Abstract
Angelman syndrome (AS) is a human neurological disorder caused by lack of maternal UBE3A expression in the brain. UBE3A is known to function as both an ubiquitin-protein ligase (E3) and a coactivator for steroid receptors. Many ubiquitin targets, as well as interacting partners, of UBE3A have been identified. However, the pathogenesis of AS, and how deficiency of maternal UBE3A can upset cellular homeostasis, remains vague. In this study, we performed a genome-wide microarray analysis on the maternal Ube3a-deficient (Ube3am−/p+) AS mouse to search for genes affected in the absence of Ube3a. We observed 64 differentially expressed transcripts (7 upregulated and 57 downregulated) showing more than 1.5-fold differences in expression (P<0.05). Pathway analysis shows that these genes are implicated in three major networks associated with cell signaling, nervous system development and cell death. Using quantitative reverse-transcription PCR, we validated the differential expression of genes (Fgf7, Glra1, Mc1r, Nr4a2, Slc5a7 and Epha6) that show functional relevance to AS phenotype. We also show that the protein level of melanocortin 1 receptor (Mc1r) and nuclear receptor subfamily 4, group A, member 2 (Nr4a2) in the AS mice cerebellum is decreased relative to that of the wild-type mice. Consistent with this finding, expression of small-interfering RNA that targets Ube3a in P19 cells caused downregulation of Mc1r and Nr4a2, whereas overexpression of Ube3a results in the upregulation of Mc1r and Nr4a2. These observation help in providing insights into the genesis of neurodevelopmental phenotype of AS and highlight specific area for future research.
Keywords: E6AP, neurological disorder, Mc1r, Nr4a2
Introduction
Angelman syndrome (AS, OMIM 105830) is a human neurogenetic disorder affecting approximately 1 in 10 000–40 000 newborns.1, 2 AS patients show distinct dysmorphic facial features, inappropriate laughter, ataxia and motor dysfunction.3 Many patients also show seizures of variable magnitude, learning deficit and hypopigmentation.3 Molecular genetic studies show that AS is caused by the lack of functional UBE3A expression.4 UBE3A encodes the E6-AP ubiquitin ligase, and is found to be imprinted in a selected brain cell population such that only the maternal inherited allele is expressed.5 UBE3A functions mainly through targeting proteins for proteosomal degradation, and is also known to function as a coactivator for steroid hormone receptors.6, 7
In the brain neurons in which UBE3A is imprinted, it was found that UBE3A is localized in the nucleus, presynaptic and postsynaptic regions.6, 8 Ube3a-deficient mice showed long-term potentiation impairment and abnormal dendritic spine number and morphology.8, 9 Similarly, in Drosophila, either deficiency or overexpression of dUBE3A leads to reduced dendritic branching.10 These reports suggested that UBE3A may exert its effect locally in the synaptic region regulating dendritic spine development, synaptic plasticity and functions. Ubiquitination targets of UBE3A, such as p53, epithelial cell-transforming sequence 2 oncogene (Ect2) and tuberous sclerosis 2 (TSC2), and interacting partners, such as HSP70, have been identified.6, 11, 12, 13, 14 However, the role of UBE3A in the brain and how its deficiency can result in AS remain unclear.
To find out which genes are affected in the absence of functional Ube3a in the brain, we performed a genome-wide microarray expression analysis on wild-type and Ube3am−/p+ mice. We identified 64 genes showing greater than 1.5-fold differences in expression (P<0.05) in the Ube3am−/p+ mouse. Pathway analysis reveals that they are involved in three major networks, including cell signaling, nervous system development and cell death. Expression of functionally relevant candidates from each pathway was validated using quantitative reverse-transcription PCR (qRT-PCR). Among them, both melanocortin 1 receptor (Mc1r), which is shown to have a neuroprotective effect in the brain, and nuclear receptor subfamily 4, group A, member 2 (Nr4a2), which is critical for survival of dopamine neurons and whose expression can be induced by Mc1r, are downregulated.15, 16, 17 We have confirmed that the decline of Mc1r and Nr4a2 at transcriptional level is reflected at the protein level in the Ube3am−/p+ mouse. Using RNA interfering approach, we have further shown that shRNA-mediated knockdown of Ube3a in P19 cells leads to downregulation of Mc1r and Nr4a2. In contrast, overexpression of Ube3a in P19 cells results in the upregulation of Mc1r and Nr4a2 mRNA levels. These results provide informative molecular insights on the pathogenesis of AS, as well as the functions of Ube3a.
Materials and methods
Ethics statement/mouse strains
All animal work was maintained, performed and approved by the School of Biological Sciences Animal Facility based on guidelines from the Institutional Animal Care and Use Committee. Ube3am−/p+ mice were generated and confirmed using PCR as previously described.9
Microarray sample preparation and hybridization
Cerebellum total RNA were extracted from 6-week-old female wild-type and Ube3am−/p+ mice using Trizol reagents (Invitrogen Inc., Carlsbad, CA, USA) according to the manufacturer's protocol. Starting total RNA (1 μg) was used from each sample for the Affymetrix GeneChip Mouse Exon Array analysis. Samples were processed according to Affymetrix GeneChip Whole Transcript Sense Target Labeling Assay (Affymetrix Inc., Santa Clara, CA, USA).
