Abstract
Purpose
The present study was undertaken to test a hypothesis that differential sensitivity of normal and cancerous human prostate cells to prooxidant effect of phenethyl isothiocyanate (PEITC) is determined by altered expression of antioxidant defense genes.
Methods
Prooxidant effect of PEITC was assessed by flow cytometry using a chemical probe and measurement of hydrogen peroxide production. Gene expression was determined by real time PCR using Human Oxidative Stress and Antioxidant Defense RT2 Profiler™. Protein expression was determined by Western blotting.
Results
The PEITC treatment resulted in generati on of reactive oxygen species and hydrogen peroxide production in PC-3 human prostate cancer cells but not in a representative normal human prostate epithelial cell line (PrEC). Basal oxidative stress-antioxidant defense gene expression signature was strikingly different between PC-3 and PrEC cells. The PEITC treatment (2.5 μM, 6 h) caused up-regulation of 29 genes and down-regulation of 2 genes in PC-3 cells. Conversely 4 genes were up-regulated and 10 genes were down-regulated by a similar PEITC treatment in the PrEC cell line.
Conclusion
Differential sensitivity of PC-3 versus PrEC cells to prooxidant effect of PEITC is likely attributable to difference in basal as well as altered expression of antioxidant defense genes.
Keywords: Phenethyl isothiocyanate, Reactive Oxygen Species, Antioxidant Defense Genes, Chemoprevention, Oxidative stress, Gene expression
INTRODUCTION
Population-based case-control studies suggest that dietary intake of cruciferous vegetables may be protective against the risk of different malignancies including cancer of the prostate (1–3), which is one of the leading causes of cancer-related death in American men (4). For example, a multicenter case-control study involving African-American, white, Japanese, and Chinese men (n= 1619) with histologically confirmed prostate cancer and matched controls (n= 1618; matched by ethnicity, age, region of residence) showed an inverse association between intake of cruciferous vegetables and the risk of prostate cancer (3). Anticancer effect of cruciferous vegetables is credited to chemicals with −N=C=S (isothiocyanate; ITC) functional group, which are produced upon cutting or chewing of these vegetables due to myrosinase-mediated hydrolysis of corresponding glucosinolates (5). Studies conducted in our laboratory and by others have shown that ITCs, including phenethyl-ITC (PEITC), benzyl-ITC (BITC), and sulforaphane (SFN), not only confer protection against chemically-induced cancer but also inhibit cancer development in transgenic mouse models (6–11). For example, oral gavage of SFN thrice per week reduced the incidence of prostatic intraepithelial neoplasia and well-differentiated carcinoma by ~23–28% (P<0.05 compared with control) in the dorsolateral prostate, which was not due to the suppression of T-antigen expression (9). Moreover, the area occupied by the well-differentiated carcinoma was ~44% lower in the dorsolateral prostate of SFN-treated mice relative to that of control mice (9). Strikingly, the SFN-treated mice exhibited an approximate 50% and 63% decrease, respectively, in pulmonary metastasis incidence and multiplicity compared with control mice (P<0.05; Ref. 9). In a separate study, we showed that dietary administration of BITC (3 mmol/kg diet) for 25 weeks markedly suppressed the incidence and/or burden of mammary hyperplasia and carcinoma in female MMTV-neu mice without causing weight loss or affecting neu protein level (10). The BITC-mediated prevention of mammary cancer development in MMTV-neu mice correlated with reduced cellular proliferation, increased apoptosis, and tumor infiltration of T cells (10).
Though the exact mechanisms responsible for the beneficial effects of ITCs are not fully understood, cancer prevention by this class of compounds in vivo correlates with apoptosis induction (10). In vitro cellular studies have revealed that ITCs can selectively kill cancer cells by causing apoptotic and/or autophagic cell death (12–21). We have shown recently that different ITCs, including PEITC, BITC, and SFN, target mitochondrial respiratory chain complexes to trigger generation of reactive oxygen species (ROS), and both apoptotic and autophagic responses to ITC treatment are intimately linked to the ROS production (15,17,19,20–22). Interestingly, normal epithelial cells (a spontaneously immortalized and non-tumorigenic MCF-10A normal mammary epithelial cell line and PrEC normal human prostate epithelial cell line) are significantly more resistant to the proapoptotic and prooxidant effect of ITCs compared with cancer cells (16,17,21,22). Despite these advances, however, the mechanism behind selectivity of ITCs for cancer cells with regards to the apoptosis induction and ROS production remains elusive.
The present study was undertaken to test a hypothesis that differential sensitivity of normal (PrEC) and cancerous human prostate cells (PC-3) to prooxidant effect of PEITC is determined by differences in basal and/or altered expression of antioxidant defense genes. We found that basal oxidative stress-antioxidant defense gene expression signature is strikingly different between PC-3 and PrEC cells. Furthermore, the PC-3 and PrEC cells respond differentially to the PEITC-mediated changes in expression of oxidative stress-antioxidative defense genes.
MATERIALS AND METHODS
Reagents
PEITC (purity >99%) was purchased from Sigma-Aldrich (St. Louis, MO). Reagents for cell culture were purchased from GIBCO-Invitrogen (Carlsbad, CA). The hydroethidine (HE) and 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate, succinimidyl ester (CDCFDA) were purchased from Molecular Probes (Eugene, OR). The antibodies against NADPH oxidase, EF hand calcium-binding domain 5 (NOX5) and Forkhead box protein M1 (FOXM1) were from Santa Cruz Biotechnology (Santa Cruz, CA). Human Oxidative Stress and Antioxidant Defense RT2 Profiler™ was obtained from SuperArray Biosciences, a QIAGEN company (Frederick, MD).
Cell Lines and Cell Culture
The PC-3 cell line was procured from the American Type Culture Collection (Manassas, VA). Monolayer cultures of PC-3 cells were maintained in F-12K Nutrient Mixture supplemented with 7% non-heat inactivated fetal bovine serum and antibiotics. The PrEC normal prostate epithelial cell line was purchased from Clonetics (now part of Lonza) and maintained in prostate epithelial basal medium (Cambrex, Walkersville, MD). Each cell line was maintained at 37°C in an atmosphere of 5% CO2 and 95% air.
