Abstract
Gene expression profiling was carried out comparing Con A elicited peritoneal macrophages from C57BL/6 and FVB/N wild-type and apolipoprotein (apo)E knockout mice. An EST, W20829, was expressed at higher levels in C57BL/6 compared with FVB/N mice. W20829 mapped to an atherosclerosis susceptibility locus on chromosome 19 revealed in an intercross between atherosclerosis-susceptible C57BL/6 and atherosclerosis-resistant FVB/N apoE knockout mice. A combination of database search and Northern analysis confirmed that W20829 corresponded to 3′-UTR of a hitherto predicted gene, named HspA12A. Blasting the National Center for Biotechnology Information database revealed a closely related homologue, HspA12B. HspA12A and -B have very close human homologues. TaqMan analysis confirmed the increased HspA12A expression (2.6-fold) in elicited peritoneal macrophages from C57BL/6 compared with FVB/N mice. TaqMan analysis also revealed increased HspA12A and HspA12B expression (87- and 6-fold, respectively) in lesional versus nonlesional portions of the thoracic aorta from C57BL/6 apoE knockout mice on a chow diet. In situ hybridization confirmed that both genes were expressed within lesions but not within nonlesional aortic tissue. Blasting of HspA12A and HspA12B against the National Center for Biotechnology Information database (NR) revealed a hit with the Conserved Domain database for Hsp70 (pfam00012.5, Hsp70). Both genes appear to contain an atypical Hsp70 ATPase domain. The BLAST search also revealed that both genes were more similar to primitive eukaryote and prokaryote than mammalian Hsp70s, making these two genes distant members of the mammalian Hsp70 family. In summary, we describe two genes that code for a subfamily of Hsp70 proteins that may be involved in atherosclerosis susceptibility.
In the last decade, induced mutant mice have become useful models of atherosclerosis. The first of these models, the apolipoprotein (apo)E knockout mouse, has been widely used to study lesion formation, test pharmacological therapies, and evaluate the influence of candidate genes on the atherosclerotic process (1, 2). In the apoE knockout mouse on a chow diet, monocyte/macrophage attachment occurs at 6–8 weeks of age followed by the appearance of subintimal lipid-laden macrophages, foam cells, at 8–10 weeks of age. Between 15 and 25 weeks of age, macrophage necrosis occurs, and the fibrous cap forms. Candidate genes that decrease macrophage number, monocyte/macrophage attachment, and homing to lesion-prone regions dramatically decrease atherosclerotic lesion formation and progression in mouse models (3–8). The morphology of the atherosclerotic plaque and the results from experiments with candidate genes all suggest a major role for macrophages in atherosclerosis.
Mouse strains have been described with differing susceptibilities to atherosclerosis based initially on responsiveness to diet (9). With the advent of induced mutant mice that develop atherosclerotic lesions on chow or more physiologically relevant Western-type diets (1, 2), these models are now being used in genetic crosses to reveal atherosclerosis modifier loci. In an intercross between an atherosclerosis-susceptible strain, C57BL/6 apoE knockout, and an atherosclerosis-resistant strain, FVB/N apoE knockout, three loci were revealed on chromosomes 10, 14, and 19 (10). The responsible genes at these loci are not currently known.
Because macrophages play such a fundamental role in the initiation and progression of atherosclerotic lesions, differentially expressed genes were sought between elicited peritoneal macrophages from C57BL/6 and FVB/N wild-type and apoE knockout mice. In the course of this analysis, one of the genes overexpressed in C57BL/6 macrophages corresponded to an EST that maps to the chromosome 19 atherosclerosis susceptibility locus. This gene, HspA12A, and another closely related mammalian homologue that maps to a different region of the genome, HspA12B, were found to be distant members of the Hsp70 family. Both genes were shown to be expressed in atherosclerotic lesions but not in nonlesional regions of the aorta. These genes now become candidates for playing a role in the atherosclerotic process, and HspA12A is also a candidate for involvement in the chromosome 19 susceptibility locus.
Materials and Methods
Animals.
Mice were housed in a specific pathogen-free humidity- and temperature-controlled room with a 12-h light/dark cycle and fed normal chow diet. Eight-week-old C57BL/6, FVB/N, and C57BL/6 apoE knockout mice were from The Jackson Laboratory. FVB/N apoE knockout mice were created by backcrossing apoE knockout heterozygous F1 (C57BL/6 × 129/Sv) mice with wild-type FVB/N mice for 10 generations and then performing brother/sister mating to derive homozygous apoE knockouts.