Analysis and statistic
Four biological replicates from each of wild-type and Ube3am−/p+ mice were included in the final analysis to detect differential gene expression derived from Partek Genomic Suite (Partek Inc., St Louis, MO, USA). The Affymetrix generated CEL files containing raw data were subjected to RMA normalization, background subtraction and summarization. One-way ANOVA was subsequently performed to detect P-values for the respective gene expression fold changes. The criteria for a gene to be considered differentially expressed were set at P≤0.05 and a minimal fold change of 1.5-fold. Gene ontology and network/pathway analyses were performed using Ingenuity Pathways Analysis software (Ingenuity System, Redwood City, CA, USA). Quality control analysis was performed using Affymetrix Expression Console software with reference to Affymetrix quality control white papers.
Semiquantitative reverse-transcription PCR
Total RNA (2 μg) from each sample was treated with RQ1 DNase (Promega, Madison, WI, USA) and then reverse-transcribed using Superscript (Invitrogen Inc.). Subsequently, 1 μl of the cDNA template generated was used for each PCR reaction using Faststart Taq Polymerase (Roche, Indianapolis, IN, USA), and cycling conditions were 95°C for 10 min, then 30–35 cycles of 30 s at 95°C, 30 s at 60°C and 30 s at 72°C, followed by a final extension of 10 min at 72°C.
qRT-PCR
A 25 μl reaction containing iTaq SYBR Green Supermix with ROX (Bio-Rad, Hercules, CA, USA) was prepared according to the manufacturer's instructions, and PCR-amplified using ABI 7500 System (Applied Biosystems, Carlsbad, CA, USA). Cycling conditions include 10 min at 50°C, 10 min at 95°C, then 45 cycles of 30 s at 95°C, 30 s at 60°C and 32 s at 72°C. The experiment was performed in three biological replicates. The CT value for each gene was determined in the linear phase of the amplification for each gene, and normalized to CT value of G3pdh to obtain the ΔCT. The fold change for each gene was obtained using 2−(mean wild-type ΔCT−mean Ube3a(m−/p+) ΔCT). A simple t-test was performed on the ΔCT for each gene to obtain a P-value for differential expression.
Western blot
Cerebellum from six-week-old female mouse was homogenized in lysis buffer (30 m Tris, 7 urea, 2 thiourea, 4% CHAPS (pH 8.5), protease inhibitor). P19 cells were lysed in lysis buffer (0.1 Tris-HCl (pH 7.4), 150 m NaCl, 1 m EDTA, 2 m MgCl2, 1% Triton X-100, 15% glycerol, 2 mg/ml phenylmethylsulfonyl fluoride and protease inhibitor). Total protein (10 μg) was separated in SDS–PAGE, and Western blotting was performed using primary anti-Ube3a (Bethyl Laboratories, Montgomery, TX, USA), anti-Mc1r and anti-Nr4a2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and detected by chemiluminescence horseradish-peroxidase-conjugated secondary antibody.
shRNA expressing vector construct
A 19-mer (5′-ctt-cgt-atg-gat-aac-aat-g-3′) against exon 5 of Ube3a (GenBank accession number: NM_011668.2) was cloned into pSUPER.puro vector (Oligoengine, Seattle, WA, USA) for the shRNA-mediated Ube3a knockdown. The targeted exon 5 corresponds to the exon 2 that was deleted in the mouse previously described.9 This construct will be referred to as pUbe3aKD hereafter.
Ube3a overexpression vector construct
Ube3a coding region was amplified from p3003 pGEM E6-AP (Addgene, Cambridge, MA, USA) using forward primer: 5′-gat-cta-ggt-acc-tat-ggc-cac-agc-ttg-taa-aag-3′ and reverse primer: 5′-act-gat-gga-tcc-tta-cag-cat-gcc-aaa-tcc-3′. The 2559 bp PCR product was then cloned into pcDNA/HisMaxB vector (Invitrogen Inc.) between the KpnI and BamHI restriction sites. To track the transfection/expression efficiency, we amplified a 1368 bp fragment containing the internal ribosomal entry site–eGFP fusion from pIGCN21 vector (NCI-Frederick, Frederick, MD, USA) using forward primer: 5′-gat-cta-gga-tcc-gcc-aag-cta-tcg-aat-tcc-gc-3′ and reverse primer: 5′-act-gat-gcg-gcc-gct-tat-gca-gaa-ttc-gaa-gct-tga-gc-3′ and cloned between the BamHI and NotI sites of the pcDNA/HisMaxB vector. This vector will be referred to as pUbe3aOE hereafter.
Cell culture and transfection
P19 cells were cultured in α-minimum essential medium supplemented with 7.5% bovine calf serum and 2.5% fetal bovine serum. P19 cells (4 × 105) were transfected with pUbe3aKD or pUbe3aOE in a six-well plate using Lipofectamine 2000 reagent (Invitrogen Inc.) according to the manufacturer's protocol. Cells were collected 24 h later.
Results
Whole-genome microarray analysis
We are interested in identifying genes that are affected in the absence of functional Ube3a. Because Ube3a is expressed from the maternal inherited allele in the cerebellum,5 we checked for differential gene expression between wild-type and Ube3am−/p+ mice. We performed microarray analysis using Affymetrix GeneChip Mouse Exon array on four biological replicates from each group. Genes are considered to be differentially expressed if they show a fold change of at least 1.5-fold and P≤0.05.