Measurement of ROS generation and Hydrogen Peroxide (H2O2) Production
Stock solution of PEITC was prepared in dimethyl sulfoxide (DMSO) and diluted with complete medium immediately before use. An equal volume of DMSO (final concentration <0.1%) was added to the controls. ROS generation was assessed by flow cytometry after staining the cells with HE and CDCFDA and colorimetric analysis of H2O2 production. Flow cytometric analysis of ROS production using chemical probes HE and CDCFDA was performed essentially as described by us previously (15,22). The H2O2 production was monitored by a colorimetric assay using a kit from BioVision (Mountain View, CA). The chemical probe reacts with H2O2 to produce a byproduct with absorption maximum at 570 nm. Briefly, PC-3 or PrEC cells (3×105) were plated and allowed to attach by overnight incubation. The cells were then treated with DMSO (control) or desired concentrations of PEITC for specified time periods. The level of H2O2 in the culture medium and cell lysate was determined by following the manufacturer’s instructions.
Gene Expression Analysis
The PC-3 and PrEC cells were plated at a density of 1×106, allowed to attach by overnight incubation, and then treated with 2.5 μM PEITC or DMSO (control) for 6 hours, or left untreated to determine basal gene expression. Cells were harvested by scraping and total RNA was extracted using the RNeasy Kit (Qiagen, Valencia, CA). Reverse transcription was performed using 3 μg of total RNA and SuperScript III First-Strand Synthesis System using standard protocol. In order to evaluate the effect of PEITC treatment on the levels of genes involved in oxidative stress and antioxidant defense and for comparison of basal expression of these genes between PrEC and PC-3 cells, the Human Oxidative Stress and Antioxidant Defense RT2 Profiler™ was used. Experimental cocktail mixture was prepared immediately before the real time analyses and contained cDNA and SYBR green probe. A 25 μL of the mixture was transferred into each well of the 96-well plate provided by the manufacturer. Two-step cycling protocol was employed using an ABI 700 cycler; 10 min at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. The data were analyzed using the web-based software provided by the manufacturer. The threshold cycle (Ct) values were calculated and the genes with Ct values above 35 were considered undetected. Baseline and threshold values were set manually at the same level for all the samples to allow the comparison of multiple plates. The Ct value of each gene was adjusted for the average Ct of the housekeeping genes to generate ΔCt values. The ΔΔCt values were calculated as the difference in ΔCt between the control and treated samples, or ΔCt in PrEC versus PC-3 cellsfor the basal gene expression levels.
Immunoblotting
Immunoblotting was performed essentially as described by us previously (13–15). Briefly, PC-3 and PrEC cells were plated at a density of 1×106 cells in 100-mm culture dishes, allowed to attach by overnight incubation, and then treated with DMSO or desired concentrations of PEITC for specified time period. To examine basal levels of protein, PrEC and PC-3 cells were plated at a density of 1×106 cells in 100-mm cultured dishes, allowed to attach overnight, and then harvested without any treatment. Immunoreactive bands were visualized by enhanced chemiluminescence method. Densitometric analysis was performed to determine change in protein expression. Actin was used as a loading control.
RESULTS AND DISCUSSION
Effect of PEITC Treatment on ROS Production
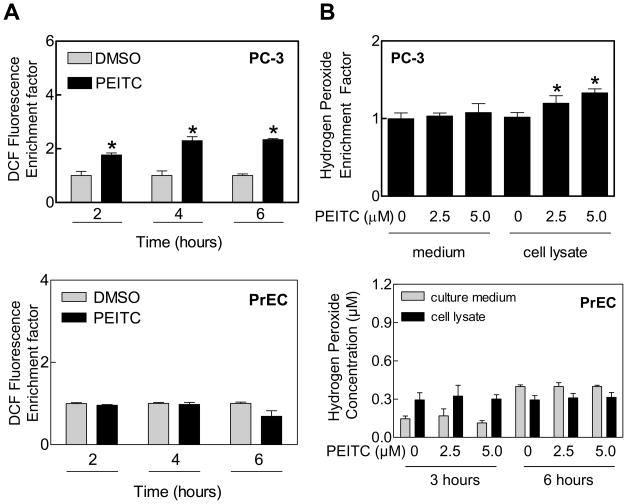
We have shown previously that the PrEC normal human prostate epithelial cell line is significantly more resistant to growth inhibition by PEITC compared with prostate cancer cells (16), and growth suppression by PEITC against prostate cancer cells is intimately linked to ROS generation (20,23). In a recently published study complementing the results shown herein, we have demonstrated that the PEITC-mediated ROS are mitochondria-derived in prostate cancer cells, including PC-3 (21). This conclusion is based on the following observations: (a) the PEITC treatment inhibits mitochondrial respiratory chain complex III and oxidative phosphorylation in PC-3 and LNCaP cells, (b) the PEITC-induced ROS production as well as histone-associated apoptotic DNA fragmentation are significantly attenuated by ectopic expression of superoxide dismutase, and (c) mitochondrial DNA deficient Rho-0 variants of PC-3 and LNCaP, which lack oxidative phosphorylation but rely on anaerobic glycolysis for survival, are significantly more resistant to PEITC-mediated ROS production, apoptotic DNA fragmentation, collapse of mitochondrial membrane potential, Bax activation, and caspse-3 activation compared with respective wild-type cells (21). In the present study, we questioned if the differential sensitivity of cancerous versus normal prostate epithelial cells to prooxidant effect of PEITC was related to differences in their antioxidant defense capacity. As shown in Fig. 1A, exposure of PC-3 cells to 5 μM PEITC resulted in ROS production, as evidenced by flow cytometric analysis of CDCFDA oxidation, in a time-dependent manner. The PEITC-mediated oxidation of CDCFDA was not observed in the PrEC cell line (Fig. 1A). Consistent with these results, 6 hour treatment with PEITC increased H2O2 levels in PC-3 cells but not in the PrEC cell line (Fig. 1B).
Figure 1.
