Abstract
The HLA-G molecule plays an important role in immune tolerance, protecting the fetus from maternal immune attack, and probably contributes to graft tolerance and tumor escape from the host immune system. HLA-G expression is tightly regulated and involves mechanisms acting in part at the transcriptional level. Nevertheless, almost all regulatory sequences that govern constitutive and inducible HLA class I gene transcription are disrupted in the HLA-G gene promoter, suggesting an unusual regulatory process. In further investigating the molecular mechanisms of HLA-G gene activation, we evaluated the influence of epigenetic mechanisms on seven HLA-G-negative cell lines that exhibit various phenotypes. Exposure of cells to histone deacetylase inhibitors, or to the demethylating agent 5-aza-2′-deoxycytidine, revealed that HLA-G gene transcription is inhibited by DNA methylation. Reversal of methylation-mediated repression may directly induce HLA-G cell-surface expression, supporting the idea that HLA-G might be activated by such a mechanism during malignancy, inflammation, and allogenic reactions.
Both classical and nonclassical HLA class I genes play a key role in the regulation of the immune response. Although HLA class I genes and products exhibit very close homologies, they acquire very specific functions, with correspondingly tight regulation. To date, numerous data concerning molecular mechanisms involved in the regulation of classical HLA class I genes have accumulated. Nevertheless, the key mechanisms controlling the constitutive and inducible expression of the nonclassical HLA class I HLA-G gene, an important molecule in the establishment of immune tolerance, remain to be elucidated, by examining alternative regulatory pathways.
Classical HLA-A, -B, and -C genes encode highly polymorphic HLA class I glycoproteins that serve as peptide presenters to cytotoxic T lymphocytes and stimulate killing of the HLA class I antigen-presenting cell. These molecules have broad tissue distribution expression that is tightly controlled at the transcriptional level by several conserved regulatory elements in the proximal promoter region. Enhancer A and IFN-stimulated regulatory element respectively, bind nuclear factor κB and IFN regulatory factor 1, mediating the constitutive and cytokine-induced expression of HLA class I genes (1, 2). The SX1X2Y module that is shared with HLA class II gene promoters binds the RFX and activating transcription factor/cAMP response element-binding protein factors, allowing their constitutive and CIITA-mediated transactivation (3).
The nonclassical HLA-G gene encodes the following quasimonomorphic molecules: four membrane-bound proteins (HLA-G1 to -G4) and three soluble proteins (HLA-G5 to -G7), generated by alternative splicing of the HLA-G primary transcript (4–7). HLA-G molecules are involved in the inhibition of both T and natural killer (NK) cell-mediated cytolysis through interaction with the ILT2, ILT4, and KIR2DL4 receptors (8–12). The constitutive expression of HLA-G proteins in extravillous cytotrophoblasts (13), and in a few other tissues (14–16), correlates with high transcriptional activity, whereas levels of HLA-G gene transcripts are generally low or absent in other tissues (17). HLA-G is also activated in virus-infected cells (18, 19), in tumoral (20–29) and inflammatory (30–32) pathologies, and during allogenic processes (33–35).
HLA-G is in part regulated at the transcriptional level (36). Nonetheless, HLA class I cis-acting regulatory elements are disrupted in the HLA-G gene promoter (37) rendering that gene unresponsive to nuclear factor κB, IFN regulatory factor 1, and CIITA factors (38). Despite the presence of an intact X1 box (39), the HLA-G gene promoter does not bind the RFX5 factor in vivo (P.R., K. Masternak, W. Reith, J.D., E.D.C., and P.M., unpublished observation). On the other hand, the HLA-G gene may be activated by stress (40) and leukemia inhibitory factor (41) treatments and is stimulated by IL-10 (42), IFNs (43, 44), GM-CSF (45), and glucocorticoids (46). More recently, three CRE/TRE elements identified in the 1,438-bp promoter region of the HLA-G gene were shown to mediate its regulation by cAMP response element-binding protein/activating transcription factors (47). Nevertheless, the binding of these factors was observed in cells that did not express HLA-G, suggesting that they do not account for tissue-specific expression.
DNA methylation and histone modification are interrelated epigenetic mechanisms known to play a key role in transcriptional control (48). Thus, they may be implicated in alternative pathways that control HLA-G gene expression. DNA methylation of CpG islands is widely used in mammals, notably in genomic imprinting and X-chromosome inactivation, and aberrant methylation patterns in CpG are also a hallmark of human cancer (49, 50). These regulatory pathways have been poorly investigated for the HLA-G gene and remain to be clarified. Indeed, DNA CpG methylation has been analyzed in the JAR choriocarcinoma cell line, revealing the activation of HLA-G transcription after treatment with the demethylating agent 5-azacytidine (51). Conversely, no correlation was observed between HLA-G gene transcriptional activity and methylation of CpG islands in the 5′ part of the HLA-G gene in cells that either express HLA-G transcripts (trophoblasts, JEG-3 cells, CD2+ lymphocytes) or do not (syncytiotrophoblasts, CD34+ cells) (51, 52).
In the present study, these aspects are reexamined using seven cell lines exhibiting negative HLA-G transcription. We show that methylation-mediated repression of the HLA-G gene is a more general mechanism than expected.
Materials and Methods
Human Cell-Line Cultures.
The following cell lines were maintained in RPMI 1640 medium with Glutamax-I (Invitrogen): choriocarcinoma [JAR; American Type Culture Collection (ATCC)]; Burkitt's B lymphoma (Raji; ATCC); lymphoblastoid B cell [LCL 721.221; ref. 53 (kindly provided by C. Munz, University of Tübingen, Tübingen, Germany)]; acute myelogenous leukemia (KG1a; ATCC); and NK cell leukemia [NKL; ref. 54 (kindly provided by E. H. Weiss, Ludwig-Maximilians-Universität München, Munich)]. The NKL cell line was supplemented with 50 units/ml Interleukin 2 (Sigma) and the JAR cell line was supplemented with 4,500 mg/liter glucose (Invitrogen). The JEG-3 (ATCC) choriocarcinoma cell line and the lung embryonic Tera-2 (ATCC) carcinoma cell line were cultivated in DMEM with Glutamax-I 4,500 mg/liter glucose (Invitrogen). The M8 melanoma cell line (55) was maintained in RPMI 1640 medium (Sigma) supplemented with l-glutamine (Sigma). All cultures were also supplemented with 10% heat-inactivated FCS, gentamicin (10 mg/liter, Invitrogen), and Fungizone (250 μg/liter, Invitrogen).
