et al. An integrated approach to management of Marfan syndrome caused by an FBN1 exon 18 mutation in an Australian Aboriginal family. Clin Genet. 2004;65:66–69. doi: 10.1111/j..2004.00186.x. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Pyeritz RE, Puffenberger EG, et al. Marfan phenotype variability in a family segregating a missense mutation in the epidermal growth factor-like motif of the fibrillin gene. J Clin Invest. 1992;89:1674–1680. doi: 10.1172/JCI115766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunol Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman TC, Goldovsky L, Brosch M, et al. Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput Biol. 2007;3:2032–2042. doi: 10.1371/journal.pcbi.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongen SV.Graph clustering by flow simulationPhD thesis, University of Utrecht, Utrecht2000
- Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CG, Appleton CT, Ulici V, Underhill TM, Beier F. Microarray analyses of gene expression during chondrocyte differentiation identifies novel regulators of hypertrophy. Mol Biol Cell. 2005;16:5316–5333. doi: 10.1091/mbc.E05-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ott KM, Yang J, Chen X, et al. Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol. 2005;16:1993–2002. doi: 10.1681/ASN.2004121127. [DOI] [PubMed] [Google Scholar]
- Li X, Madison BB, Zacharias W, Kolterud A, States D, Gumucio DL. Deconvoluting the intestine: molecular evidence for a major role of the mesenchyme in the modulation of signaling cross talk. Physiol Genomics. 2007;29:290–301. doi: 10.1152/physiolgenomics.00269.2006. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Forrest AR, van Nimwegen E, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta L, Pereira L, Arteaga-Solis E, et al. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J Biol Chem. 2006;281:8016–8023. doi: 10.1074/jbc.M511599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeda CL, Ranish JA, Hauschka SD. Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol Cell Biol. 2008;28:6521–6535. doi: 10.1128/MCB.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Ito Y, Ueno-Kudoh H, et al. A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev Cell. 2009;17:836–848. doi: 10.1016/j.devcel.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- Hayata T, Nakamoto T, Ezura Y, Noda M. Ciz, a transcription factor with a nucleocytoplasmic shuttling activity, interacts with C-propeptides of type I collagen. Biochem Biophys Res Commun. 2008;368:205–210. doi: 10.1016/j.bbrc.2008.01.040. [DOI] [PubMed] [Google Scholar]
- Summers KM, Bokil NJ, Baisden JM, et al. Experimental and bioinformatic characterisation of the promoter region of the Marfan syndrome gene, FBN1. Genomics. 2009;94:233–240. doi: 10.1016/j.ygeno.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Wells CA, Ravasi T, Faulkner GJ, et al. Genetic control of the innate immune response. BMC Immunol. 2003;4:5. doi: 10.1186/1471-2172-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L, Mihalik D, Babelova A, et al. Regulation of fibrillin-1 by biglycan and decorin is important for tissue preservation in the kidney during pressure-induced injury. Am J Pathol. 2004;165:383–396. doi: 10.1016/S0002-9440(10)63305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Bianco P, Fisher LW, et al. Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice. Nat Genet. 1998;20:78–82. doi: 10.1038/1746. [DOI] [PubMed] [Google Scholar]
- Wagenseil JE, Mecham RP. New insights into elastic fiber assembly. Birth Defects Res C Embryo Today. 2007;81:229–240. doi: 10.1002/bdrc.20111. [DOI] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Nicolau M, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Christiansen HE, Schwarze U, Pyott SM, et al. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86:389–398. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA, Summers KM, Raza S, Baillie JK, Freeman TC. Functional clustering and lineage markers: insights into cellular differentiation and gene function from large-scale microarray studies of purified primary cell populations. Genomics. 2010;95:328–338. doi: 10.1016/j.ygeno.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev Genet. 1998;22:43–55. doi: 10.1002/(SICI)1520-6408(1998)22:1<43::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Mahoney WM, Jr, Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–225. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Meng G, Suzuki K, et al. RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J Biol Chem. 1998;273:26698–26704. doi: 10.1074/jbc.273.41.26698. [DOI] [PubMed] [Google Scholar]
- Song J, Murakami H, Tsutsui H, et al. Structural organization and expression of the mouse gene for Pur-1, a highly conserved homolog of the human MAZ gene. Eur J Biochem. 1999;259:676–683. doi: 10.1046/j.1432-1327.1999.00081.x. [DOI] [PubMed] [Google Scholar]
- Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrousos GP, Kino T. Ikaros transcription factors: flying between stress and inflammation. J Clin Invest. 2005;115:844–848. doi: 10.1172/JCI24886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzat S, Yu S, Asa SL. The zinc finger Ikaros transcription factor regulates pituitary growth hormone and prolactin gene expression through distinct effects on chromatin accessibility. Mol Endocrinol. 2005;19:1004–1011. doi: 10.1210/me.2004-0432. [DOI] [PubMed] [Google Scholar]
- Thunyakitpisal P, Alvarez M, Tokunaga K, et al. Cloning and functional analysis of a family of nuclear matrix transcription factors (NP/NMP4) that regulate type I collagen expression in osteoblasts. J Bone Miner Res. 2001;16:10–23. doi: 10.1359/jbmr.2001.16.1.10. [DOI] [PubMed] [Google Scholar]
- Chang DH, Cattoretti G, Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev. 2002;117:305–309. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.