Generation of reactive oxygen species by PEITC treatment in PC-3 cells. A) Flow cytometric analysis of DCF fluorescence (a measure of ROS production) in PC-3 and PrEC cells treated with DMSO (control) or 5 μM PEITC for the indicated time periods. B) Hydrogen peroxide production in medium and lysate of PC-3 and PrEC cells treated with the indicated concentrations of PEITC for 6 hour (PC-3) or 3 and 6 hours (PrEC). Data represent mean ±SD (n=3). *Significantly different compared with corresponding DMSO-treated control by Student’s t-test.
Basal Expression of Redox Genes in PC-3 and PrEC Cells
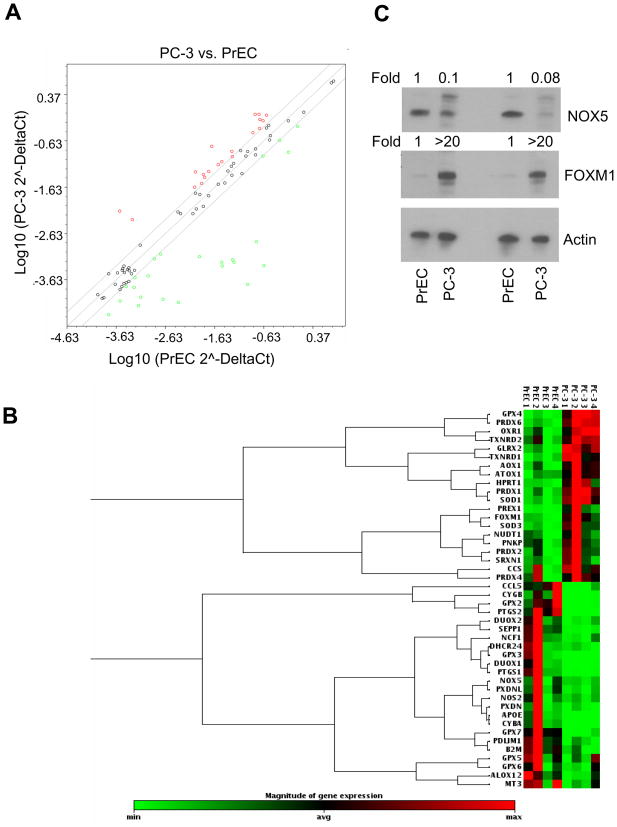
Next, we explored the possibility whether resistance of PrEC cells to prooxidant effect of PEITC compared with PC-3 was due to differences in basal expression of antioxidant defense genes. We tested this possibility by real time PCR for a set of 84 selected genes involved in the oxidative stress and antioxidant defense. This analysis revealed that 44 genes were differentially expressed between PC-3 and PrEC cells (Fig. 2A,B). Of the 84 genes, 20 were up-regulated and 24 were down-regulated in PC-3 cells compared with PrEC (Table 1). Genes statistically significantly down-regulated in PC-3 cells included Apolipoprotein E, dual oxidase 1 and 2, some members of the glutathione peroxidase family to name a few. On the other hand, genes that were up-regulated in the PC-3 cell line compared with PrEC included FOXM1, superoxide dismutase 1 (soluble) and 2 (mitochondrial), among others (Table 1). Gene expression differences were confirmed by Western blotting for selected proteins. In agreement with real-time PCR results, basal expression of NOX5 protein was significantly higher in the PrEC cell line compared with PC-3 (Fig. 2C). Conversely, higher constitutive expression of FOXM1 protein was clearly visible in the PC-3 relative to the PrEC cells (Fig. 2C).
Figure 2.
Comparative expression of oxidative stress response and antioxidant defense genes in PC-3 and PrEC cells. A) Relative expression of genes involved in oxidative stress response and antioxidant defense between PrEC and PC-3 cells. Genes up- and down-regulated are represented by red and green dots, respectively. Scatter plot shows a log transformation of the relative expression level of each gene between PC-3 and PrEC cells. B) Cluster analysis demonstrating differences in gene expression between PC-3 and PrEC cells. Genes represented in the gene cluster analysis are limited to those whose expression differs between the cells by at least 2-fold. Two independently prepared samples of each cell line in duplicate were used for gene expression profiling (n=4). C) Immunoblotting for NOX5 and FOXM1 using two lysates from PrEC and PC-3 cells. Membranes were stripped and re-probed with anti-actin antibody to ensure equal protein loading.
Table 1.