Cell Treatments.
Histone deacetylase (HDAC) inhibitory treatment was carried out for 24 h with sodium butyrate (1 M, Calbiochem) at a final concentration of 3 mM, or with trichostatin A (TSA) (1 mM, Calbiochem) at final concentrations of 0.1, 1, and 10 μM. Demethylating treatment was carried out for 72 h with 5-aza-2′-deoxycytidine (5-Aza-dC) (10 mM, Sigma) at final concentrations of 1, 10, and 100 μM. Cell lines, including an untreated culture, were cultured for 8 h before treatment (Table 1).
Table 1.
HLA-G activation in cell lines exposed to inhibitors of HDAC or to 5-Aza-dC
| Inhibitor | Conc. | JAR
|
LCL 721.221
|
Raji
|
NKL
|
KG1a
|
Tera-2
|
M8
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | WB | IC | FC | PCR | WB | IC | FC | PCR | WB | IC | FC | PCR | WB | IC | PCR | WB | IC | PCR | WB | IC | PCR | WB | IC | ||
| TSA | 0 μM | − | − | − | − | − | − | − | |||||||||||||||||
| 0.1 μM | − | − | − | − | − | − | + | ||||||||||||||||||
| 1 μM | − | − | − | − | − | − | + | ||||||||||||||||||
| 10 μM | − | − | − | − | − | − | + | ||||||||||||||||||
| Butyrate | 0 mM | − | − | − | − | − | − | − | |||||||||||||||||
| 3 mM | − | − | − | − | − | − | + | ||||||||||||||||||
| 5-Aza-dC | 0 μM | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 1 μM | + | + | + | + | + | + | ± | + | |||||||||||||||||
| 10 μM | + | + | + | + | + | + | + | + | ± | + | |||||||||||||||
| 100 μM | + | + | + | + | + | + | + | + | + | + | − | − | + | − | − | ± | − | − | + | − | − | ||||
PCR, RT-PCR; WB, Western blotting; IC, immunocytochemistry; FC, flow cytometry; −, HLA-G-negative; +, HLA-G-positive; ±, low HLA-G expression.
Standard and Real-Time RT-PCR Analysis.
Total RNA was extracted from 5–10 million cells by adding 1 ml of RNAwiz reagent (Ambion) according to the manufacturer's recommendations. Residual DNA was eliminated by DNase I treatment (10–20 units/100 μg, Roche Molecular Biochemicals) for 1 h at 25°C. Retrotranscription was carried out for 1 h at 42°C on 5 μg of RNA, which was denatured for 5 min at 65°C, using oligo(dT) primer (0.5 μg, Invitrogen) and Moloney murine leukemia virus (MMLV)-reverse transcriptase (200 units, Invitrogen) in a final volume of 20 μl. The reaction was stopped by heating at 95°C for 8 min. PCR was carried out according to the 13th HLA Workshop (56) procedure with a Perkin–Elmer DNA thermal 2400 cycler in a total volume of 100 μl containing 2 μl of the reverse transcription reaction, 200 μM each dNTP (Invitrogen), 100 ng of each primer, 10 μl of 10× Taq buffer (Perkin–Elmer), and 2.5 units of Taq DNA polymerase (Perkin–Elmer). Coamplification of HLA-G cDNAs (pan-HLA-G primers: G.257F/G.1004R and HLA-G5-specific primers: G.526F/G.i4b; ref. 23), and β-actin was used as an internal standard (forward primer: 5′-ATCTGGCACCACACCTTCTACAATGAGCTGCG; reverse primer: 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC) was carried out for 30 sec at 94°C, for 45 sec at 61°C, and for 1 min at 72°C for 35 cycles, and β-actin primers were added for the last 16 amplification cycles. After amplification, PCR products were separated on a 1.5% agarose gel, denatured for 20 min by 0.4 M NaOH, then vacuum-transferred onto Hybond-N+ membranes (Amersham Pharmacia). Successive membrane hybridizations were carried out with the 32P-labeled HLA-G-specific GR probe or with the 32P-labeled β-actin probe (56). Dehybridizations were done in boiling 0.5% SDS solution. Hybridized membranes were exposed to a Biomax film (Kodak).
Real-time RT-PCR (ABI Prism 7000 SDS, Applied Biosystems) was used to quantify variations in the amounts of HLA-G transcripts after cell treatment compared with those of JEG-3. Duplex PCR was carried out for 40 amplification rounds in the presence of TaqMan Universal PCR Master Mix, using the predeveloped TaqMan assay reagent GAPDH as an endogenous control [probe with VIC reporter and 6-carboxytetramethylrhodamine (TAMRA) quencher (Applied Biosystems)], HLA-G-specific probe located in exon 5 [200 nM; Applied Biosystems: 5′-CACTGGAGCTGCGGTCGCTGCT; 6-carboxyfluorescein (FAM) reporter and TAMRA quencher] and HLA-G-specific primers [300 nM (Qbiogene, Illkirch, France): forward 5′-CTGGTTGTCCTTGCAGCTGTAG; reverse 5′-CCTTTTCAATCTGAGCTCTTCTTTCT] we designed, using primer express software. The primers were selected to amplify all alternative forms of HLA-G transcripts, including those deleted in the 3′UTR because of the presence of a 14-bp polymorphism (57). The specificity of HLA-G cDNA amplifications was assessed on various cell lines that did not express classical and nonclassical HLA class I genes and that did or did not express HLA-class II genes (not shown). Quantification relative to JEG-3 was carried out in duplicate, using the comparative CT method: ΔCT = CT HLA-G − CT GAPDH; ΔΔCT = ΔCT sample − ΔCT JEG-3; relative HLA-G expression = 2−ΔΔCT.
Monoclonal Antibodies (mAbs).
The following mAbs were used: 4H84, IgG1 anti-denatured HLA-G heavy chain (kindly provided by M. McMaster, Department of Stomatology, University of California, San Francisco), and MemG/09 (Exbio, Prague) which is the IgG1 conformational antibody against HLA-G1 and HLA-G5.
