Abstract
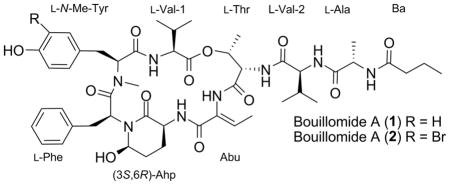
Bouillomides A (1) and B (2) are two depsipeptide analogues of dolastatin 13. Isolated from a Guamanian sample of Lyngbya bouillonii, the planar structures were elucidated on the basis of HR-ESI-MS and NMR data, while the absolute configurations were determined by employing functional group conversions, modified Marfey’s analysis, and detailed analyses of ROESY correlations. Compounds 1 and 2 selectively inhibited serine proteases elastase (IC50 = 1.9 μM for both) and chymotrypsin (IC50 = 0.17 and 9.3 μM, respectively) while showing no inhibition of trypsin (IC50 > 100 μM).
A number of cyanobacterial compounds inhibit serine proteases, which are enzymes central to the regulation of digestion in most animals.1,2 The family of inhibitory depsipeptides that are exemplified by dolastatin 133 are one of the most widely distributed in Nature. While the patriarchal compound was originally isolated from the sea hare Dolabella auricularia, subsequent studies showed that the metabolite was actually produced by cyanobacteria upon which the nudibranch fed.4,5 Dolastatin 13 analogues have now been isolated from marine,6–11 freshwater,12–14 and terrestrial15 cyanobacterial strains collected all over the world. The distinguishing features of these depsipeptides are a 3-amino-6-hydroxy-2-piperidone (Ahp) residue contained within a 20-membered macrocycle, which is formed via an ester linkage between a threonine hydroxyl group and the hydrophobic C-terminus of the precursor peptide.4 The remaining units are highly variable though, particularly the residues that cap the threonine N-terminus. The responsible biosynthetic gene clusters, therefore, have a high degree of similarity in their non-ribosomal peptide synthetase (NRPS) domains. Since stringent substrate specificity is not always observed with NRPS domains, even analogues from the same strain will display remarkable structural diversity. While the actual ecological role of these compounds remain uncertain,16 the active sites of several proteases (namely chymotrypsin, trypsin and elastase) are fairly well conserved.2 Production of these suites of structurally diverse serine protease inhibitors is therefore proposed to be primarily for their anti-predation effect.1
As part of a larger program examining the unique chemical diversity inherent to marine cyanobacteria, two new members of the dolastatin 13 family, bouillomides A (1) and B (2) were discovered. Discussed herein are the isolation, structure elucidation and protease screening of these two compounds, which were isolated from a Guamanian sample of Lyngbya bouillonii (L. Hoffman and V. Demoulin).
A mass guided approach was used to purify bouillomides A (1) and B (2) from the crude extract of the L. bouillonii sample. First detected by LC-MS analyses, the compounds exhibited pseudomolecular ions with isotopic patterns that indicated the two compounds differed by the substitution of a bromine. Accordingly, the total crude extract was subjected to a modified Kupchan protocol, in which the extract was partitioned successively17 between hexanes, CH2Cl2, CH3OH and H2O. Compounds 1 and 2 were isolated from the CH2Cl2 partition. Three rounds of fractionation using RP-HPLC afforded the amorphous white powders, bouillomides A (1) and B (2).
The planar structure of bouillomide A (1) was elucidated using the HR-ESI-MS and NMR data. The HR-ESI-MS spectrum for 1 displayed a pseudomolecular ion at m/z = 983.4843 [M + Na]+ consistent with a molecular formula of C49H68N8O12. The 13C NMR data showed signals for eight amide carbons (δC = 165.8 to 172.6 ppm) and another carbon resonance attributed to an ester carbonyl, based on the carbon chemical shift of δC 171.9 and the low-field resonance observed for an acyloxy proton at δH = 5.52 ppm in the 1H NMR spectrum. Taken together these data suggested a depsipeptide structure for 1. All peaks in the 1H and 13C spectra were quickly assigned from analyses of the TOCSY, COSY, HSQC, HMBC, and ROESY data (Table 1).
Table 1.