We analyzed the gene expression profiles between the two groups using the Core Probeset, which is based on highly confident supporting evidence from RefSeq and GenBank full-length mRNAs. This yielded a total of 64 differentially expressed genes (7 upregulated and 57 downregulated) that were statistically significant (Table 1). The most heavily represented downregulated genes in the Ube3am−/p+ mice appear to encode receptors for neurogenic functions, such as neurotransmitter receptors (eg Glra1/3, Chrna4 and Drd2). Another substantial group of genes that were downregulated involves transcription regulation, functioning in neurogenesis and other physiological aspects (eg Mc1r, Nr4a2 and Npas).
Table 1. Microarray gene expression analysis (Core Probeset): wild-type (Ube3am+/p+) vs Ube3a knockout (Ube3am−/p+) mice.
| No. | Affymetrix transcript ID | NCBI accession no. | Gene symbol | Gene name | P-value | Fold change |
|---|---|---|---|---|---|---|
| Upregulation in mutant | ||||||
| 1 | 6880900 | NM_008008 | Fgf7 | Fibroblast growth factor 7/ | 0.022 | 2.091 |
| 2 | 6927341 | NM_080445 | B3galt6 | UDP-Gal:βGal β 1,3-galactosyltransferase | 0.036 | 1.907 |
| 3 | 6817951 | AK029771 | 9330180L21Rik | RIKEN cDNA 9330180L21 | 0.015 | 1.750 |
| 4 | 6985703 | NM_026758 | Mphosph6 | M-phase phosphoprotein 6 | 0.039 | 1.701 |
| 5 | 6786954 | AK085965 | 2010316F05Rik | RIKEN cDNA 2010316F05 | 0.027 | 1.587 |
| 6 | 6935555 | NM_019647 | Rpl21 | Ribosomal protein L21 | 0.032 | 1.542 |
| 7 | 6935197 | NM_001038703 | Gpr146 | G-protein-coupled receptor 146 | 0.035 | 1.541 |
| Downregulation in mutant | ||||||
| 8 | 6992946 | NM_178676 | Entpd3 | Ectonucleoside triphosphate diphosphohydrolase 3 | 0.008 | −1.501 |
| 9 | 6976233 | NM_080438 | Glra3 | Glycine receptor, α-3 subunit | 0.045 | −1.502 |
| 10 | 6967109 | NM_013643 | Ptpn5 | Protein tyrosine phosphatase, nonreceptor type 5 | 0.002 | −1.507 |
| 11 | 6963197 | NM_007627 | Cckbr | Cholecystokinin B receptor | 0.012 | −1.517 |
| 12 | 6786049 | NM_172496 | Cobl | Cordon-bleu | 0.040 | −1.531 |
| 13 | 6898010 | NM_008604 | Mme | Membrane metallo-endopeptidase | 0.027 | −1.532 |
| 14 | 6925872 | NM_008154 | Gpr3 | G-protein-coupled receptor 3 | 0.029 | −1.542 |
| 15 | 6846576 | NM_007938 | Epha6 | Eph receptor A6 | 0.004 | −1.555 |
| 16 | 6979704 | NM_008559 | Mc1r | Melanocortin 1 receptor | 0.001 | −1.556 |
| 17 | 6942379 | NM_010717 | Limk1 | LIM-domain containing, protein kinase | 0.043 | −1.561 |
| 18 | 6870979 | BC023699 | AI790298 | Expressed sequence AI790298 | 0.024 | −1.566 |
| 19 | 6819928 | NM_175498 | Pnma2 | Paraneoplastic antigen MA2 | 0.044 | −1.575 |
| 20 | 6960931 | NM_001033962 | Ube3a | Ubiquitin protein ligase E3A | 0.016 | −1.579 |
| 21 | 6971996 | NM_021302 | Stk32c | Serine/threonine kinase 32C | 0.029 | −1.587 |
| 22 | 6764046 | BC126965 | Pcp4l1 | Purkinje cell protein 4-like 1 | 0.029 | −1.589 |
| 23 | 6819244 | NM_009947 | Cpne6 | Copine VI | 0.014 | −1.601 |
| 24 | 6873187 | NM_145123 | Crtac1 | Cartilage acidic protein 1 | 0.002 | −1.613 |
| 25 | 6966324 | NM_010758 | Mag | Myelin-associated glycoprotein | 0.038 | −1.615 |
| 26 | 6833516 | NM_008800 | Pde1b | Phosphodiesterase 1B, Ca2+-calmodulin dependent | 0.049 | −1.620 |
| 27 | 6930606 | NM_178804 | Slit2 | Slit homologue 2 (Drosophila) | 0.034 | −1.622 |
| 28 | 6820282 | NM_172812 | Htr2a | 5-Hydroxytryptamine (serotonin) receptor 2A | 0.010 | −1.625 |
| 29 | 6760417 | NM_021306 | Ecel1 | Endothelin-converting enzyme-like 1 | 0.004 | −1.631 |