RNA Isolation from Elicited Peritoneal Macrophages.
Male mice of each genotype were used to isolate peritoneal macrophages. Each mouse was injected in the peritoneum with 0.5 ml of 80 μg/ml Con A (Sigma) in PBS. After 72 h, mice were euthanized by asphyxiation in CO2 and the peritoneal cavity immediately lavaged with 9 ml of PBS at 4°C. The lavagates from 5–15 mice were then combined and spun at 500 × g and pelleted cells resuspended in RPMI 1640 medium (American Type Culture Collection). The cells were then plated in P100 plastic tissue culture dishes (Corning), incubated at 37°C for 2 h, and washed three times with cold PBS to remove unattached cells. By using the RNeasy Mini kit (Qiagen, Chatsworth, CA), attached cells were lysed and total RNA isolated.
cDNA Microarray Expression Analysis.
Two micrograms of total RNA was used to synthesize fluorescently labeled cDNA probes with a 3DNA submicro kit (Genisphere, Montvale, NJ). Glass slides printed with ≈9,000 cDNAs or ESTs were a generous gift from Raju Kucherlapati at Albert Einstein College of Medicine (11). Arrays were hybridized according to their recommended protocol (http://sequence.aecom.yu.edu/bioinf/microarray/protocol4.html). Hybridized arrays were scanned with a ScanArray4000 (General Scanning, Watertown, MA) and analyzed by SCANALYZE software (http://rana.stanford.edu/software).
Creation of cDNA Probes and Northern Blot Analysis.
cDNAs corresponding to the EST W20829, the midpoint, and the beginning of the predicted exon 12 of HspA12A were made by PCR. The primers used corresponded to the sequences indicated in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org. The PCR reaction used High Fidelity Taq (Invitrogen) and the template was mouse brain/heart cDNA (Ambion, Austin, TX). Twenty-five nanograms of each cDNA was radiolabeled by nick-translation ECA prime II (Ambion) and 32P-dATP (Perkin–Elmer). Nitrocellulose blots containing RNA from multiple mouse tissues of BALB/c strain were purchased from CLONTECH and hybridized with Express Hyb Rapid Hybridization Buffer (CLONTECH) according to the manufacturer's directions. After each hybridization, the blot was stripped by incubation in 1 liter of 0.5% SDS solution at 100°C for 15 min and exposed to x-ray film for 24 h to ensure that there were no residues from previous hybridizations. Northern blot analysis of human HspA12A and HspA12B was carried out by a similar method, and the primer sequences used are specified in Table 3.
TaqMan (Real-Time Quantitative RT-PCR) Analysis.
Total RNA from peritoneal macrophages was isolated as above. Total RNA was isolated from aortic tissue as follows: four 10-mo-old C57BL/6 apoE knockout mice fed a chow diet were anesthetized, the chest was opened, the right atrium nicked, and perfusion carried out through the left ventricle with 10 ml of PBS. The thoracic aorta was dissected free of adventitial fat, opened longitudinally, and lesional and nonlesional portions separated by dissection under a dissecting microscope. Total RNA was isolated by using the RNeasy Mini kit (Qiagen). HspA12A mRNA was quantified in peritoneal macrophages and aortic tissue, whereas HspA12B was quantified only in aortic tissue. In all cases, 3 μg of total RNA was treated with DNaseI (Ambion DNA-free), the DNaseI removed, and RNA reverse transcribed in 20 μl by using oligo(dT)15 primers (Promega) and Superscript II (Invitrogen). After adding 1 ml of water and mixing, 5-μl aliquots were used for each TaqMan reaction. HspA12A, HspA12B, and cyclophilin, a control gene, were amplified and detected with specific primers and probes specified in Table 3. These were designed by using Primer Express (Applied Biosystems). TaqMan reactions were carried out in a total volume of 50 μl containing 1× Jumpstart PCR buffer, 1 unit Jumpstart Taq (Sigma), 3.5 mM MgCl2, 200 μM dNTPs, 300 nM forward primer, 300 nM reverse primer, 100 nM 6FAM/TAMRA TaqMan probe, and 1× ROX reference dye (Invitrogen). In each TaqMan experiment, a standard was run that consisted of a mixture of cDNAs made from mouse brain and heart RNA. An Applied Biosystems Prism 7700 Sequence Detection System (Applied Biosystems) was used with the default thermal cycling program (95°C for 10 min followed by 40 cycles of 95°C, 15 sec, 60°C, 1 min). The threshold was set at 0.05 units of normalized fluorescence; the threshold cycle (Cτ) was measured for each well. A standard curve was plotted for each gene, and the mean Cτ for each sample was expressed as an arbitrary value relative to the standard. For HspA12A and HspA12B, the arbitrary values were divided by the corresponding value for cyclophilin and expressed as a ratio. Groups were compared by a two-tailed type 2 Student's t test.