Gene expression in prostate cancer cells (PC-3) and in normal prostate epithelial cells (PrEC)
| Gene name | Symbol | UniGene | PC-3/PrEC | P-value |
|---|---|---|---|---|
| Aldehyde oxidase 1 | AOX1 | Hs.406238 | 5.233 | 0.00639 |
| ATX1 antioxidant protein 1 homolog (yeast) | ATOX1 | Hs.125213 | 2.491 | 0.00602 |
| Copper chaperone for superoxide dismutase | CCS | Hs.502917 | 2.241 | 0.11227 |
| Forkhead box M1 | FOXM1 | Hs.239 | 8.622 | 0.01064 |
| Glutaredoxin 2 | GLRX2 | Hs.458283 | 2.554 | 0.00029 |
| Glutathione peroxidase 4 (phospholipid hydroperoxidase) | GPX4 | Hs.433951 | 2.837 | 0.00017 |
| Nudix (nucleoside diphosphate linked moiety X)-type motif 1 | NUDT1 | Hs.534331 | 2.535 | 0.12080 |
| Oxidation resistance 1 | OXR1 | Hs.148778 | 3.199 | 0.00400 |
| Polynucleotide kinase 3′-phosphatase | PNKP | Hs.78016 | 2.870 | 0.11285 |
| Peroxiredoxin 1 | PRDX1 | Hs.180909 | 2.005 | 0.00075 |
| Peroxiredoxin 2 | PRDX2 | Hs.432121 | 3.516 | 0.05590 |
| Peroxiredoxin 4 | PRDX4 | Hs.83383 | 2.266 | 0.18258 |
| Peroxiredoxin 6 | PRDX6 | Hs.120 | 5.692 | 0.00005 |
| Phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | PREX1 | Hs.153310 | 25.880 | 0.05518 |
| Superoxide dismutase 1, soluble | SOD1 | Hs.443914 | 3.166 | 0.00044 |
| Superoxide dismutase 2, mitochondrial | SOD3 | Hs.487046 | 9.249 | 0.01570 |
| Sulfiredoxin 1 homolog (S. cerevisiae) | SRXN1 | Hs.516830 | 2.595 | 0.09924 |
| Thioredoxin reductase 1 | TXNRD1 | Hs.708065 | 4.641 | 0.00324 |
| Thioredoxin reductase 2 | TXNRD2 | Hs.443430 | 2.562 | 0.01016 |
| Hypoxanthine phosphoribosyltransferase 1 | HPRT1 | Hs.412707 | 2.647 | 0.00857 |
| Arachidonate 12-lipoxygenase | ALOX12 | Hs.654431 | 0.4583 | 0.14090 |
| Apolipoprotein E | APOE | Hs.654439 | 0.0116 | 0.00794 |
| Chemokine (C-C motif) ligand 5 | CCL5 | Hs.514821 | 0.1343 | 0.10734 |
| Cytochrome b-245, alpha polypeptide | CYBA | Hs.513803 | 0.0017 | 0.05172 |
| Cytoglobin | CYGB | Hs.95120 | 0.0139 | 0.11042 |
| 24-dehydrocholesterol reductase | DHCR24 | Hs.498727 | 0.1795 | 0.10203 |
| Dual oxidase 1 | DUOX1 | Hs.272813 | 0.0169 | 0.05656 |
| Dual oxidase 2 | DUOX2 | Hs.71377 | 0.0971 | 0.00523 |
| Glutathione peroxidase 2 (gastrointestinal) | GPX2 | Hs.2704 | 0.0097 | 0.10246 |
| Glutathione peroxidase 3 (plasma) | GPX3 | Hs.386793 | 0.0094 | 0.23506 |
| Glutathione peroxidase 5 (epididymal androgen-related protein) | GPX5 | Hs.248129 | 0.2942 | 0.24582 |
| Glutathione peroxidase 6 (olfactory) | GPX6 | Hs.448570 | 0.4428 | 0.00320 |
| Glutathione peroxidase 7 | GPX7 | Hs.43728 | 0.0421 | 0.07772 |
| Metallothionein 3 | MT3 | Hs.73133 | 0.2954 | 0.09047 |
| Neutrophil cytosolic factor 1 | NCF1 | Hs.647047 | 0.2756 | 0.18396 |
| Nitric oxide synthase 2, inducible | NOS2 | Hs.709191 | 0.4796 | 0.17236 |
| NADPH oxidase, EF-hand Ca binding domain 5 | NOX5 | Hs.657932 | 0.4679 | 0.14090 |
| PDZ and LIM domain 1 | PDLIM1 | Hs.368525 | 0.4975 | 0.01117 |
| Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | Hs.201978 | 0.0176 | 0.06976 |
| Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | PTGS2 | Hs.196384 | 0.002 | 0.00293 |
| Peroxidasin homolog (Drosophila) | PXDN | Hs.332197 | 0.4788 | 0.22395 |
| Peroxidasin homolog (Drosophila)-like | PXDNL | Hs.444882 | 0.2552 | 0.10323 |
| Selenoprotein P, plasma, 1 | SEPP1 | Hs.275775 | 0.0646 | 0.01386 |
| Beta-2-microglobulin | B2M | Hs.534255 | 0.4284 | 0.01100 |
Cancer cells are characterized by increased level of ROS and reduced ability to remove these deleterious species (24,25). It has been postulated that persistent oxidative stress promotes tumor cell survival, proliferation, migration/invasion, and angiogenesis and inhibits apoptosis by activating certain redox-sensitive transcription factors such as NF-κB and AP-1 (26–28). The present study reveals that the basal expression of glutathione peroxidases and NOX5 is significantly lower in PC-3 cells than in the PrEC cell line. Glutathione peroxidases are responsible for reduction of H2O2, as well as soluble fatty acid hydroperoxides (29). The glutathione peroxidases function as antioxidant enzymes and their activity has been inversely associated with the development of various types of malignancies including prostate cancer. This hypothesis is supported by the findings from several clinical studies which reveal that prostate cancer patients exhibit lower levels of glutathione peroxide activity in the plasma as well as in prostate tissue when compared to men with benign hyperplasia or healthy controls (30–32). Levels of glutathione peroxidase 1 were 5-fold lower in PC-3 (R), a variant that is more resistant to several anticancer drugs as compared to the wild type PC-3 cells (33). In accordance with our findings, glutathione peroxidase 2 was previously shown to be down-regulated in Nlkx3.1 mutant mice, which demonstrated prostatic intraepithelial neoplasia (34). Additionally, Nlkx3.1 and PTEN double knock out mice showed down-regulation of both glutathione peroxidase 2 and glutathione peroxidase 3, which would indicate that in this model glutathione peroxidases were being progressively down-regulated as the disease developed. The glutathione peroxidase 3 was also shown to be affecting metastatic ability of prostate cancer cells and its down-regulation correlated with CpG methylation (35,36). The glutathione peroxidase 5, which is expressed in the male reproductive organs, was previously shown to protect immature spermatozoa from oxidative damage (37). Even though this isoform has not been directly linked to prostate cancer, its down-regulation in our model may further contribute to environment promoting oxidative stress and prostate carcinogenesis. Glutathione peroxidase 6 (expressed in olfactory epithelium) and glutathione peroxidase 7 (putative) have not directly been linked to prostate cancer development, however it is of interest that glutathione peroxidase 7 is hypermethylated in Barrett’s adenocarcinoma (38).
Another gene down-regulated in PC-3 cells was the NOX5, which has been shown to regulate growth and apoptosis in DU145 human prostate cancer cells (39). The NOX5 has been shown to contribute to the ROS generation by generating large amounts of superoxide anion (40). It would seem counterintuitive for prostate cancer cells in our system to down-regulate expression of this enzyme. However, it is possible that PC-3 cells rely mostly on mitochondria for generation of ROS and additional superoxide production by NOX5 would actually lead to dangerous increases in ROS and to apoptosis. This speculation is plausible considering hydrogen peroxide-mediated down-regulation of NOX5 has been observed in MCF-7 human breast cancer cells (41).