| 30 | 6933072 | NM_009263 | Spp1 | Secreted phosphoprotein 1 | 0.047 | −1.633 |
| 31 | 6994790 | NM_178737 | AW551984 | Expressed sequence AW551984 | 0.025 | −1.634 |
| 32 | 6982725 | BC111102 | 4930431L04Rik | RIKEN cDNA 4930431L04 gene | 0.032 | −1.636 |
| 33 | 6762197 | NM_008795 | Pctk3 | PCTAIRE-motif protein kinase 3 | 0.029 | −1.638 |
| 34 | 6805200 | NM_145451 | Gpx6 | Glutathione peroxidase 6 | 0.026 | −1.639 |
| 35 | 6832276 | NM_172610 | Mpped1 | Metallophosphoesterase domain containing 1 | 0.017 | −1.658 |
| 36 | 6947131 | NM_028880 | Lrrtm1 | Leucine-rich repeat transmembrane neuronal 1 | 0.044 | −1.669 |
| 37 | 6810961 | NM_033269 | Chrm3 | Cholinergic receptor, muscarinic 3 | 0.020 | −1.679 |
| 38 | 6864813 | NM_011898 | Spry4 | Sprouty homologue 4 (Drosophila) | 0.022 | −1.687 |
| 39 | 6750314 | NM_177164 | A830006F12Rik | RIKEN cDNA A830006F12 gene | 0.005 | −1.698 |
| 40 | 6931001 | NM_018764 | Pcdh7 | Protocadherin 7 | 0.003 | −1.725 |
| 41 | 6856133 | NM_022025 | Slc5a7 | Solute carrier family 5 (choline transporter) | 0.039 | −1.738 |
| 42 | 6854467 | XM_989487 | LOC671855 | Similar to Rho GDP-dissociation inhibitor 3 | 0.005 | −1.753 |
| 43 | 6901119 | NM_022565 | Ndst4 | N-Deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 | 0.042 | −1.762 |
| 44 | 6808279 | NM_013628 | Pcsk1 | Proprotein convertase subtilisin/kexin type 1 | 0.033 | −1.765 |
| 45 | 6854844 | NM_010831 | Snf1lk | SNF1-like kinase | 0.015 | −1.775 |
| 46 | 6801807 | NM_172805 | Kcnh5 | Potassium voltage-gated channel, subfamily H (eag-related) | 0.007 | −1.787 |
| 47 | 6803891 | NM_178915 | Tmem179 | Transmembrane protein 179 | 0.005 | −1.788 |
| 48 | 6988976 | NM_010077 | Drd2 | Dopamine receptor 2 | 0.017 | −1.794 |
| 49 | 6815027 | NM_009027 | Rasgrf2 | RAS protein-specific guanine nucleotide-releasing factor 2 | 0.044 | −1.815 |
| 50 | 6931355 | NM_011670 | Uchl1 | Ubiquitin C-terminal hydrolase L1 | 0.041 | −1.822 |
| 51 | 6906620 | NM_011839 | Mab21l2 | Mab-21-like 2 (C. elegans) | 0.031 | −1.979 |
| 52 | 6862816 | NM_144946 | Neto1 | Neuropilin (NRP) and tolloid (TLL)-like 1 | 0.016 | −1.981 |
| 53 | 6785684 | NM_010904 | Nefh | Neurofilament, heavy polypeptide | 0.043 | −2.088 |
| 54 | 6894253 | NM_015730 | Chrna4 | Cholinergic receptor, nicotinic, α-polypeptide 4 | 0.004 | −2.093 |
| 55 | 6886908 | NM_013613 | Nr4a2 | Nuclear receptor subfamily 4, group A, member 2 | 0.028 | −2.148 |
| 56 | 6844649 | NM_009215 | Sst | Somatostatin | 0.041 | −2.211 |
| 57 | 6796691 | NM_010234 | Fos | FBJ osteosarcoma oncogene | 0.015 | −2.242 |
| 58 | 6889978 | NM_010825 | Mrg1 | Myeloid ecotropic viral integration site-related gene 1 | 0.023 | −2.334 |
| 59 | 6871062 | NM_153553 | Npas4 | Neuronal PAS domain protein 4 | 0.012 | −2.479 |
| 60 | 6967593 | NM_176942 | Gabra5 | γ-Aminobutyric acid (GABA-A) receptor | 0.017 | −2.548 |
| 61 | 6943974 | NM_009311 | Tac1 | Tachykinin 1 | 0.013 | −2.615 |
| 62 | 6833311 | NM_010444 | Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 0.016 | −2.741 |
| 63 | 6788423 | NM_020492 | Glra1 | Glycine receptor, α-1 subunit | 0.031 | −4.946 |
| 64 | 6881459 | NM_029530 | 6330527O06Rik | RIKEN cDNA 6330527O06 gene | 0.010 | −6.616 |
Differentially expressed genes with a fold change ≥1.5 (P≤0.05) from Ube3am−/p+ mice compared with the wild-type littermates. List is shown in the order of the most upregulated, to the most downregulated gene (as shown by ‘−' sign).