NMR Data of Bouillomide A (1) in DMSO-d6 a
| unit | position | δC, typeb | δH, mult. (J in Hz) | COSY | HMBC (H to C)c | ROESYc |
|---|---|---|---|---|---|---|
| Val-1 | 1 | 171.9, qC | - | - | - | - |
| 2 | 56.1, CH | 4.63, m | 3, NH | 1 | 3, 4, 5, NH | |
| 3 | 30.7, CH | 2.06, m | 2, 4, 5 | 1 | 2, 4, 5 | |
| 4 | 19.2, CH3 | 0.86, d (7.0) | 3 | 2, 3, 5 | 2, 3, 5, 4(Thr) | |
| 5 | 17.4, CH3 | 0.74, d (7.0) | 3 | 2, 3, 4 | 2, 3, 4, 4(Thr) | |
| NH | - | 7.52, d (8.0) | 2 | 2, 2(N-Me-Tyr) | ||
| N-Me-Tyr | 1 | 169.4, qC | - | - | - | - |
| 2 | 60.8, CH | 4.87, dd (11.5, 2.0) | 3a, 3b | 1 | 3a, NH(Val-1), 2(Phe) | |
| 3a | 32.8, CH2 | 3.08, dd (14.0, 12.0) | 2 | 4, 5/9 | 2, 3b, 5/9 | |
| 3b | 2.70, dd (12.0, 2.0) | 2 | 4, 5/9 | 3a, 5/9 | ||
| 4 | 127.3, qC | - | - | - | - | |
| 5/9 | 130.4, CH | 6.97, d (8.5) | 6/8 | 3a, 3b, 6/8, 7 | 6/8, 2(Phe) | |
| 6/8 | 115.3, CH | 6.76, d (8.5) | 5/9 | 4, 7 | 5/9 | |
| 7 | 156.4, qC | - | - | - | - | |
| N-Me | 30.4, CH3 | 2.74, s | 2, 1(Phe) | |||
| Phe | 1 | 170.5, qC | - | - | - | - |
| 2 | 50.2, CH | 4.72, dd (11.5, 4.5) | 3a, 3b | 1, 2(Ahp), 6(Ahp) | 3b, 2(N-Me-Tyr), 5/9(N-Me-Tyr) | |
| 3a | 35.3, CH2 | 2.86, dd (14.0, 12.0) | 2, 3b | 4, 5/9 | 3b, 5/9, 6(Ahp) 5(Ahp) | |
| 3b | 1.81, dd (14.0, 3.0) | 2, 3a | 4, 5/9 | 2, 3a, 5/9 | ||
| 4 | 136.7, qC | - | - | - | - | |
| 5/9 | 129.4, CH | 6.82, d (7.0) | 6/8 | 3a, 3b, 7 | 3a, 3b, 6/8, 5b (Ahp), 6(Ahp) | |
| 6/8 | 127.8, CH | 7.14, t (7.5) | 5/9, 7 | 4, 7 | 5/9 | |
| 7 | 126.2, CH | 7.14, t (7.5) | 6/8 | 4, 5/9 | ||
| Ahp | 2 | 168.7, qC | - | - | - | - |
| 3 | 48.2, CH | 3.77, m | 4a, 4b, NH | 2 | 5b, NH | |
| 4a | 21.9, CH2 | 2.40, m | 3, 4b, 5a | 4b, NH | ||
| 4b | 1.56, m | 3, 4a, | 4a, 5a, NH | |||
| 5a | 29.4, CH2 | 1.71, m | 4a, 5b, 6 | 4b, 5b, 6 | ||
| 5b | 1.52, m | 5a, 6 | 3, 5a, 6, 5/9 (Phe) | |||
| 6 | 73.7, CH | 5.06, s | 5a, 5b, OH | 2, OH | 5a, 5b, OH, 3a(Phe), 5/9(Phe) | |
| NH | - | 7.19, m | 3 | 3, 4a, 4b, NH(Abu) | ||
| OH | - | 6.20, br s | 6 | 6 | ||
| Abu | 1 | 165.8, qC | - | - | - | - |
| 2 | 130.1, qC | - | - | - | - | |
| 3 | 131.6, CH | 6.49, q (7.0) | 4 | 1, 4 | 4 | |
| 4 | 13.1, CH3 | 1.47, d (7.0) | 3 | 2, 3, 1(Thr) | 3, NH | |
| NH | - | 9.25, br s | 4, 2(Thr), 3(Thr), NH(Ahp) | |||
| Thr | 1 | 172.6, qC | - | - | - | - |
| 2 | 55.3, CH | 4.62, br s | NH | 3, 4, NH, NH(Abu) | ||
| 3 | 71.7, CH | 5.52, br s | 4 | 2, 4, NH, NH(Abu) | ||
| 4 | 18.0, CH3 | 1.21, d (6.5) | 3 | 2, 3 | 2, 3, 4(Val-1), 5(Val-1) | |
| NH | - | 7.93, d (6.5) | 2 | 2, 2(Val-2), 3(Val-2) | ||
| Val-2 | 1 | 171.8, qC | - | - | - | - |
| 2 | 57.1, CH | 4.36, t (7.0) | 3, NH | 1 | 3, 4, 5, NH(Thr) | |