Interestingly, expression of the FOXM1 is markedly higher in the PC-3 cell line compared with PrEC (Table 1). FOXM1 is expressed in a variety of cancers including prostate (42–44). It was previously shown that increased levels of FOXM1 accelerated prostate cancer development and progression in TRAMP and LADY mouse models (44). Additionally, inhibition of FOXM1 expression with FOXM1-specific siRNA in DU145, PC-3 and LNCaP prostate cancer cells led to reduction in proliferation in association with reduction in cyclinA2 and cyclinB1 proteins (44). The FOXM1 was also up-regulated in metastatic prostate caner tissue samples, which suggests that FOXM1 is involved in regulation of tumor progression and the development of metastasis (45). The idea that FOXM1 regulates metastasis was supported by a study in breast cancer which revealed that down-regulation of FOXM1 attenuated not only cell proliferation but also migration and invasion of MDA-MB-231 breast cancer cell (46). Simultaneously, reduced FOXM1 levels cause reduced secretion of factors such as MMP-2, MMP-9, and VEGF, all of which promote extracellular matrix remodeling and angiogenesis (46). The FOXM1 was also implicated in oxidative stress response (47). Specifically, Ras-induced ROS overexpressed FOXM1 which in turn modulated the ROS levels by inducing expression of genes such as Mn-SOD (47). This may explain why we saw moderate upregulation of SOD in our system and provide a mechanism by which prostate cancer cells modulate ROS levels to escape cell death.
PEITC Treatment Differentially Altered Expression of Genes Involved in Oxidative Stress Response and Antioxidant Defense in PC-3 andPrEC cells
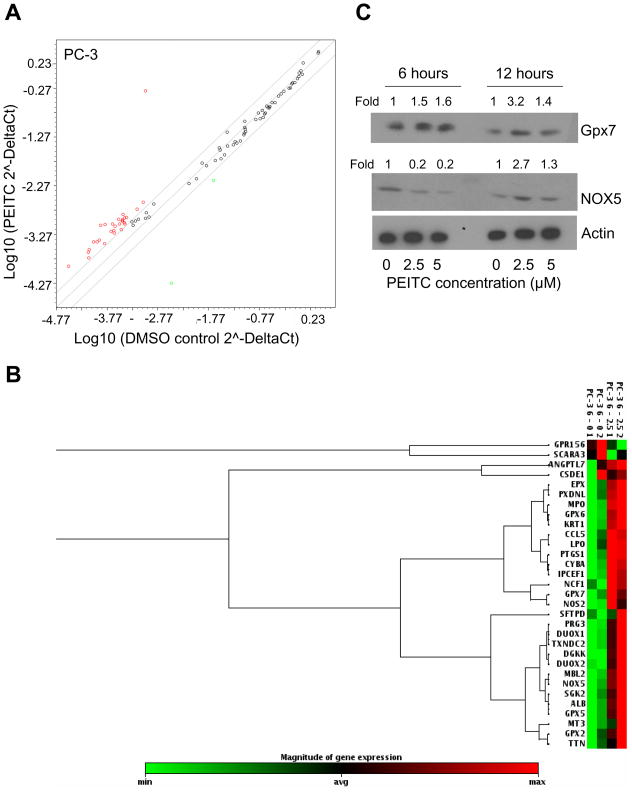
Next, we proceeded to determine the effect of PEITC treatment on genes associated with oxidative stress response and antioxidant defense in these cells. Exposure of PC-3 cells to growth suppressive and proapoptotic concentration of PEITC (2.5 μM for 6 hours) resulted in up-regulation of 29 genes and down-regulation of only 2 genes at a 2-fold level (Fig. 3A,B). Genes up-regulated in response to the PEITC treatment in PC-3 cells included some glutathione peroxidases (2, 5, 6, and 7), myeloperoxidase, and lactoperoxidase to name a few. The two genes down-regulated included G protein-coupled receptor 156 and scavenger receptor class A, member 3 (Table 2). We selected a few proteins to confirm our initial findings. As can be seen in Fig. 3C, we observed increased protein levels of glutathione peroxidase 7 at 6 and 12 hour time points in response to 2.5 and 5 μM PEITC treatments. Additionally, NOX5 protein level was slightly up-regulated after 12 hour exposure to PEITC (Fig. 3C).
Figure 3.
Effect of PEITC treatment on expression of oxidative stress response and antioxidant defense genes in PC-3 cells. A) Scatter plot shows a log transformation of the relative expression level of each gene between PC-3 cells treated with DMSO (control) and 2.5 μM PEITC for 6 hours. B) Cluster analysis demonstrating differences in gene expression in PC-3 cells in response to PEITC treatment. Cluster analysis shows only those genes which have a minimum of 2-fold change in expression in response to PEITC treatment. The PC-3 cells were treated with DMSO or 2.5 μM PEITC for 6 hours. Data are from duplicate measurements (n=2). C) Immunoblotting for glutathione peroxidase 7 and NOX5 using lysates from PC-3 cells treated with 2.5 and 5 μM PEITC or DMSO (control) for 6 or 12 hours. In order to ensure equal lysate protein loading, membranes were stripped and re-probed with anti-actin antibody. Change in protein level is expressed relative to DMSO-treated control.
Table 2.