In Situ Hybridization.
cDNAs corresponding to distinct portions of the 3′-UTR of mouse HspA12A and HspA12B were made by using primers specified in Table 3 on a template consisting of a mixture of cDNAs from mouse brain and heart RNA. These were cloned into pBST19. Antisense digoxigenin (DIG)-labeled RNA probes (ribo-probes) were generated by in vitro transcription with T7 RNA polymerase (Roche Diagnostics). Sense ribo-probes were generated by in vitro transcription with SP6 RNA polymerase from the same plasmids. After perfusion as above, the thoracic aorta was removed from 6-mo-old chow-fed C57BL/6 apoE knockout mice, fixed in 4% paraformaldehyde at 4°C for 72 h, immersed in 25% sucrose at 4°C for 96 h, immersed in OCT, and frozen at −80°C until sectioning. Sections at 8-μm intervals were made with a cryostat at −23°C, collected onto silanized glass slides (PGC Scientific, Gaithersburg, MD), and stored at –80°C until hybridized. Before hybridization, sections were fixed in 4% paraformaldehyde, permeabilized with 0.01 mg/ml proteinase K, treated with 0.2 M HCl, and acetylated in 1.3% triethanolamine and 0.25% acetic anhydride (Sigma). After each of these steps, the slides were washed once with 0.1 M phosphate buffer (pH 7.4). Sections were then hybridized with antisense or sense ribo-probes in Hybriwell chambers (PGC Scientific) at 55°C for 24 h. Sections were then washed in 5× SSC at 60°C for 20 min, 2× SSC, and 50% formamide at 60°C for 30 min, treated with 1 mg/ml RNaseA at 37°C for 30 min, and washed with 2× SSC and 50% formamide at 60°C for 30 min. The sections were then treated with anti-DIG antibody–alkaline phosphatase conjugate (Roche) at room temperature for 18 h. After washing with 0.1 M phosphate buffer (pH 7.4), nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrates (Roche Diagnostics) were added and blue color developed in the dark for 3 h. Sense-strand ribo-probes were used as negative controls in adjacent tissue sections.
Results
Identification of the Gene HspA12A.
Gene expression profiling comparing Con A elicited peritoneal macrophages from C57BL/6 and FVB/N apoE knockout and C57BL/6 and FVB/N wild-type mice was carried out by using cDNA microarrays. A total of eight comparisons were done and in each, the gene corresponding to EST W20829 was expressed at a higher level in C57BL/6 compared with FVB/N macrophages (average 1.6-fold) (data not shown). This gene mapped to an atherosclerosis susceptibility locus on chromosome 19 identified in an intercross between C57BL/6 (atherosclerosis prone) and FVB/N (atherosclerosis resistant) apoE knockout mice (10).
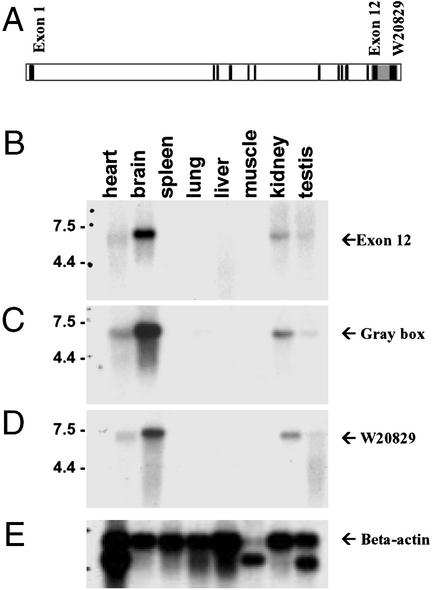
In the Unigene database, the EST W20829 is 988 nt long and contains no ORFs greater than 68 codons, suggesting it corresponds to the 3′-UTR of a gene. However, when the nucleotide sequence was blasted against the Celera database, a putative gene (mCG10682) containing 12 exons was predicted to be ≈3.5 kb upstream of W20829 (Fig. 1A). To determine whether this gene is expressed and whether EST W20829 is part of it, a mouse multiple tissue Northern blot was probed with sequences corresponding to exon 12 of the putative gene (Fig. 1B), the region intermediate between exon 12 and W20829 (gray area in Fig. 1A; Fig. 1C), and W20829 (Fig. 1D). Each of these probes gave a single band of message length 6.5 kb expressed in brain > kidney > heart, but whose expression was low to absent in other tissues. This provided proof that the putative gene exists and W20829 is located in its 3′-UTR. As will be clear from later results, this gene has been named mHspA12A.