| 3 | 30.7, CH | 2.04, m | 2, 4, 5 | 1 | 2, 4, 5, NH(Thr) | |
| 4 | 19.2, CH3 | 0.85, d (7.0) | 3 | 1, 2, 3 | 2, 3, 5 | |
| 5 | 17.7, CH3 | 0.80, d (7.0) | 3 | 1, 2, 3, 4 | 2, 3, 4, NH | |
| NH | - | 7.71, br s | 2 | 3, 5, 2(Ala) | ||
| Ala | 1 | 172.5, qC | - | - | - | - |
| 2 | 48.0, CH | 4.34, dd (7.5, 7.0) | 3, NH | 1, 3 | 3, NH (Val-2) | |
| 3 | 18.0, CH3 | 1.18, d (7.0) | 2 | 1, 2 | 2, NH | |
| NH | - | 8.03, d (7.5) | 2 | 2, 3, 1(Ba) | 2, 3, 2(Ba) | |
| Ba | 1 | 171.9, qC | - | - | - | - |
| 2 | 37.0, CH2 | 2.07, t (7.0) | 3 | 1, 3, 4 | 3, NH(Ala) | |
| 3 | 18.7, CH2 | 1.48, sxt (7.0) | 2, 4 | 1, 2, 4 | 2, 4 | |
| 4 | 13.1, CH3 | 0.83, t (7.0) | 3 | 2, 3 | 3 |
Measured at 500 MHz (1H) and 125 MHz (13C).
Determined from HSQC and/or HMBC spectra.
Refers to correlations on the same unit unless otherwise indicated.
The 1D and 2D NMR data showed that 1 was an assemblage of eight amino acid subunits (alanine, 3-amino-6-hydroxy-2-piperidone (Ahp), threonine, 2-amino-2-butenoic acid (Abu), N-Me-tyrosine, phenylalanine and two valines) and one butanoic acid (Ba) subunit (Figure 1). The Ala, Val-1, Val-2, and Ba units were elucidated through analyses of the spin systems observed in a TOCSY experiment, after excitation of their respective alpha protons, and identification of their respective amide carbonyl resonance on the basis of HMBC correlations observed. Due to the small magnitude of the 3JH,H between the stereogenic centers, the entire proton spin system corresponding to the Thr units was not observed after irradiation of the corresponding amide proton in the 1D TOCSY experiment. However, this unit could be easily identified based on COSY and HMBC correlations from the terminal methyl group to H-3 and C-2, respectively. N-Me-Tyr and Phe units were deduced based on the distinctive chemical shifts of their diastereotopic β-protons and the HMBC correlations observed to the aryl rings. In the former case, the standard para-substitution of the Tyr ring evident on the basis by the coupling pattern from the aromatic protons (δ H-5/H-9 6.97, d (8.5 Hz); δ H-6/H-8 6.76, d (8.5 Hz)). The sole N-methylation was assigned to the Tyr unit due to an HMBC correlation from the N-CH3 to the alpha carbon of Tyr. The two most distinctive residues, which focused the elucidation on the dolastatin 13 family of depsipeptides,3 were the Ahp and Abu residues. The latter was deduced based on HMBC correlations from the vinyl methyl group to corresponding sp2 carbons of that residue, while the network of proton signals observed TOCSY experiment when H-3 proton was irradiated could be ascribed to an Ahp unit, which completed the identification of the subunits of 1.