Western Blotting.
Dry cell pellets (0.4 million) were incubated for 1 h at 4°C in 10 μl of lysis buffer containing 1% Nonidet P-40, 0.05 M Tris⋅HCl, and Complete (Roche Diagnostics). Proteins were denatured by heat and 2-mercaptoethanol, then separated by electrophoresis in SDS/12% polyacrylamide gels. The proteins on the gels were electroblotted onto Hybond-C extra membrane (Amersham Pharmacia). The membranes were blocked for 30 min in PBS containing 0.2% Tween-20 and 5% nonfat dry milk. Membranes were then incubated with the 4H84 mAb diluted 1/10,000 for 12 h, and after washing, were incubated with F(ab′)2 goat anti-mouse IgG antibody conjugated with horseradish peroxidase diluted 1/10,000 for 30 min. After washing, membranes were treated with enhanced chemiluminescence reagent (ECL Plus Western Blotting Detection Systems, Amersham Biosciences) and exposed to Biomax film (Kodak) for 1–10 min.
Immunocytochemistry.
Cells (0.5 million) were cytocentrifuged on Superfrost/plus slides by Cytospin-3 (Shandon, Pittsburgh) and fixed in cold acetone for 10 min. Immunochemistry analysis was carried out by using the Streptavidin-Biotin Universal Detection system (Immunotech, Westbrook, ME) according to the manufacturer's instructions. Nuclei were counterstained in Mayer's hematoxylin (Sigma) for 5 min. Each cell line was stained by using the IgG1 isotype control diluted 1/200, 4H84 mAb diluted 1/500, and the anti-tubulin positive-control antibody diluted 1/100.
Flow Cytometry.
Cells (0.5 million) were stained with MemG/09 mAb diluted 1/500 in PBS containing 2% heat-inactivated FCS for 20 min at 4°C. After washing, cells were secondarily stained with a F(ab′)2 goat anti-mouse IgG antibody conjugated with phycoerythrin (Immunotech) for 20 min at 4°C. Fluorescence was detected by an EPICS XL flow cytometer (Beckman Coulter). Control aliquots were stained with the isotype-matched IgG1 antibody diluted 1/50. Specific fluorescence indexes (SFIs) were calculated by dividing the mean fluorescence obtained with MemG/09 by the mean fluorescence obtained with the IgG1 isotype control. SFI values >1.3 were considered positive.
Results
5-Aza-dC Activates HLA-G Gene Transcription in Cell Lines of Different Tissue Origins.
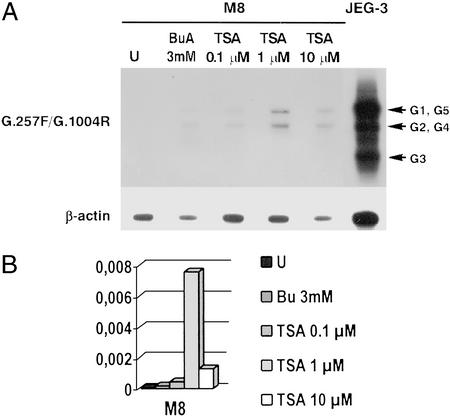
In an attempt to identify the molecular mechanisms participating in HLA-G gene expression, we exposed HLA-G-negative cell lines that did or did not express HLA class I and HLA class II molecules established from choriocarcinoma (JAR), embryonal carcinoma (Tera-2), melanoma (M8), acute myelogenous leukemia (KG1a), NKL, Burkitt's lymphoma (Raji), and lymphoblastoid B cell (LCL 721.221) to inhibitors of HDACs, sodium butyrate and TSA, for 24 h, and to the demethylating reagent 5-Aza-dC for 72 h. Total RNA was extracted from both treated and untreated cells, and from the JEG-3 choriocarcinoma cell line, used as a positive control for HLA-G expression. To visualize the alternative mRNA forms, HLA-G transcripts were analyzed by semiquantitative RT-PCR and Southern blot hybridization, using the procedures validated at the 13th International HLA Workshop (56). The quantities of HLA-G transcripts were evaluated in comparison to those of the JEG-3 cell, by using real-time RT-PCR. Cell exposure to 3 mM sodium butyrate and to 0.1, 1, or 10 μM TSA resulted in the induction of HLA-G transcription in only the M8 human melanoma cell line (Fig. 1). Activation of the HLA-G gene generates low levels of HLA-G transcripts in comparison to those of JEG-3 cells, and the appearance of at least the HLA-G1 and HLA-G2, G4 mRNA forms. The absence of HLA-G transcripts in other cell lines after both treatments (not shown) suggests that HDAC-mediated repression of the HLA-G gene is not a general, but a cell-specific, regulatory mechanism.
Figure 1.
Activation of HLA-G gene transcription in the M8 melanoma cell line exposed to HDAC inhibitors. (A) Representative Southern blot of RT-PCR products obtained by amplification with the G.257F/G.1004R pan-HLA-G primers and β-actin as an internal control on the M8 melanoma cell line, either untreated (U) or treated for 24 h with 3 mM sodium butyrate (BuA) or TSA at three concentrations. HLA-G and β-actin transcripts are respectively revealed by successive hybridizations with 32P-labeled GR (exon 2) and β-actin oligonucleotides. JEG-3 corresponds to the HLA-G-positive control. (B) Results of real-time RT-PCR analysis showing relative quantities of HLA-G transcripts in treated and untreated cell lines, compared with those of JEG-3 (assigned a value of 1). Bu, sodium butyrate.