Gene expression changes in PC-3 cells treated with 2.5 μM PEITC for 6 hrs.
| Gene name | Symbol | UniGene | PEITC/Control |
|---|---|---|---|
| Albumin | ALB | Hs.418167 | 3.9945 |
| Angiopoietin-like 7 | ANGPTL7 | Hs.146559 | 2.0677 |
| Chemokine (C-C motif) ligand 5 | CCL5 | Hs.514821 | 2.2548 |
| Cold shock domain containing E1, RNA-binding | CSDE1 | Hs.69855 | 492.1656 |
| Cytochrome b-245, alpha polypeptide | CYBA | Hs.513803 | 3.2221 |
| Diacylglycerol kinase, kappa | DGKK | Hs.631770 | 4.1068 |
| Dual oxidase 1 | DUOX1 | Hs.272813 | 2.9445 |
| Dual oxidase 2 | DUOX2 | Hs.71377 | 3.4895 |
| Eosinophil peroxidase | EPX | Hs.279259 | 2.8939 |
| Glutathione peroxidase 2 (gastrointestinal) | GPX2 | Hs.2704 | 2.5193 |
| Glutathione peroxidase 5 (epididymal androgen-related protein) | GPX5 | Hs.248129 | 3.0483 |
| Glutathione peroxidase 6 (olfactory) | GPX6 | Hs.448570 | 4.0222 |
| Glutathione peroxidase 7 | GPX7 | Hs.43728 | 3.1667 |
| Keratin 1 (epidermolytic hyperkeratosis) | KRT1 | Hs.80828 | 3.8053 |
| Lactoperoxidase | LPO | Hs.234742 | 6.4666 |
| Mannose-binding lectin (protein C) 2, soluble | MBL2 | Hs.499674 | 3.1777 |
| Myeloperoxidase | MPO | Hs.458272 | 3.394 |
| Metallothionein 3 | MT3 | Hs.73133 | 2.8343 |
| Neutrophil cytosolic factor 1, (chronic granulomatous disease, autosomal 1) | NCF1 | Hs.647047 | 5.4945 |
| Nitric oxide synthase 2A (inducible, hepatocytes) | NOS2 | Hs.706746 | 2.7856 |
| NADPH oxidase, EF-hand calcium binding domain 5 | NOX5 | Hs.657932 | 2.3506 |
| Interaction protein for cytohesin exchange factors 1 | IPCEF1 | Hs.146100 | 3.1998 |
| Proteoglycan 3 | PRG3 | Hs.251386 | 3.0168 |
| Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | Hs.201978 | 3.4895 |
| Peroxidasin homolog (Drosophila)-like | PXDNL | Hs.444882 | 4.0362 |
| Surfactant, pulmonary-associated protein D | SFTPD | Hs.253495 | 2.3751 |
| Serum/glucocorticoid regulated kinase 2 | SGK2 | Hs.472793 | 2.6445 |
| Titin | TTN | Hs.654592 | 2.4334 |
| Thioredoxin domain-containing 2 (spermatozoa) | TXNDC2 | Hs.98712 | 2.9241 |
| G protein-coupled receptor 156 | GPR156 | Hs.333358 | 0.0171 |
| Scavenger receptor class A, member 3 | SCARA3 | Hs.128856 | 0.3226 |
We have previously described that PEITC can induce cell cycle arrest as well as apoptotic and autophagic cell death in prostate cancer cells (14,16,18,21,23). Results of the present study indicate that prostate cancer cells do attempt to counteract this additional ROS threat by up-regulating a variety of peroxidases including: glutathione peroxidases 6 and 7, lactoperoxiadase, and myeloperoxidase. All of these enzymes are able to reduce oxidative stress and may function to protect the cancer cell from apoptosis. Simultaneously though, PEITC increases expression of dual oxidase 2, which is responsible for generation of hydrogen peroxide, and down-regulates scavenger receptor class A, member 3, which in turn function to protect from ROS-induced damage.
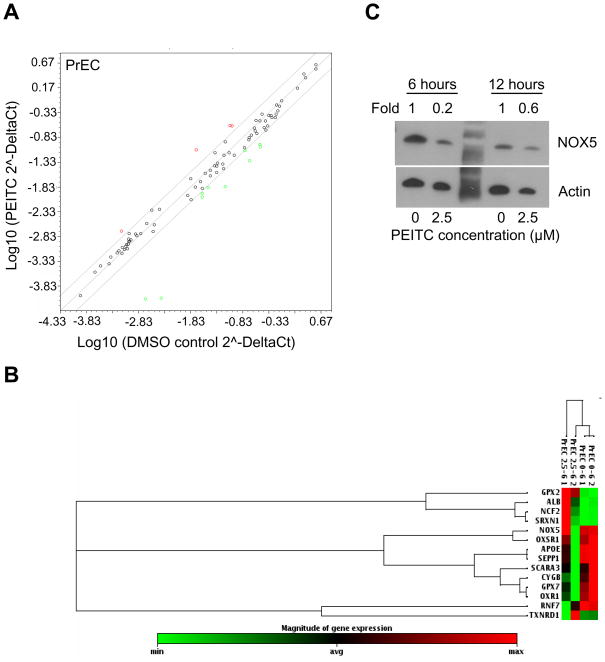
Strikingly, a similar PEITC treatment produced a different response in the PrEC cell line (Fig. 4A,B). Specifically, treatment of PrEC cells with 2.5 μM PEITC for 6 hour resulted in change in expression of only 14 genes (Table 3). In contrast to PC-3 cells, more genes were down-regulated by PEITC treatment in the PrEC cell line (Table 3). Among the genes affected by the PEITC treatment 10 were down-regulated and four were up-regulated. These results indicate that the normal prostate epithelial cells respond by changing expression of several genes. In fact, it is possible that this state of readiness exhibited by untreated PrEC cells is the cause of the resistance of these cells to PEITC-induced ROS generation. Specifically, in our system PrEC cells responded by up-regulating glutathione peroxidase 2 and down-regulating genes such as NOX5. Our results suggest that PEITC does not induce a dramatic change in PrEC cells, and these cells only require modest change in their status to balance their redox status. It is not clear why Apolipoprotein E and glutathione peroxidase 7 were down-regulated in our system. It is possible that these proteins do not play a major role in the regulation of oxidative stress in normal prostate epithelium, or that their levels are high enough in the basal state and no additional gene expression is required to protect the cells.
Figure 4.
Effect of PEITC treatment on expression of oxidative stress response and antioxidant defense genes in PrEC cells. A) Scatter plot shows a log transformation of the relative expression level of each gene between the PrEC cells treated with DMSO and 2.5 μM PEITC for 6 hours. B) Cluster analysis demonstrating differences in gene expression in PrEC cells in response to PEITC treatment. Cluster analysis shows only those genes whose expression was changed by a minimum of 2-fold in response to PEITC treatment. C) Immunoblotting for NOX5 using lysates from PrEC cells treated with DMSO or the indicated concentrations of PEITC for 6 or 12 hour. Membranes was stripped and re-probed with anti-actin antibody to ensure equal protein loading. Change in the protein level is expressed relative to DMSO-treated control.