Figure 1.
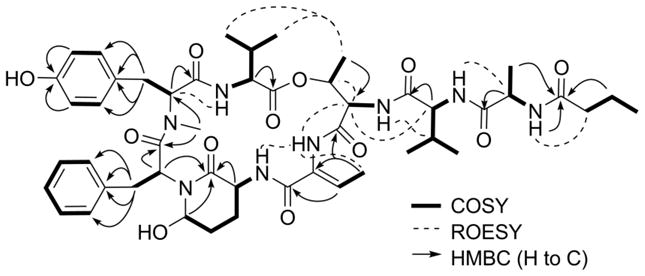
Key 2D NMR correlations for 1.
The sequence of these nine subunits was determined based on detailed analyses of the HMBC and ROESY correlations (Figure 1). Starting from the N-Me-Tyr residue, a clear HMBC correlation was observed from proton signal of the N-CH3 group to the amide carbonyl of the adjacent Phe unit. This nascent unit was connected to the Ahp group through incorporation of the amino terminus on the basis of HMBC correlations from the α-proton resonance of Phe across the tertiary nitrogen to C-2 of the Ahp group. This unit was then expanded to include the Abu and Thr units sequentially, based on a suite of ROESY correlations observed between the resonances for the amide protons of Ahp, Abu and the α-proton of Thr. That these units formed a macrocycle was finally deduced based on ROESY correlations observed between the methyl groups of Thr and Val-1. The nitrogen of this Thr unit served as an anchor point for the remaining acyclic subunits of 1. Specifically, a ROESY correlation was observed between the amide proton resonance of Thr and the alpha proton of Val-2, thus establishing their order. The remaining 2 residues, Ala and Butanoic acid (Ba), were appended sequentially to the amino terminus of Val-2 to complete the structure determination of 1. Evidence in support of this order included a ROESY correlation between the amino proton resonance of Val-2 and the alpha proton of Ala and an HMBC correlation from the latter proton and the amide carbonyl of Ba. Additional support for our proposed structure also comes from comparison of the NMR data (13C, ROESY and HMBC) with those observed for several recent dolastatin 13 analogues.7,10,18 In particular molassamide,18 isolated from Dichothrix utahensis, bears the most closest structural resemblance to 1, as they possess identical macrocycles, and the reported 13C NMR signals from this compounds are in good agreement with those observed for 1.
The planar structure of bouillomide B (2) was deduced in a similar manner to that described above. The HR-ESI-MS data showed a pseudomolecular ion cluster at m/z = 1061.3960 [M + Na]+ with an isotopic pattern suggestive of a brominated analogue (C49H67N8O12Br). The observed bathochromic shift in the UV spectrum of 2 (λmax 283 and 290 nm), compared to 1 (λmax 279 and 286 nm), indicated that one of the aromatic chromophores was halogenated. Analyses of the 1D and 2D NMR data (Supplementary Data, Table S1) verified this hypothesis as 2 was clearly comprised of the same nine basic amino acid residues as 1, with the exception of a brominated N-Me-Tyr unit. The observed 1H NMR proton-proton coupling patterns for this aryl unit indicated a 1,2,4-trisubstituted ring with bromination at the 2-position (δ H-5 = 7.25, d (1.5 Hz); δ H-8 = 6.99, d (8.0 Hz); δ H-9 = 6.95, dd (8.0, 1.5 Hz)). Further analyses of the spectral data in a similar manner to that described for 1 defined the planar sequence depicted.7,10,18
The absolute configuration of the individual amino acid subunits was determined using a combination of functional group interconversions, ROESY correlations and the modified Marfey method. Bouillomides A (1) and B (2) were first oxidized under classic Jones’ conditions with K2Cr2O3.11 This transformation converted the Ahp unit into a 5-carboxy-δ-lactam, which after acid hydrolysis, was liberated as glutamic acid.19 The hydrolysate was then derivatized with L- fluoro-2,4-dinitrophenyl-5-L-leucinamide (L-FDLA) and resulting mixture analyzed by HRESI-LC-MS. Standards of the respective L- and D- amino acids were coupled with L-FDLA and analyzed under the same conditions to confirm the identity of the components in the hydroylzate. For both 1 and 2, these analyses established the L- configurations of the Phe, Ala, Thr and Val residues after comparison with the appropriate standard. Peaks with the correct retention times and pseudomolecular ions consistent with L-glutamic acid, which must have originated from the oxidation of the L-Ahp units, were also present in the LC-MS chromatograms of 1 and 2. With the absolute configuration of the amino-bearing center of the Ahp units defined, ROESY correlation within these units were used to define the relative stereochemistry with respect to the C-6 hydroxyl groups. Specifically, the resonance for H-3 displayed a ROESY crosspeak with H-5, suggestive of a 1,3-diaxial relationship. This proton resonance in turn displayed a small 3JH,H with H-6 was equatorial. Thus a 3S, 6R absolute configuration was assigned to the stereocenters in the Ahp unit. Furthermore, ROESY correlations in both 1 and 2 between the CH3 and NH of the Abu unit assigned a (Z)-configuration to the double bond.