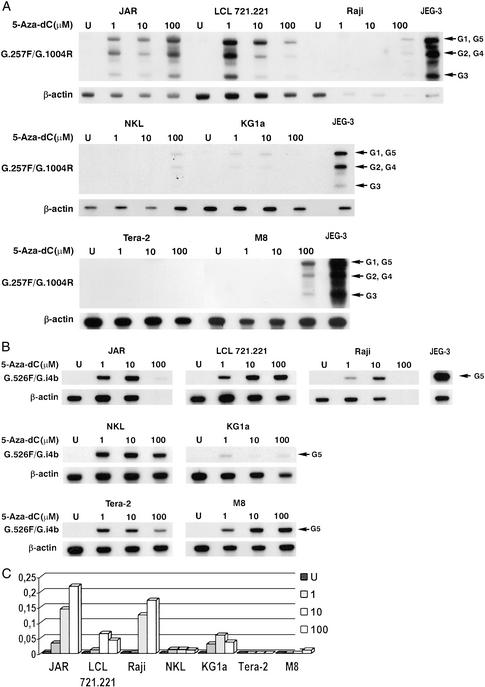
When cells were subjected to the demethylating agent 5-Aza-dC at 1, 10, and 100 μM, RT-PCR carried out with pan-HLA-G and HLA-G5-specific primers revealed HLA-G mRNA induction in the JAR cell line, as described (51), as well as in all treated cell lines (Fig. 2). In the Tera-2 cell line, HLA-G expression seems to be limited to the HLA-G5 mRNA form (Fig. 2B), whereas in other cells the hybridization patterns of HLA-G expression obtained with pan-HLA-G primers are very similar to JEG-3, with three major bands corresponding to HLA-G5, -G1; HLA-G2, -G4; and HLA-G3 mRNA forms (Fig. 2A). Real-time RT-PCR analysis showed that after 5-Aza-dC exposure, HLA-G transcripts levels were lower than those observed in JEG-3 cells and varied according to cell type. The highest HLA-G mRNA levels were observed in the JAR choriocarcinoma cell line, whereas the lowest were found in Tera-2 cells (Fig. 2C). Demethylation treatment with 5-Aza-dC therefore appears to have a more pleiotropic effect on HLA-G gene activation than HDAC inhibitors.
Figure 2.
Repression of the HLA-G gene is reversed by demethylating treatment in several cell lines (A and C). Representative Southern blots of RT-PCR product obtained by amplification with pan-HLA-G G.257F/G.1004R (A) or HLA-G5-specific G.526F/G.i4b (B) primer sets on the JAR, LCL 721.221, Raji, NKL, KG1a, Tera-2, and M8 cell lines, either untreated (U) or treated with increasing concentrations of 5-Aza-dC (1, 10, or 100 μM). GR and GI4F oligonucleotide probes were used to detect all HLA-G transcripts and the HLA-G transcripts that encode soluble HLA-G, respectively. (C) Results of real-time RT-PCR analysis showing relative quantities of HLA-G transcripts in treated (1, 10, or 100 μM 5-Aza-dC) and untreated (U) cell lines, compared with those of JEG-3 (assigned a value of 1).
5-Aza-dC Treatment Induces HLA-G Protein Expression in JAR and B-Type Cell Lines.
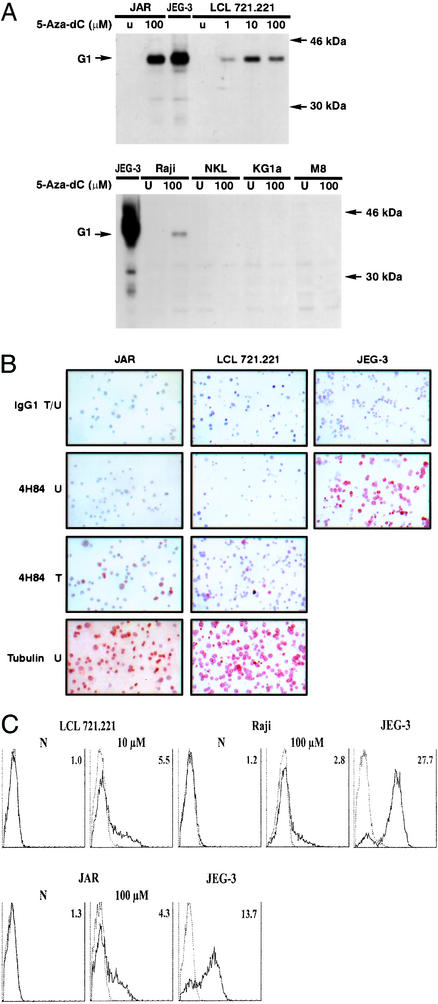
To further investigate the impact of demethylating treatments on the HLA-G protein expression, we first carried out Western blot analysis of cell lysates obtained from JAR, LCL 721.221, Raji, NKL, KG1a, and M8 cell lines previously treated with 5-Aza-dC at concentrations allowing transcriptional activation. They were compared with cell lysates obtained from untreated cells and from the JEG-3 cell line. The use of the 4H84 antibody, which specifically recognized all denatured HLA-G isoforms, did not reveal any HLA-G proteins in untreated cells, whereas it clearly revealed the presence of the HLA-G1 isoform in JEG-3 cells and in the JAR, LCL 721.221, and Raji cell lines that had been exposed to 5-Aza-dC (Fig. 3A). Although HLA-G transcription was activated in the NKL, KG1a, and M8 cell lines after 5-Aza-dC exposure (Fig. 2), the absence of HLA-G protein synthesis observed by Western blotting was confirmed by immunocytochemistry using the same anti-HLA-G antibody (data not shown). This finding suggests that the regulation of HLA-G expression is acting at the transcriptional and posttranscriptional levels.
Figure 3.
HLA-G protein expression is induced in the HLA-G-negative JAR, LCL 721.221, and Raji cell lines after 5-Aza-dC treatment. (A) Western blot analysis of HLA-G protein expression in the JAR, LCL 721.221, Raji, NKL, KG1a, and M8 cell lines, using the 4H84 antibody, which recognizes all intracellular HLA-G isoforms. HLA-G protein is induced in JAR, LCL 721.221, and Raji. U, untreated cells. G1 is the band corresponding to the HLA-G1 isoform. (B) Control experiments for immunocytochemical analysis showing HLA-G induction in JAR and LCL 721.221 after 5-Aza-dC treatment and heterogeneous HLA-G expression in JEG-3. IgG1 is the isotypic control antibody and 4H84 is the HLA-G-specific antibody. T indicates treatment with 100 and 10 μM 5-Aza-dC for JAR and LCL 721.221 cells, respectively, U indicates untreated, and T/U indicates that JAR and LCL721.221 cells were exposed to 5-Aza-dC, whereas JEG-3 cells were not. HLA-G expression was not observed in treated or untreated NKL, KG1a, and M8 cells (not shown). (C) Representative flow-cytometry analysis carried out with the anti-HLA-G1/G5 MemG/09 mAb (represented by a thick line) and an IgG1 isotype control antibody (represented by a thin line), showing HLA-G induction at the cell surface of LCL 721.221, Raji, and JAR after treatment with 10 or 100 μM 5-Aza-dC. Specific fluorescence indexes (on the right of the histograms) were calculated by dividing mean fluorescence values obtained with MemG/09 by mean fluorescence values obtained with the isotype control.