Figure 1.
Multiple-tissue Northern blots of mHspA12A. (A) Schematic representation of EST W20829 in relation to mCG10682, an upstream putative gene predicted by the Celera database. Black-filled boxes represent exons, and open boxes represent introns (in scale). A mouse multiple-tissue Northern blot was hybridized with probes representing exon 12 of mCG10682 (B), gray box sequences (3′UTR) (C), W20829 (D), and β-actin (E).
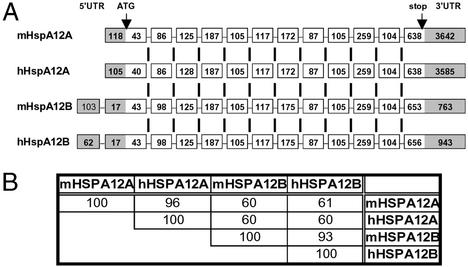
mHspA12A encodes a protein of 675 aa and when this sequence was blasted against the National Center for Biotechnology Information database, a single mouse homolog was identified, now named mHspA12B. Both of these mouse genes have human homologues, hHspA12A and hHspA12B, respectively. For each of the mouse and human genes, the GenBank accession no., Unigene cluster, chromosomal location, transcript, and protein length are listed in Table 1 and the organization of the genes and amino acid identities between them shown in Fig. 2 A and B. The HspA12A and HspA12B genes are dispersed in the genome in both mouse and human; however, they are quite similar in structure. The HspA12A genes have 12 exons and long 3′-UTRs, whereas the HspA12Bs have 13 exons and relatively short 3′-UTRs. The extra exon in the HspA12B genes results from an intron in the 5′-UTR, but the rest of the exons in terms of number, size, and exon/intron organization are strikingly similar between the HspA12A and HspA12B genes of both species. The amino acid identity between HspA12A and HspA12B in both mouse and human is 60%, whereas between mice and humans it is 96% for HspA12A and 93% for HspA12B. Aside from these two closely related genes that have remained highly conserved in evolution, blasting did not reveal any other related mammalian gene.
Table 1.
General information of mouse and human HspA12A and HspA12B
| Genes | mHspA12A | hHspA12A | mHspA12B | hHspA12B |
|---|---|---|---|---|
| GenBank accession no. | BC030362 | AB007877 | BC011103 | AK056712 |
| Unigene cluster(s) | Mm.39739 | Hs.12385 | Mm.158850 | Hs.302110 |
| Celera gene designation | mCG10682 | hCG1792255 | mCG18511 | hCG37961 |
| Chromosome position | 19–59 Mb | 10–117 Mb | 2–130 Mb | 20–4 Mb |
| Northern mRNA length, kb | ∼6.5 | ∼6.5 | ∼3.5 | ∼3.5 |
| Protein length, aa | 675 | 675 | 685 | 686 |
Figure 2.
Mouse and human HPA12A and HPA12B exon/intron organization and amino acid identity. (A) The exon/intron organization of mHP12A and mHP12B and their human homologues, hHPA12A and hHPA12B. Exons are represented as boxes, and the number inside each box denotes its length in nucleotides. Open areas of boxes are coding regions, whereas gray areas are UTRs. Vertical bars indicate conserved splice junctions. The exon/intron boundaries were determined by aligning the cDNA with genomic DNA sequence. The 5′ transcription initiation site for mHspA12A was determined by 5′-RACE (Ambion; data not shown), and the rest were deduced from compilations of all known ESTs and cDNA sequences. Notice the extraordinary conservation of exon/intron organization between HspA12A and HspA12B, despite the fact that the gene length of the former was >65 kb and the latter only ≈18 kb, for both human and mouse genes (see Table 4, which is published as supporting information on the PNAS web site, for details on intron). (B) Percentage amino acid identity in pair-wise comparisons among mouse and human HspA12A and HspA12B. Sequence alignments were performed by using CLUSTAL W from the DNASTAR package with the default parameters.