Under this oxidization and hydrolysis sequence, no signals were observed for the tyrosine units. Therefore, a portion of 1 was hydrolyzed, in the presence of 0.1% w/v of phenol, without prior oxidation with Jones’ reagent. These conditions have been shown to preserve easily oxidizable aromatic units.20 Under these modified conditions, the L-FDLA coupling successfully yielded di-L-FDLA-L-N-Me-Tyr, which could be identified by HR-ESI-LC-MS after comparison with standards. These data established the presence of adjacent L-N-Me-Tyr and L-Phe residues; a configuration that is conserved in nearly all members of the dolastatin 13 family.4 In solution, a ROESY correlation is typically observed between the alpha protons of these residues, which was also the case for 1. This observation proved useful, as while no standards for the Br-N-Me-Tyr unit in 2 were available, a ROESY correlation between the alpha protons of the L-Phe and Br-N-Me-Tyr residues suggested this latter residue had an L-configuration in 2. Careful comparison of the carbon chemical shifts between the backbone carbons in 1 and 2, along with other related members of this structural family, verified this stereochemical assignment. As mention earlier, the closest structural relative was molassamide,18 which has an L-Thr unit in place of the L-Val-2 unit in 1. No other brominated molassamide cogeners are known though.
Given the SAR trends previously noted for this series of Ahp containing molecules, bouillomides A (1) and B (2) were screened for against common serine proteases. Dolastatin 13 analogues have consistently been reported as inhibitors of serine proteases. The specificity of this inhibition, for chymotrypsin or trypsin, depends strongly on the hydrophobicity or hydrophilicity, respectively, of the subunits neighboring the Ahp moiety.11,21 Regardless of the identity of the neighboring subunits, these Ahp-containing compounds should inhibit elastase.7 Compounds 1 and 2 were no exception to these trends, inhibiting chymotrypsin (IC50 = 0.17 and 9.3 μM, respectively) while displaying no inhibition of trypsin at 100 μM, the highest concentration tested. Furthermore, both of these compounds demonstrated the same elastase inhibition with IC50 values of 1.9 μM. A similar, though more potent, serine protease activity profile was reported for molassamide.18 The observed inhibition appears to be specific to serine protease though as 1 does not inhibit the aspartic protease BACE1 at concentrations up to 30 μM.
Supplementary Material
Acknowledgments
This work was funded by grants from the Alzheimer’s Association NIRG-08-90880, Alzheimer’s Drug Discovery Foundation (281204), and the National Institute of Aging (R20072671). Funds for the upgrades of the NMR instrumentation were provided by the CRIF program of the National Science Foundation (CH E9974921) and the Elsa Pardee Foundation. The purchase of the Agilent LC-MS was funded by grant W911NF-04-1-0344 from the Department of Defense. P.J.S. acknowledges NIH MBRS SCORE grant S06-GM-44796 for support. This is contribution #661 of the University of Guam Marine Laboratory.