Notably, 5-Aza-dC-treated LCL 721.221 or JAR cells used as controls of demethylation efficiency showed that HLA-G protein induction is heterogeneous (Fig. 3B). Induction of the HLA-G protein after exposure to 5-Aza-dC was also monitored by flow cytometry analysis with the MEMG/09 antibody, showing that HLA-G1 is induced and expressed at the cell surface of ≈30% of treated JAR, LCL 721.221, and Raji cells (Fig. 3C).
Discussion
In our search to identify pathways that control HLA-G gene transcription and protein expression, we investigated the influence of epigenetic mechanisms, showing that the demethylation process may be an important mechanism in reversing HLA-G gene silencing in tumor and B-EBV cell lines exhibiting various phenotypes. Although this work has been done in cell lines that may have abnormal gene regulation at some levels, experiments do not seem to support that this is the case for HLA-G. Until now, the effect of demethylating treatment has mainly been analyzed in the JAR choriocarcinoma cell line (51), but was not expected to activate HLA-G genes in all of the cell lines tested in this study. Although methylation-mediated repression is known to target other HLA genes, for example, the HLA-A and HLA class II genes in trophoblasts (51, 58–60), HLA-G silencing occurs independently of silencing of HLA-class I and II molecules, because the latter may be detected in cells in which HLA-G is repressed. Such a repression mechanism might thus account for tissue-specific expression of the HLA-G molecule and might explain the lack of HLA-G gene activation after cytokine exposure in cells devoid of basal HLA-G transcriptional activity (61).
Several studies have shown that the quantity of HLA-G gene transcripts is an important parameter in the induction of HLA-G protein synthesis. We observed that HLA-G protein was expressed in 5-Aza-dC-treated JAR, LCL 721.221, and Raji cells, despite lower quantities of HLA-G mRNA, compared with JEG-3. We also noted that the induction of HLA-G protein expression was heterogeneous (≈30% positive cells), suggesting that HLA-G mRNAs were not induced in HLA-G-negative cells. We thus propose that in the absence of cell cycle synchronization, it is possible that the efficiency of 5-Aza-dC treatment could vary according to the number of replication cycles.
Conversely, despite the enhancement of HLA-G transcription in M8, NKL, and KG1, we did not observe any protein expression after 5-Aza-dC treatment. The following hypothesis is proposed: (i) There are specific regulatory mechanisms acting at a posttranscriptional level, as well as one that does not imply protein assembly, because HLA-G molecules can be expressed in HLA-G-transfected M8 cells (55). Such a process has been previously suggested with respect to trophoblast cell differentiation, where TAP, tapasin, and β2-microglobulin were shown to accumulate in advance of the HLA-G protein, but were present at the same time as its mRNA (62). (ii) A nonsense mutation or a mutation that prevents translation of the HLA-G mRNA is present. Interestingly, despite HLA-G transcriptional activation, the absence of induction of HLA-G protein expression in the M8 cell line was also reported in an earlier study, after heat-shock treatment or arsenic exposure (E. C. Ibrahim, P. Paul, and E.D.C., unpublished observations), and has also been described in some tumors (23, 63, 64). (iii) The secreted cytokine pattern, which may differ according to the cell type, does not favor the enhancement of HLA-G gene transcription and/or of HLA-G transcript stabilization, thus avoiding HLA-G protein expression.
Standard RT-PCR analysis using pan-HLA-G primers and HLA-G-specific Southern blot hybridization did not allow detection of HLA-G transcripts in 5-Aza-dC-treated Tera-2 cells. Nevertheless, very low levels of HLA-G mRNA were detected by real-time PCR, and probably corresponded to synthesis of the HLA-G5 mRNA form. If reversal of HLA-G silencing first involves HLA-G5 transcription, this is a hypothesis that needs to be investigated. Similarly, specific activation of the HLA-G5 isoform was observed during allogenic reactions in vitro (33). Moreover, it was shown that in M8 cells treated with arsenite, HLA-G6 was the first transcript induced (40). It thus appears that HLA-G transcripts that contain intron 4 might be preferentially expressed after HLA-G gene activation in some cell lines, suggesting tight control of HLA-G alternative splicing, which in part may be linked to the HLA-G allelic form (65). For example, we can postulate that mutations in HLA-G gene splicing sites might favor the expression of intron 4-containing transcripts in some cell lines. On the other hand, intron 4 retention might be involved in the stabilization of these transcripts, thus allowing the HLA-G5 detection in Tera-2 cells.
Demonstration of the effect of demethylation treatment leads us to wonder about the mechanisms that govern HLA-G gene repression observed in untreated LCL 721.221, Raji, Tera-2, M8, NKL, and KG1a cell lines. Cytosine methylation at the dinucleotide CpG island is an important regulator of gene expression in higher eukaryotes, which has been shown to directly affect gene expression either by preventing binding of necessary transcription factors to the promoter, or more frequently, by involving methylated DNA-binding proteins that recruit accessory factors for transcriptional silencing (66). This latter mechanism can lead to local recruitment of HDAC and thus may be hypothesized in the repression of the HLA-G gene in M8 melanoma cells, since both inhibitors of HDAC and demethylant treatments activate the HLA-G gene. Accordingly, the silencing of tumor-suppressor genes by hypermethylation is well described in human cancer and may involve the transcriptional repressor MeCP2, which is associated in vivo with chromatin modifiers and transcriptional corepressors (67). Conversely, the absence of HLA-G activation in the other cell lines incubated with HDAC inhibitors suggests that such a process does not account for HLA-G silencing in these cells. Different mechanisms involving the methylation process may thus account for HLA-G silencing, according to the cell type.