Pathway analysis
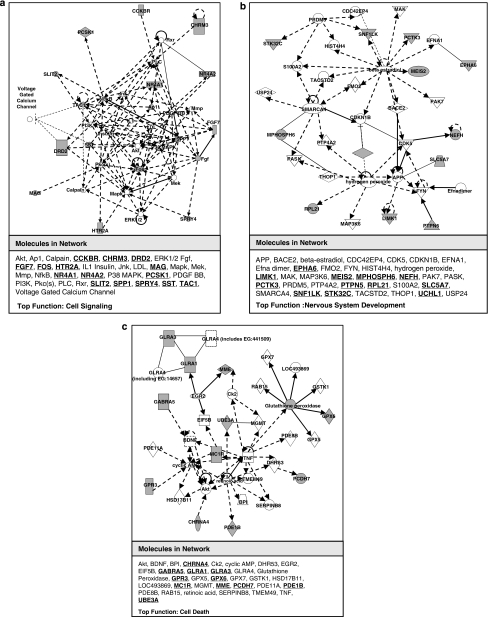
Pathway analysis shows that the differentially expressed genes are implicated in three major pathways/networks including cell signaling, nervous system development and cell death. Fifteen genes are involved in the first network associated with cell signaling (Figure 1a), including Fgf7 and Nr4a2. In the brain, the orphan receptor Nr4a2 supports dopaminergic neurons to survive and differentiate.15 Twelve genes are associated with the nervous system development and functions (Figure 1b). Among them are Epha6, a tyrosine kinase receptor important for axon guidance, as well as Slc5a7, which encodes choline transporter responsible for proper choline uptake along the synapse.18, 19 Eleven genes are associated with cellular development/death (Figure 1c). Among them, downregulation of Mc1r and downregulation of Glra1 are two examples that show functional relevance to AS.
Figure 1.
Pathway analysis on differentially expressed genes. (a) Network 1 is associated with cell signaling and involves 15 of our reported differentially expressed genes (in bold). Representative genes such as Fgf7 and Nr4a2 are qRT-PCR validated, which are up- and downregulated in the Ube3am−/p+ mice, respectively. (b) Network 2 is associated with nervous system development and involves 12 of our reported microarray hits. Epha6 and Slc5a7 in this network are qRT-PCR validated showing both downregulations in the Ube3am−/p+ mice. (c) Network 3 is associated with cell death and involves 11 genes, such as Glra1 and Mc1r. Both are down-regulated in the Ube3am−/p+ mice and validated using qRT-PCR.
Differential expression validation
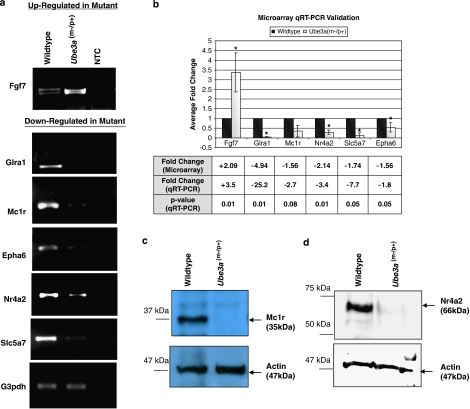
We have validated and confirmed the differential expression status of two genes per network described above, using semiquantitative reverse-transcription PCR (Figure 2a) and qRT-PCR in biological triplicates (Figure 2b). These validated genes (ie Fgf7, Glra1, Mc1r, Nr4a2, Slc5a7 and Epha6) were chosen because their functions are relevant to the AS phenotype.
Figure 2.
Semiquantitative reverse-transcription PCR, qRT-PCR and Western blot validation confirming on a selection of differentially expressed genes identified by microarray. (a) Semiquantitative reverse-transcription PCR validation: Fgf7 is upregulated in Ube3am−/p+ mice, whereas the rest of the genes, including Glra1, Mc1r, Nr4a2, Epha6 and Slc5a, are confirmed to be downregulated. NTC: no template control. (b) qRT-PCR validation showing the normalized mean fold change from the biological triplicates. The fold change is calculated using 2−(mean wild-type ΔCT−mean Ube3a (m−/p+) ΔCT); ‘+' and ‘−' represent upregulation and downregulation of transcript, respectively; *P<0.05. (c and d) Total protein (10 μg) extracted from mouse cerebellum was analyzed by SDS–PAGE using 6% acrylamide gel. Western blot analyses using antibody against Mc1r and Nr4a2 show that the 35 kDa Mc1r (c) and the 66 kDa Nr4a2 (d) proteins, respectively, are downregulated in the Ube3am−/p+ mice. β-Actin is used as endogenous internal control in the Western blot analyses.
A recent report shows that Mc1r signaling induces the expression of Nr4a2.17 Because both mRNAs are downregulated in the AS mice, we extended our differential expression analysis to the protein level for these two genes. To determine if downregulation of Mc1r and Nr4a2 can be reflected at the protein levels, we performed Western blot comparing cerebellum total protein extract from wild-type and Ube3am−/p+ mice. We found that both Mc1r (Figure 2c) and Nr4a2 proteins (Figure 2d), like its relative transcript, are downregulated in the AS mouse.
Ube3a knockdown in P19 cell line
We were interested in finding out if knockdown of Ube3a in P19 cell line will lead to downregulation of Mc1r and Nr4a2 similar to what we have observed in the Ube3am−/p+ mice. It is possible that the constitutive loss of Ube3a activity during mouse development may result in adaptive change in gene expression (to cope with loss of Ube3a activity), and thus many of the changes observed in the transcriptional level may represent indirect, rather than direct, consequences on loss of Ube3a activity.