Footnotes
Supplementary Data. NMR data table for 2 and spectra/data for NMR, MS, UV, IR, IC50 curves and general experimentation associated with this article can be found, in the online version, at (insert doi)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lesk AM, Fordham WD. J Mol Biol. 1996;258:501–537. doi: 10.1006/jmbi.1996.0264. [DOI] [PubMed] [Google Scholar]
- 2.Baptista AM, Jonson PH, Hough E, Petersen SB. J Mol Evol. 1998;47:353–362. doi: 10.1007/pl00006393. [DOI] [PubMed] [Google Scholar]
- 3.Pettit GR, Kamano Y, Herald CL, Dufresne C, Cerny RL, Herald DL, Schmidt JM, Kizu H. J Am Chem Soc. 1989;111:5015–5017. [Google Scholar]
- 4.Weckesser J, Martin C, Jakobi C. Syst Appl Microbiol. 1996;19:133–138. [Google Scholar]
- 5.Harrigan GG, Luesch K, Yoshida WY, Moore RE, Nagle DG, Paul VJ. J Nat Prod. 1999;62:655–658. doi: 10.1021/np980553b. [DOI] [PubMed] [Google Scholar]
- 6.Harrigan GG, Yoshida WY, Moore RE, Nagle DG, Park PU, Biggs J, Paul VJ, Mooberry SL, Corbett TH, Valeriote FA. J Nat Prod. 1998;61:1221–1225. doi: 10.1021/np9801211. [DOI] [PubMed] [Google Scholar]
- 7.Taori K, Paul VJ, Luesch H. J Nat Prod. 2008;71:1625–1629. doi: 10.1021/np8002172. [DOI] [PubMed] [Google Scholar]
- 8.Luesch H, Yoshida WY, Moore RE, Paul VJ. J Nat Prod. 1999;62:1702–1706. doi: 10.1021/np990310z. [DOI] [PubMed] [Google Scholar]
- 9.Williams PG, Moore RE, Paul VJ. J Nat Prod. 2003;66:1356–1363. doi: 10.1021/np0302145. [DOI] [PubMed] [Google Scholar]
- 10.Matthew S, Ross C, Rocca JR, Paul VJ, Luesch H. J Nat Prod. 2007;70:124–127. doi: 10.1021/np060471k. [DOI] [PubMed] [Google Scholar]
- 11.Taori K, Matthew S, Rocca JR, Paul VJ, Luesch H. J Nat Prod. 2007;70:1593–1600. doi: 10.1021/np0702436. [DOI] [PubMed] [Google Scholar]
- 12.Zafrir E, Carmeli S. J Nat Prod. 2010;73:352–358. doi: 10.1021/np900546u. [DOI] [PubMed] [Google Scholar]
- 13.Okino T, Qi S, Matsuda H, Murakami M, Yamaguchi K. J Nat Prod. 1997;60:158–161. doi: 10.1021/np970146k. [DOI] [PubMed] [Google Scholar]
- 14.von Elert E, Oberer L, Merkel P, Huhn T, Blom JF. J Nat Prod. 2005;68:1324–1327. doi: 10.1021/np050079r. [DOI] [PubMed] [Google Scholar]
- 15.Matern U, Oberer L, Falchetto RA, Erhard M, Konig WA, Herdman M, Weckesser J. Phytochemistry. 2003;64:1175–1175. doi: 10.1016/s0031-9422(01)00400-9. [DOI] [PubMed] [Google Scholar]
- 16.Rounge TB, Rohrlack T, Kristensen T, Jakobsen KS. BMC Microbiol. 2008;8:141–151. doi: 10.1186/1471-2180-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kupchan SM, Streelman DR, Sneden AT. J Nat Prod. 1980;43:296–301. doi: 10.1021/np50008a010. [DOI] [PubMed] [Google Scholar]
- 18.Gunasekera SP, Miller MW, Kwan JC, Luesch H, Paul VJ. J Nat Prod. 2010;73:459–462. doi: 10.1021/np900603f. [DOI] [PubMed] [Google Scholar]
- 19.Harada K, Fujii K, Mayumi T, Hibino Y, Suzuki M, Ikai Y, Oka H. Tetrahedron Lett. 1995;36:1515–1518. [Google Scholar]
- 20.Muramoto K, Kamiya H. Anal Biochem. 1990;189:223–230. doi: 10.1016/0003-2697(90)90112-m. [DOI] [PubMed] [Google Scholar]
- 21.Perona JJ, Craik CS. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