The relationship between the methylation status of the HLA-G gene/promoter and HLA-G transcriptional activity has been previously investigated in a few cell types, and these investigations led to the conclusion that gene/promoter methylation was not the sole mechanism that achieves the repression (51, 52). Nevertheless, these studies were limited to the proximal promoter region of the HLA-G gene, and questions remain concerning potential subsets of CpG sites that might be important in gene silencing. A trans-acting demethylating process could also be envisaged that reverses repression of specific transcriptional factors, which in turn would activate HLA-G transcription. Such a mechanism is well documented in the activation of HLA-class II genes after 5-Aza-dC treatment, which restores IFN-γ inducibility to CIITA (58). In such case, CpG methylation targets promoter IV in choriocarcinoma cell lines, and has been shown to be critical for transcriptional control of these genes (58, 60). Whether DNA demethylation is a cis-acting and/or trans-acting process involved in HLA-G gene activation is not yet known. This preliminary step will allow to direct a mapping of the methylated CpG, in the presence and absence of 5-Aza-dC, focusing on either the HLA-G promoter (distal and proximal regions) and/or the promoter of a putative transcriptional factor induced by demethylating treatment.
In conclusion, the work reported here shows that epigenetic mechanisms may play an important role in the regulation of HLA-G expression, suggesting it may be an important step in the activation of the HLA-G gene in some pathological situations. Genome-wide demethylation is an event that occurs frequently in cancer cells, leading to gene activation, as it does for members of the MAGE family (68). Consistent with this observation is the demonstration of the activation of the HLA-G gene in tumor cell lines treated with the DNA-demethylating drug 5-Aza-dC. Therefore, treatment blocking HDAC and/or reversing DNA methylation (69, 70), and the presence of a specific cytokine microenvironment such as IL-10 (28) or IFNs (26), might favor enhancement of HLA-G transcription and protein expression in tumors and in some inflammatory processes. This finding suggests that the immunogenicity and tumorigenicity of various tumors may be potentially changed according to treatment.
Furthermore, it is becoming apparent that the mechanisms that control HLA-G gene activation have evolved to limit HLA-G expression, to control very specific functions in immune tolerance. Therefore, it is understandable that the HLA-G transcriptional-regulatory pathways shared with classical HLA-class I genes are very restricted. In some cases, such as those observed with the JAR and B-type cell lines Raji and LCL 721.221, it appears that the activation of HLA-G protein expression does not involve many steps, despite the absence of transcriptional activity. Reversal of HLA-G silencing thus appears to be favored in some HLA-G-negative cell types to promote a rapid protective immune response.
Acknowledgments
We thank Staëlle Corvo for excellent technical assistance, Noah Hardy for editing the manuscript, and the Service Photo de l'Institut d'Hématologie. This work was supported by the Commissariat à l'Energie Atomique and by the Association de la Recherche Contre le Cancer. P.R. is a recipient of a grant from the French Ministère de l'Education Nationale, de la Recherche, et de la Technologie.
Abbreviations
- HDAC
histone deacetylase
- TSA
trichostatin A
- 5-Aza-dC
5-aza-2′-deoxycytidine
- NK
natural killer
- NKL
NK cell leukemia
References
- 1.Gobin S J, Keijsers V, van Zutphen M, van den Elsen P J. J Immunol. 1998;161:2276–2283. [PubMed] [Google Scholar]
- 2.Gobin S J, van Zutphen M, Woltman A M, van den Elsen P J. J Immunol. 1999;163:1428–1434. [PubMed] [Google Scholar]
- 3.Gobin S J, Peijnenburg A, van Eggermond M, van Zutphen M, van den Berg R, van den Elsen P J. Immunity. 1998;9:531–541. doi: 10.1016/s1074-7613(00)80636-6. [DOI] [PubMed] [Google Scholar]
- 4.Ishitani A, Geraghty D E. Proc Natl Acad Sci USA. 1992;89:3947–3951. doi: 10.1073/pnas.89.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul P, Cabestre F A, Ibrahim E C, Lefebvre S, Khalil-Daher I, Vazeux G, Quiles R M, Bermond F, Dausset J, Carosella E D. Hum Immunol. 2000;61:1138–1149. doi: 10.1016/s0198-8859(00)00197-x. [DOI] [PubMed] [Google Scholar]
- 6.Moreau P, Carosella E, Teyssier M, Prost S, Gluckman E, Dausset J, Kirszenbaum M. Hum Immunol. 1995;43:231–236. doi: 10.1016/0198-8859(95)00009-s. [DOI] [PubMed] [Google Scholar]
- 7.Fujii T, Ishitani A, Geraghty D E. J Immunol. 1994;153:5516–5524. [PubMed] [Google Scholar]