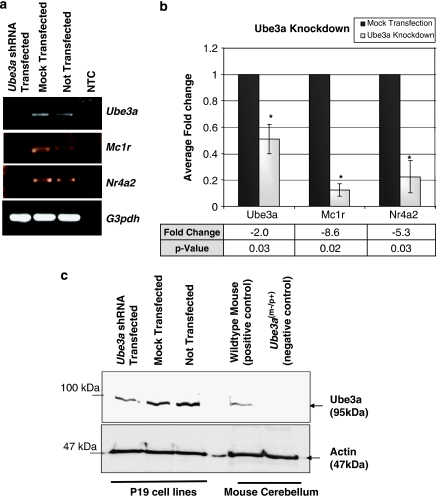
To address this problem, we have generated an RNAi system with target sequence against Ube3a, in which immediate effect of loss of Ube3a activity on respective genes/proteins can be evaluated. The P19 cells transfected with shRNA expression plasmid show downregulation of Ube3a at both transcription and protein levels. There is a two-fold reduction in the Ube3a mRNA level after knockdown (Figure 3a and b). Consistent with this result, Ube3a protein in P19 cells was reduced after transfection with the Ube3a shRNA expression plasmid as determined by Western blot analysis (Figure 3c). We then check for relative transcript expression of Mc1r and Nr4a2 in the Ube3a knockdown and control cells. Semiquantitative reverse-transcription PCR (Figure 3a) and biological triplicates of qRT-PCR (Figure 3b) show that Mc1r and Nr4a2 mRNA levels decreased by 8.6- and 5.3-fold, respectively, in the Ube3a knockdown cells. These results suggest that functional Ube3a is perhaps required for Mc1r and Nr4a2 gene expression.
Figure 3.
Validation of downregulation of Mc1r and Nr4a2 by shRNA-mediated knockdown of Ube3a in P19 cells. (a) Semiquantitative reverse-transcription PCR showing the downexpression of Ube3a, Mc1r and Nr4a2 transcript in the Ube3a shRNA-transfected cells. NTC: no template control. (b) Biological triplicates of qRT-PCR analyses showing the normalized mean fold change. ‘−' represents a downregulation in the Ube3a shRNA-transfected cells; *P<0.05. (c) Total protein (10 μg) extracted from the Ube3a shRNA-transfected and control P19 cells was analyzed by SDS–PAGE using 6% acrylamide gel. Western blot analyses using antibody against Ube3a show the knockdown of the 95 kDa Ube3a on transfection with the Ube3a shRNA plasmid (pUbe3aKD). Total protein extracted from wild-type and the Ube3am−/p+ mice cerebellum was used as positive and negative control in the same Western blot analyses. β-Actin is used as endogenous internal control in the Western blot analysis.
Overexpression of Ube3a in P19 cell line
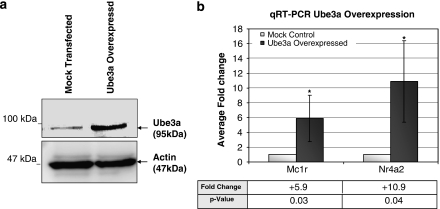
Because downregulation of Mc1r and Nr4a2 is observed in the Ube3am−/p+ mice and Ube3a knockdown P19 cells, we were interested in determining if the levels of Mc1r and Nr4a2 will be affected when Ube3a is overexpressed. We constructed an Ube3a expression vector, pUbe3aOE, and transfection of P19 cells with this plasmid resulted in higher level of Ube3a expression as determined by Western blot (Figure 4a). Subsequently, the mRNA level of Mc1r and Nr4a2 was determined using qRT-PCR. We observed a 5.9- and 10.9-fold increase in the mRNA level of Mc1r and Nr4a2, respectively (Figure 4b).
Figure 4.
Ube3a overexpression results in an upregulation of Mc1r and Nr4a2. (a) Total protein (10 μg) extracted from P19 cells transfected with the Ube3a expression plasmid (pUbe3aOE) and control cells was analyzed by SDS–PAGE using 6% acrylamide gel. Western blot analyses using antibody against Ube3a show the increase protein level of the 95 kDa Ube3a on overexpression of Ube3a. β-Actin is used as endogenous internal control in the Western blot analysis. (b) Biological triplicates of qRT-PCR analyses showing the normalized mean fold change of Mc1r and Nr4a2 on Ube3a overexpression in P19 cells. ‘+' represents an upregulation in the Ube3a-overexpressed cells; *P<0.05.