Table 3.
Gene expression changes in PrEC cells treated with 2.5 μM PEITC for 6 hrs.
| Gene name | Symbol | UniGene | PEITC/Control |
|---|---|---|---|
| Albumin | ALB | Hs.418167 | 2.7435 |
| Glutathione peroxidase 2 (gastrointestinal) | GPX2 | Hs.2704 | 3.108 |
| Neutrophil cytosolic factor 2 (65kDa, chronic granulomatous disease, autosomal 2) | NCF2 | Hs.587558 | 4.426 |
| Sulfiredoxin 1 homolog (S. cerevisiae) | SRXN1 | Hs.516830 | 2.7151 |
| Apolipoprotein E | APOE | Hs.654439 | 0.2996 |
| Cytoglobin | CYGB | Hs.95120 | 0.2208 |
| Glutathione peroxidase 7 | GPX7 | Hs.43728 | 0.0207 |
| NADPH oxidase, EF-hand calcium binding domain 5 | NOX5 | Hs.657932 | 0.0403 |
| Oxidation resistance 1 | OXR1 | Hs.148778 | 0.4478 |
| Oxidative-stress responsive 1 | OXSR1 | Hs.475970 | 0.4969 |
| Ring finger protein 7 | RNF7 | Hs.134623 | 0.2537 |
| Scavenger receptor class A, member 3 | SCARA3 | Hs.128856 | 0.374 |
| Selenoprotein P, plasma, 1 | SEPP1 | Hs.275775 | 0.4417 |
| Thioredoxin reductase 1 | TXNRD1 | Hs.680369 | 0.3441 |
CONCLUSIONS
Based on the results shown herein we speculate that the differences in basal as well as altered expression of oxidative stress-related and antioxidant defense genes result in increased susceptibility of PC-3 cells to PEITC-induced ROS generation and apoptosis compared to normal prostate epithelial cell line PrEC. However, as a note of caution, our conclusion is based on a single pair of cell line. Additional studies with multiple normal and cancerous cell pairs are needed to firmly establish the validity of our conclusions.
Acknowledgments
The authors thank Dong Xiao and Eun-Ryeong Hahm for technical assistance.
Grant support: This investigation was supported by the USPHS grant CA101753-07, awarded by the National Cancer Institute.
References
- 1.Verhoeven DT, Goldbohm RA, van Poppel G, Verhagen H, van den Brandt PA. Epidemiological studies on brassica vegetables and cancer risk. Cancer Epidemiol Biomarkers Prev. 1996;5:733–48. [PubMed] [Google Scholar]
- 2.Ambrosone CB, McCann SE, Freudenheim JL, Marshall JR, Zhang Y, Shields PG. Breast cancer risk in premenopausal women is inversely associated with consumption of broccoli, a source of isothiocyanates, but is not modified by GST genotype. J Nutr. 2004;134:1134–8. doi: 10.1093/jn/134.5.1134. [DOI] [PubMed] [Google Scholar]
- 3.Kolonel LN, Hankin JH, Whittemore AS, et al. Vegetables, fruits, legumes and prostate cancer: a multiethnic case-control study. Cancer Epidemiol Biomarkers Prev. 2000;9:795–804. [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 5.Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochem. 2001;56:5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 6.Hecht SS, Trushin N, Rigotty J, et al. Complete inhibition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced rat lung tumorigenesis and favorable modification of biomarkers by phenethyl isothiocyanate. Cancer Epidemiol Biomarkers Prev. 1996;5:645–52. [PubMed] [Google Scholar]
- 7.Pereira MA. Chemoprevention of diethylnitrosamine-induced liver foci and hepatocellular adenomas in C3H mice. Anticancer Res. 1995;15:1953–6. [PubMed] [Google Scholar]
- 8.Stoner GD, Morrissey DT, Heur YH, Galati AJ, Daniel EM, Wagner SA. Inhibitory effects of Phenethyl isothiocyanate on N-nitrosobenzylmethylamine carcinogenesis in the rat esophagus. Cancer Res. 1991;51:2063–8. [PubMed] [Google Scholar]
- 9.Singh SV, Warin R, Xiao D, et al. Sulforaphane inhibits prostate carcinogenesis and pulmonary metastasis in TRAMP mice in association with increased cytotoxicity of natural killer cells. Cancer Res. 2009;69:2117–25. doi: 10.1158/0008-5472.CAN-08-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warin R, Chambers WH, Potter DM, Singh SV. Prevention of mammary carcinogenesis in MMTV-neu mice by cruciferous vegetable constituent benzyl isothiocyanate. Cancer Res. 2009;69:9473–80. doi: 10.1158/0008-5472.CAN-09-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khor TO, Cheung WK, Prawan A, Reddy BS, Kong AN. Chemoprevention of familial adenomatous polyposis in Apc(Min/+) mice by phenethyl isothiocyanate (PEITC) Mol Carcin. 2008;47:321–5. doi: 10.1002/mc.20390. [DOI] [PubMed] [Google Scholar]
- 12.Chen YR, Han J, Kori R, Kong AN, Tan TH. Phenylethyl isothiocyanate induces apoptotic signaling via suppressing phosphatase activity against c-Jun N-terminal kinase. J Biol Chem. 2002;277:39334–42. doi: 10.1074/jbc.M202070200. [DOI] [PubMed] [Google Scholar]
- 13.Xiao D, Singh SV. Phenethyl isothiocyanate-induced apoptosis in p53-deficient PC-3 human prostate cancer cell line is mediated by extracellular signal-regulated kinases. Cancer Res. 2002;62:3615–9. [PubMed] [Google Scholar]
- 14.Xiao D, Johnson CS, Trump DL, Singh SV. Proteasome-mediated degradation of cell division cycle 25C and cyclin-dependent kinase 1 in phenethyl isothiocyanate-induced G2-M-phase cell cycle arrest in PC-3 human prostate cancer cells. Mol Cancer Ther. 2004;3:567–75. [PubMed] [Google Scholar]
- 15.Singh SV, Srivastava SK, Choi S, et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J Biol Chem. 2005;280:19911–24. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- 16.Xiao D, Zeng Y, Choi S, Lew KL, Nelson JB, Singh SV. Caspase-dependent apoptosis induction by phenethyl isothiocyanate, a cruciferous vegetable-derived cancer chemopreventive agent, is mediated by Bak and Bax. Clin Cancer Res. 2005;11:2670–9. doi: 10.1158/1078-0432.CCR-04-1545. [DOI] [PubMed] [Google Scholar]