- 8.Colonna M, Samaridis J, Cella M, Angman L, Allen R L, O'Callaghan C A, Dunbar R, Ogg G S, Cerundolo V, Rolink A. J Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- 9.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu M L. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 10.Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, Angman L, Cella M, Lopez-Botet M. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponte M, Cantoni C, Biassoni R, Tradori-Cappai A, Bentivoglio G, Vitale C, Bertone S, Moretta A, Moretta L, Mingari M C. Proc Natl Acad Sci USA. 1999;96:5674–5679. doi: 10.1073/pnas.96.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajagopalan S, Long E O. J Exp Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMaster M T, Librach C L, Zhou Y, Lim K H, Janatpour M J, DeMars R, Kovats S, Damsky C, Fisher S J. J Immunol. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 14.Hammer A, Hutter H, Blaschitz A, Mahnert W, Hartmann M, Uchanska-Ziegler B, Ziegler A, Dohr G. Am J Reprod Immunol. 1997;37:161–171. doi: 10.1111/j.1600-0897.1997.tb00208.x. [DOI] [PubMed] [Google Scholar]
- 15.Blaschitz A, Lenfant F, Mallet V, Hartmann M, Bensussan A, Geraghty D E, Le Bouteiller P, Dohr G. Eur J Immunol. 1997;27:3380–3388. doi: 10.1002/eji.1830271237. [DOI] [PubMed] [Google Scholar]
- 16.Crisa L, McMaster M T, Ishii J K, Fisher S J, Salomon D R. J Exp Med. 1997;186:289–298. doi: 10.1084/jem.186.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onno M, Guillaudeux T, Amiot L, Renard I, Drenou B, Hirel B, Girr M, Semana G, Le Bouteiller P, Fauchet R. Hum Immunol. 1994;41:79–86. doi: 10.1016/0198-8859(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 18.Onno M, Pangault C, Le Friec G, Guilloux V, Andre P, Fauchet R. J Immunol. 2000;164:6426–6434. doi: 10.4049/jimmunol.164.12.6426. [DOI] [PubMed] [Google Scholar]
- 19.Lozano J M, Gonzalez R, Kindelan J M, Rouas-Freiss N, Caballos R, Dausset J, Carosella E D, Pena J. AIDS. 2002;16:347–351. doi: 10.1097/00002030-200202150-00005. [DOI] [PubMed] [Google Scholar]
- 20.Ugurel S, Rebmann V, Ferrone S, Tilgen W, Grosse-Wilde H, Reinhold U. Cancer (Philadelphia) 2001;92:369–376. doi: 10.1002/1097-0142(20010715)92:2<369::aid-cncr1332>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 21.Ugurel S, Reinhold U, Tilgen W. Onkologie. 2002;25:129–134. doi: 10.1159/000055222. [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre S, Adrian F, Moreau P, Gourand L, Dausset J, Berrih-Aknin S, Carosella E D, Paul P. Hum Immunol. 2000;61:1095–1101. doi: 10.1016/s0198-8859(00)00192-0. [DOI] [PubMed] [Google Scholar]
- 23.Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal F A, Avril M F, Dausset J, Guillet J G, Carosella E D. Proc Natl Acad Sci USA. 1998;95:4510–4515. doi: 10.1073/pnas.95.8.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul P, Cabestre F A, Le Gal F A, Khalil-Daher I, Le Danff C, Schmid M, Mercier S, Avril M F, Dausset J, Guillet J G, Carosella E D. Cancer Res. 1999;59:1954–1960. [PubMed] [Google Scholar]
- 25.Lefebvre S, Antoine M, Uzan S, McMaster M, Dausset J, Carosella E D, Paul P. J Pathol. 2002;196:266–274. doi: 10.1002/path.1039. [DOI] [PubMed] [Google Scholar]
- 26.Wiendl H, Mitsdoerffer M, Hofmeister V, Wischhusen J, Bornemann A, Meyermann R, Weiss E H, Melms A, Weller M. J Immunol. 2002;168:4772–4780. doi: 10.4049/jimmunol.168.9.4772. [DOI] [PubMed] [Google Scholar]
- 27.Urosevic M, Willers J, Mueller B, Kempf W, Burg G, Dummer R. Blood. 2002;99:609–617. doi: 10.1182/blood.v99.2.609. [DOI] [PubMed] [Google Scholar]
- 28.Urosevic M, Kurrer M O, Kamarashev J, Mueller B, Weder W, Burg G, Stahel R A, Dummer R, Trojan A. Am J Pathol. 2001;159:817–824. doi: 10.1016/S0002-9440(10)61756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibrahim E C, Guerra N, Lacombe M J, Angevin E, Chouaib S, Carosella E D, Caignard A, Paul P. Cancer Res. 2001;61:6838–6845. [PubMed] [Google Scholar]
- 30.Aractingi S, Briand N, Le Danff C, Viguier M, Bachelez H, Michel L, Dubertret L, Carosella E D. Am J Pathol. 2001;159:71–77. doi: 10.1016/S0002-9440(10)61675-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khosrotehrani K, Le Danff C, Reynaud-Mendel B, Dubertret L, Carosella E D, Aractingi S. J Invest Dermatol. 2001;117:750–752. doi: 10.1046/j.0022-202x.2001.01487.x. [DOI] [PubMed] [Google Scholar]
- 32.Carosella E D, Moreau P, Aractingi S, Rouas-Freiss N. Trends Immunol. 2001;22:553–555. doi: 10.1016/s1471-4906(01)02007-5. [DOI] [PubMed] [Google Scholar]
- 33.Lila N, Rouas-Freiss N, Dausset J, Carpentier A, Carosella E D. Proc Natl Acad Sci USA. 2001;98:12150–12155. doi: 10.1073/pnas.201407398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lila N, Carpentier A, Amrein C, Khalil-Daher I, Dausset J, Carosella E D. Lancet. 2000;355:2138. doi: 10.1016/S0140-6736(00)02386-2. (lett.). [DOI] [PubMed] [Google Scholar]
- 35.Lila N, Amrein C, Guillemain R, Chevalier P, Latremouille C, Fabiani J N, Dausset J, Carosella E D, Carpentier A. Circulation. 2002;105:1949–1954. doi: 10.1161/01.cir.0000015075.89984.46. [DOI] [PubMed] [Google Scholar]
- 36.Solier C, Mallet V, Lenfant F, Bertrand A, Huchenq A, Le Bouteiller P. Immunogenetics. 2001;53:617–625. doi: 10.1007/s00251-001-0373-0. [DOI] [PubMed] [Google Scholar]
- 37.van den Elsen P J, Peijnenburg A, van Eggermond M C, Gobin S J. Immunol Today. 1998;19:308–312. doi: 10.1016/s0167-5699(98)01287-0. [DOI] [PubMed] [Google Scholar]
- 38.Gobin S J, van den Elsen P J. Hum Immunol. 2000;61:1102–1107. doi: 10.1016/s0198-8859(00)00198-1. [DOI] [PubMed] [Google Scholar]