Discussion
Lack of functional maternal Ube3a expression in imprinted brain tissue can result in the accumulation of target proteins that are meant to be degraded through the ubiquitin proteosomal system, as well as dysregulation of genes expression due to the lack of the coactivation function of Ube3a. We used a genome-wide approach to detect differential genes expression between wild-type and Ube3am−/p+ mice cerebellum. The mouse cerebellum was used because previous studies show that Ube3a is imprinted in the cerebellum and electrophysiology recording reveal abnormal oscillatory activity in the AS mice.5, 20 In addition, the cerebellum controls motor movements, which most AS patient lack.3 P19 cells were used to investigate the knockdown/overexpression effect of Ube3a because these pluripotent embryonic cells can be induced to differentiate into neurons.21 In addition, neurons and skin are derived from the same lineage progenitor cells.22
In this study, we have shown that Mc1r is downregulated in AS mice at mRNA and protein levels. Mc1r, a G-protein-coupled receptor, is widely studied in peripheral tissues, such as the skin, for its regulation of pigment production.23 OCA2, an autosomal recessive gene that is responsible for type 2 oculocutaneous albinism, is currently attributed toward the hypopigmentation phenotype seen in type I (deletion) AS patients as it lies within the AS deletion region along 15q11–q13.24, 25 However, this does not explain why other AS patients with UBE3A mutations, imprinting defect along 15q11–q13 or paternal uniparental disomy (type II–IV), which have intact OCA2, still show to a certain extent, the hypopigmentation phenotype.26 Gene expression in AS mouse cerebellum used in this study may be different from gene expression in other tissues such as the skin, even though neurons and skin are derived from the same lineage cells.22 However it would be interesting to determine if Ube3a mutation also causes downregulation of Mc1r in the skin, resulting in the perturbation of normal pigment production seen in type II–IV patients. Co-deletion of OCA2 and UBE3A, which results in the decrease of Mc1r, observed in type I deletion patients could perhaps lead to a synergistic effect, resulting in the stable and full-blown hypopigmentation phenotype.
In the brain, Mc1r was shown to prevent inflammation, as well as to provide a neuroprotective effect on the brain cell population.16 On top of that, a recent report shows that Mc1r signaling rapidly, yet transiently, induces transcription of the Nr4a subfamily receptors.17 The Nr4a subfamily receptors, Nr4a1/Nurr77, Nr4a2/Nurr1, and Nr4a3/NOR-1 are orphan receptors, well known for their close ligand-binding sites.27 We have shown that in the absence of maternal Ube3a in the brain, Nr4a2 transcript is reduced. Taken together, it is conceivable that Ube3a might have a direct role in stimulating the synthesis of Mc1r, which in turn, regulates Nr4a2 gene expression.
Reducing the expression of Nr4a2 in the brain might explain certain AS phenotype, including poor learning/memory, and motor incoordination. Nr4a2 knockdown in rat hippocampus was reported to affect spatial discrimination, learning and memory.28 In the AS mouse model, where Nr4a2 is downregulated, the mice show severe long-term potentiation and learning impairment.9 In contrast, Nr4a2 mRNA expression was found to be increased during learning in the rat models.29 In a recent report, Nr4a2 has been shown to interact with Wnt signaling via β-catenin in the establishment and development of the nervous system.30 More importantly, Nr4a2 was reported to be critical for induction and survival of dopaminergic neurons.15 Nr4a2+/− mice appear normal at birth, but show motor abnormality as a result of reduce numbers of dopaminergic neurons.31 Hence, the motor dysfunction observed in AS patients as a result of loss of maternal Ube3a could possibly be related to the decrease levels of Nr4a2, which mediates the induction and survival of dopaminergic neurons.15
In addition, we have identified many neurotransmitter receptors that are differentially expressed in the AS mice, including glycine receptor (Glra1), γ-aminobutyric acid receptor (Gabra5) and cholinergic receptor (Chrna4). The localization of UBE3AYFP fusion gene at the pre/postsynaptic regions of cultured hippocampal neurons led to the speculation that UBE3A may directly regulate the development and/or synaptic functions.8 Downregulation of neurotransmitter receptors could affect proper neuro-signal transduction and normal neuronal and motor functions. For example, mutations in Glra1 result in hyperekplexia, where patients show ‘drop seizure' phenotype.32 These symptoms show overlapping phenotype to the AS, creating a possibility that the cause of some of the AS phenotype, such as seizure, might be associated with lack of Glra1 expression. The reason why Gabra5 is downregulated in Ube3a-deficient mouse is unclear. Besides direct or indirect mechanisms involving the loss of the coactivator and/or ubiquitin ligase functions of Ube3a, it is possible that a chromatin structure alteration, or the loss of a positive regulatory element caused by Ube3a knockout may lead to downregulation of Gabra5, given that Gabra5 is located adjacent to Ube3a in mouse (7c) and human (15q12). A similar effect is seen for upregulation of Irak1 in the Mecp2-deficient mouse model.33, 34
Altogether, we had performed a genome-wide gene expression profiling of Ube3am−/p+ mice with the intention of identifying genes that are affected in the absence of functional Ube3a. We have observed 64 genes that are differentially expressed in the AS mice. These genes fit into three major networks associated with cell signaling, nervous system development and cell death. We validated the expression of six representative genes (Fgf7, Glra1, Mc1r, Nr4a2, Epha6, and Slc5a7) using qRT-PCR. Using an shRNA knockdown system, we have shown that Ube3a knockdown causes downregulation of Mc1r and Nr4a2 in the embryonic P19 cell, whereas overexpression of Ube3a results in upregulation of Mc1r and Nr4a2, suggesting that Ube3a is involved in regulating their expression. These results can provide a step forward toward a better understanding of AS development and narrows down some critical genes affected in the event of Ube3a mutations that might contribute to the onset of the disease.
Acknowledgments
This work was supported by the Academic Research Fund (M52080025) from Nanyang Technological University awarded to Ken-Shiung Chen.
The authors declare no conflict of interest.