- 17.Xiao D, Powolny AA, Singh SV. Benzyl isothiocyanate targets mitochondrial respiratory chain to trigger ROS-dependent apoptosis in human breast cancer cells. J Biol Chem. 2008;283:30151–63. doi: 10.1074/jbc.M802529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bommareddy A, Hahm ER, Xiao D, et al. Atg5 regulates phenethyl isothiocyanate-induced autophagic and apoptotic cell death in human prostate cancer cells. Cancer Res. 2009;69:3704–12. doi: 10.1158/0008-5472.CAN-08-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao D, Powolny AA, Antosiewicz J, et al. Cellular responses to cancer chemopreventive agent D,L-sulforaphane in human prostate cancer cells are initiated by mitochondrial reactive oxygen species. Pharm Res. 2009;26:1729–38. doi: 10.1007/s11095-009-9883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao D, Singh SV. p66shc is indispensable for phenethyl isothiocyanate-induced apoptosis in human prostate cancer cells. Cancer Res. 2010;70:3150–8. doi: 10.1158/0008-5472.CAN-09-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao D, Powolny A, Moura MB, et al. Phenethyl isothiocyanate inhibits oxidative phosphorylation to trigger reactive oxygen species-mediated death of human prostate cancer cells. J Biol Chem. 2010;285:26558–69. doi: 10.1074/jbc.M109.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao D, Vogel V, Singh SV. Benzyl isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by Bax and Bak. Mol Cancer Ther. 2006;5:2931–45. doi: 10.1158/1535-7163.MCT-06-0396. [DOI] [PubMed] [Google Scholar]
- 23.Xiao D, Lew KL, Zeng Y, et al. Phenethyl isothiocyanate-induced apoptosis in PC-3 human prostate cancer cells is mediated by reactive oxygen species-dependent disruption of the mitochondrial membrane potential. Carcinogenesis. 2006;27:2223–34. doi: 10.1093/carcin/bgl087. [DOI] [PubMed] [Google Scholar]
- 24.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–8. [PubMed] [Google Scholar]
- 25.Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 26.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Wu WS. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 2006;25:695–705. doi: 10.1007/s10555-006-9037-8. [DOI] [PubMed] [Google Scholar]
- 28.Cerutti PA. Prooxidant states and tumor promotion. Science. 1985;227:375–81. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 29.Brigelius-Flohé R, Kipp A. Glutathione peroxidases in different stages of carcinogenesis. Biochim Biophys Acta. 2009;1790:1555–68. doi: 10.1016/j.bbagen.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Zachara BA, Szewczyk-Golec K, Tyloch J, et al. Blood and tissue selenium concentrations and glutathione peroxidase activities in patients with prostate cancer and benign prostate hyperplasia. Neoplasma. 2005;52:248–54. [PubMed] [Google Scholar]
- 31.Arsova-Sarafinovska Z, Eken A, Matevska N, et al. Increased oxidative/nitrosative stress and decreased antioxidant enzyme activities in prostate cancer. Clin Biochem. 2009;42:1228–35. doi: 10.1016/j.clinbiochem.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Aydin A, Arsova-Sarafinovska Z, Sayal A, et al. Oxidative stress and antioxidant status in non-metastatic prostate cancer and benign prostatic hyperplasia. Clin Biochem. 2006;9:176–9. doi: 10.1016/j.clinbiochem.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki H, Schneider E, Myers CE, Sinha BK. Oncogene overexpression and de novo drug-resistance in human prostate cancer cells. Biochim Biophys Acta. 1994;1226:89–96. doi: 10.1016/0925-4439(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 34.Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. 2005;65:6773–9. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
- 35.Yu YP, Yu G, Tseng G, et al. Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 2007;67:8043–50. doi: 10.1158/0008-5472.CAN-07-0648. [DOI] [PubMed] [Google Scholar]
- 36.Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–27. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- 37.Aitken RJ. Gpx5 protects the family jewels. J Clin Invest. 2009;119:1849–51. doi: 10.1172/JCI39688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng DF, Razvi M, Chen H, et al. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinoma. Gut. 2009;58:5–15. doi: 10.1136/gut.2007.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brar SS, Corbin Z, Kennedy TP, et al. NOX5 NAD(P)H oxidase regulates growth and apoptosis in DU145 prostate cancer cells. Am J Physiol Cell Physiol. 2003;285:C353–69. doi: 10.1152/ajpcell.00525.2002. [DOI] [PubMed] [Google Scholar]
- 40.Ushio-Fukai M, Nakamura Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008;266:37–52. doi: 10.1016/j.canlet.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chua PJ, Yip GW, Bay BH. Cell cycle arrest induced by hydrogen peroxide is associated with modulation of oxidative stress related genes in breast cancer cells. Exp Biol Med. 2009;234:1086–94. doi: 10.3181/0903-RM-98. [DOI] [PubMed] [Google Scholar]
- 42.Madureira PA, Varshochi R, Constantinidou D, et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281:25167–76. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- 43.Waseem A, Ali M, Odell EW, Fortune F, Teh MT. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010;46:536–42. doi: 10.1016/j.oraloncology.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 44.Kalin TV, Wang IC, Ackerson TJ, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–20. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandran UR, Ma C, Dhir R, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmad A, Wang Z, Kong D, et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2010;122:337–46. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- 47.Park HJ, Carr JR, Wang Z, et al. FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. 2009;28:2908–18. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]