- 39.Rousseau P, Paul P, O'Brien M, Dausset J, Carosella E D, Moreau P. Hum Immunol. 2000;61:1132–1137. doi: 10.1016/s0198-8859(00)00199-3. [DOI] [PubMed] [Google Scholar]
- 40.Ibrahim E C, Morange M, Dausset J, Carosella E D, Paul P. Cell Stress Chaperones. 2000;5:207–218. doi: 10.1379/1466-1268(2000)005<0207:hsaaie>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bamberger A M, Jenatschke S, Schulte H M, Loning T, Bamberger M C. J Clin Endocrinol Metab. 2000;85:3932–3936. doi: 10.1210/jcem.85.10.6849. [DOI] [PubMed] [Google Scholar]
- 42.Moreau P, Adrian-Cabestre F, Menier C, Guiard V, Gourand L, Dausset J, Carosella E D, Paul P. Int Immunol. 1999;11:803–811. doi: 10.1093/intimm/11.5.803. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Chu W, Geraghty D E, Hunt J S. J Immunol. 1996;156:4224–4231. [PubMed] [Google Scholar]
- 44.Lefebvre S, Berrih-Aknin S, Adrian F, Moreau P, Poea S, Gourand L, Dausset J, Carosella E D, Paul P. J Biol Chem. 2001;276:6133–6139. doi: 10.1074/jbc.M008496200. [DOI] [PubMed] [Google Scholar]
- 45.Amiot L, Onno M, Drenou B, Monvoisin C, Fauchet R. Hum Immunol. 1998;59:524–528. doi: 10.1016/s0198-8859(98)00041-x. [DOI] [PubMed] [Google Scholar]
- 46.Moreau P, Faure O, Lefebvre S, Ibrahim E C, O'Brien M, Gourand L, Dausset J, Carosella E D, Paul P. Transplant Proc. 2001;33:2277–2280. doi: 10.1016/s0041-1345(01)01990-x. [DOI] [PubMed] [Google Scholar]
- 47.Gobin S J, Biesta P, De Steenwinkel J E, Datema G, van den Elsen P J. J Biol Chem. 2002;277:39525–39531. doi: 10.1074/jbc.M112273200. [DOI] [PubMed] [Google Scholar]
- 48.El-Osta A, Wolffe A P. Gene Expression. 2000;9:63–75. doi: 10.3727/000000001783992731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolffe A P. Oncogene. 2001;20:2988–2990. doi: 10.1038/sj.onc.1204322. [DOI] [PubMed] [Google Scholar]
- 50.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 51.Guillaudeux T, Rodriguez A M, Girr M, Mallet V, Ellis S A, Sargent I L, Fauchet R, Alsat E, Le Bouteiller P. J Immunol. 1995;154:3283–3299. [PubMed] [Google Scholar]
- 52.Onno M, Amiot L, Bertho N, Drenou B, Fauchet R. Tissue Antigens. 1997;49:356–364. doi: 10.1111/j.1399-0039.1997.tb02763.x. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu Y, Geraghty D E, Koller B H, Orr H T, DeMars R. Proc Natl Acad Sci USA. 1988;85:227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robertson M J, Cochran K J, Cameron C, Le J M, Tantravahi R, Ritz J. Exp Hematol (Charlottesville, VA) 1996;24:406–415. [PubMed] [Google Scholar]
- 55.Riteau B, Moreau P, Menier C, Khalil-Daher I, Khosrotehrani K, Bras-Goncalves R, Paul P, Dausset J, Rouas-Freiss N, Carosella E D. Transplant Proc. 2001;33:2360–2364. doi: 10.1016/s0041-1345(01)02021-8. [DOI] [PubMed] [Google Scholar]
- 56.Paul P, Rouas-Freiss N, Moreau P, Cabestre F A, Menier C, Khalil-Daher I, Pangault C, Onno M, Fauchet R, Martinez-Laso J, et al. Hum Immunol. 2000;61:1177–1195. doi: 10.1016/s0198-8859(00)00154-3. [DOI] [PubMed] [Google Scholar]
- 57.Hiby S E, King A, Sharkey A, Loke Y W. Tissue Antigens. 1999;53:1–13. doi: 10.1034/j.1399-0039.1999.530101.x. [DOI] [PubMed] [Google Scholar]
- 58.Morris A C, Spangler W E, Boss J M. J Immunol. 2000;164:4143–4149. doi: 10.4049/jimmunol.164.8.4143. [DOI] [PubMed] [Google Scholar]
- 59.van den Elsen P J, Gobin S J, van der Stoep N, Datema G, Vietor H E. J Reprod Immunol. 2001;52:129–145. doi: 10.1016/s0165-0378(01)00115-2. [DOI] [PubMed] [Google Scholar]
- 60.Morris A C, Beresford G W, Mooney M R, Boss J M. Mol Cell Biol. 2002;22:4781–4791. doi: 10.1128/MCB.22.13.4781-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lefebvre S, Moreau P, Guiard V, Ibrahim E C, Adrian-Cabestre F, Menier C, Dausset J, Carosella E D, Paul P. J Reprod Immunol. 1999;43:213–224. doi: 10.1016/s0165-0378(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 62.Copeman J, Han R N, Caniggia I, McMaster M, Fisher S J, Cross J C. Biol Reprod. 2000;62:1543–1550. doi: 10.1095/biolreprod62.6.1543. [DOI] [PubMed] [Google Scholar]
- 63.Polakova K, Russ G. Neoplasma (Bratisl) 2000;47:342–348. [PubMed] [Google Scholar]
- 64.Real L M, Cabrera T, Collado A, Jimenez P, Garcia A, Ruiz-Cabello F, Garrido F. Int J Cancer. 1999;81:512–518. doi: 10.1002/(sici)1097-0215(19990517)81:4<512::aid-ijc2>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 65.Rebmann V, van der Ven K, Passler M, Pfeiffer K, Krebs D, Grosse-Wilde H. Tissue Antigens. 2001;57:15–21. doi: 10.1034/j.1399-0039.2001.057001015.x. [DOI] [PubMed] [Google Scholar]
- 66.Wade P A. BioEssays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen C T, Gonzales F A, Jones P A. Nucleic Acids Res. 2001;29:4598–4606. doi: 10.1093/nar/29.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lurquin C, De Smet C, Brasseur F, Muscatelli F, Martelange V, De Plaen E, Brasseur R, Monaco A P, Boon T. Genomics. 1997;46:397–408. doi: 10.1006/geno.1997.5052. [DOI] [PubMed] [Google Scholar]
- 69.Della Ragione F, Criniti V, Della Pietra V, Borriello A, Oliva A, Indaco S, Yamamoto T, Zappia V. FEBS Lett. 2001;499:199–204. doi: 10.1016/s0014-5793(01)02539-x. [DOI] [PubMed] [Google Scholar]
- 70.Marks P A, Richon V M, Breslow R, Rifkind R A. Curr Opin Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]