References
- Clayton-Smith J, Driscoll DJ, Waters MF, et al. Difference in methylation patterns within the D15s9 region of chromosome 15q11–13 in 1st cousins with Angelman syndrome and Prader–Willi syndrome. Am J Med Genet. 1993;47:683–686. doi: 10.1002/ajmg.1320470519. [DOI] [PubMed] [Google Scholar]
- Thomson AK, Glasson EJ, Bittles AH. A long-term population-based clinical and morbidity profile of Angelman syndrome in Western Australia: 1953–2003. Disabil Rehabil. 2006;28:299–305. doi: 10.1080/09638280500190631. [DOI] [PubMed] [Google Scholar]
- Williams CA, Beaudet AL, Clayton-Smith J, et al. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140A:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Sutcliffe JS, Cattanach BM, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The Hpv-16 E6 and E6-Ap complex functions as a ubiquitin-protein ligase in the ubiquitination of P53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DH, Smith CL, et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Lu YB, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet. 2009;18:454–462. doi: 10.1093/hmg/ddn373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with P53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Seagroves TN, Bowers M, Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum Mol Genet. 2006;15:2825–2835. doi: 10.1093/hmg/ddl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Ding HR, Lu ZM, et al. E3 ubiquitin ligase E6AP-mediated TSC2 turnover in the presence and absence of HPV16 E6. Genes Cells. 2008;13:285–294. doi: 10.1111/j.1365-2443.2008.01162.x. [DOI] [PubMed] [Google Scholar]
- Mishra A, Godavarthi SK, Maheshwari M, Goswami A, Jana NR. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J Biol Chem. 2009;284:10537–10545. doi: 10.1074/jbc.M806804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, et al. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania A. Neuroprotective actions of melanocortins: a therapeutic opportunity. Trends Neurosci. 2008;31:353–360. doi: 10.1016/j.tins.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Smith AG, Luk N, Newton RA, Roberts DW, Sturm RA, Muscat GEO. Melanocortin-1 receptor signaling markedly induces the expression of the NR4A nuclear receptor subgroup in melanocytic cells. J Biol Chem. 2008;283:12564–12570. doi: 10.1074/jbc.M800480200. [DOI] [PubMed] [Google Scholar]
- Apparsundaram S, Ferguson SM, George AL, Blakely RD. Molecular cloning of a human, hemicholinium-3-sensitive choline transporter. Biochem Biophys Res Commun. 2000;276:862–867. doi: 10.1006/bbrc.2000.3561. [DOI] [PubMed] [Google Scholar]
- Yue Y, Chen ZY, Gale NW, et al. Mistargeting hippocampal axons by expression of a truncated Eph receptor. Proc Natl Acad Sci USA. 2002;99:10777–10782. doi: 10.1073/pnas.162354599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheron G, Servais L, Wagstaff J, Dan B. Fast cerebellar oscillation associated with ataxia in a mouse model of Angelman syndrome. Neuroscience. 2005;130:631–637. doi: 10.1016/j.neuroscience.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Jonesvilleneuve EMV, McBurney MW, Rogers KA, Kalnins VI. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and gial cells. J Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroffio A, Dupin E, Ledouarin NM. Common precursors for neural and mesectodermal derivatives in the cephalic neural crest. Development. 1991;112:301–305. doi: 10.1242/dev.112.1.301. [DOI] [PubMed] [Google Scholar]
- Luger TA, Paus R, Lipton JM, Slominski AT.(eds):: The melanocortin-1 receptor and human pigmentation Conference on Cutaneous Neuroimmunomodulation – The Proopiomelanocortin System11–13 Sep; Munster, Germany.
- Oetting WS, King RA. Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat. 1999;13:99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Fridman C, Hosomi N, Varela MC, Souza AH, Fukai K, Koiffmann CP. Angelman syndrome associated with oculocutaneous albinism due to an intragenic deletion of the P gene. Am J Med Genet A. 2003;119A:180–183. doi: 10.1002/ajmg.a.20105. [DOI] [PubMed] [Google Scholar]
- Lossie AC, Whitney MM, Amidon D, et al. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Cesario WI, Martinez-Montemayor MM, Morales S, et al. Knockdown of Nurr1 in the rat hippocampus: implications to spatial discrimination learning and memory. Learn Mem. 2006;13:734–744. doi: 10.1101/lm.407706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ortiz SP, Maldonado-Vlaar CS, Carrasquillo Y. Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiol Learn Mem. 2000;74:161–178. doi: 10.1006/nlme.1999.3952. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Ray WJ, Glantschnig H, et al. A regulatory circuit mediating convergence between Nurr1 transcriptional regulation and Wnt signaling. Mol Cell Biol. 2007;27:7486–7496. doi: 10.1128/MCB.00409-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jiang CT, Wan XH, He Y, Pan TH, Jankovic J, Le WD. Age-dependent dopaminergic dysfunction in Nurr1 knockout mice. Exp Neurol. 2005;191:154–162. doi: 10.1016/j.expneurol.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Kirstein L, Silfverskiold BP. A family with emotionally precipitated drop seizures. Acta Psychiatr Neurol. 1958;33:471–476. doi: 10.1111/j.1600-0447.1958.tb03533.x. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Lopez-Serra L, Lopez-Nieva P, et al. Mecp2-null mice provide new neuronal targets for Rett syndrome. PLoS One. 2008;3:e3669. doi: 10.1371/journal.pone.0003669. [DOI] [PMC free article] [PubMed] [Google Scholar]