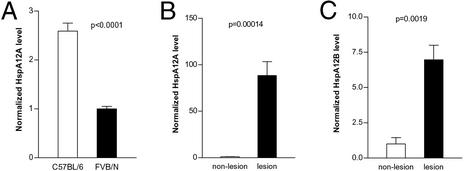
To confirm the results of the cDNA microarray experiments and to see whether the gene is expressed in atherosclerotic lesions, mRNA levels were measured by TaqMan analysis by using total RNA isolated from elicited peritoneal macrophages from C57BL/6 and FVB/N apoE knockout mice, and from lesional and nonlesional portions of the thoracic aorta from apoE knockout mice. As shown in Fig. 3A, C57BL/6 apoE knockout mice had 2.6-fold more HspA12A mRNA in their elicited peritoneal macrophages than FVB/N apoE knockout mice. As shown in Fig. 3 B and C, portions of the thoracic aorta from 10-mo-old chow-fed C57BL/6 apoE knockout mice containing lesions had 87- and 6-fold more HspA12A and HspA12B, respectively, than nonlesional areas. Thus HspA12A appears to be more highly expressed in elicited peritoneal macrophages from the atherosclerosis sensitive strain as well as preferentially expressed in lesional portions of the aorta.
Figure 3.
mHspA12A and mHspA12B expression in macrophages and atherosclerotic lesions. (A) mHspA12A mRNA level, normalized to cyclophilin A, in elicited peritoneal macrophages from male, 8-wk-old C57BL/6 and FVB/N apoE knockout mice by TaqMan assay (see Table 3 for primer and probe sequences). The FVB/N mHspA12A mRNA level is normalized to one. (B and C) mHspA12A and mHspA12B mRNA levels normalized to cyclophilin A, respectively, in nonlesional and lesional portion of the thoracic aorta from 10-mo-old C57BL/6 mice (n = 2). The nonlesional mRNA level is normalized to one.
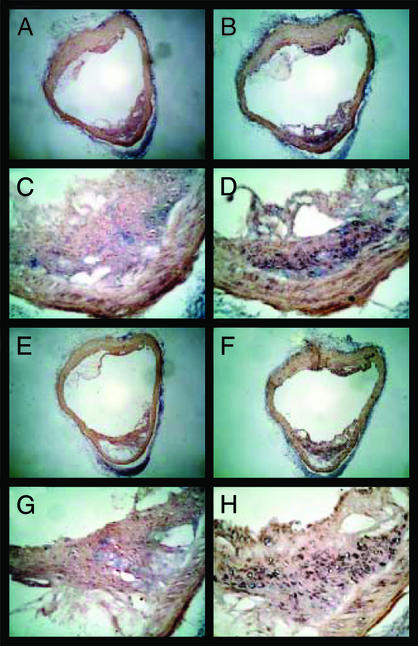
To confirm that HspA12A and Hsp12B are expressed in lesions, in situ hybridization was carried out by using the aortic arch from 6-mo-old apoE knockout mice and is shown in Fig. 4. To distinguish between HspA12A and HspA12B, probes to the 3′-UTRs of each gene were used (Fig. 4 A–D and E–H, respectively). The sense probes are shown in Fig. 4 A, C, E, and G, and the antisense probes in Fig. 4 B, D, F, and H. As shown at both low- and high-power magnification, specific hybridization in lesions occurs for both HspA12A and HspA12B (Fig. 4 B and D and F and H, respectively). Specific hybridization was not observed in normal nonlesional aortic tissue. This confirms the results obtained by TaqMan analysis of RNA extracted from lesions and suggests a major increase in the expression of these genes in the presence of atherosclerosis.
Figure 4.
In situ hybridization of mHspA12A and mHspA12B mRNA in atherosclerotic lesions from the thoracic aorta of C57BL/6 mice The magnifications are ×400 for A, B, E, and F; ×200 for C, D, G, and H. (A) Lesional cross section hybridized to a sense strand of mHspA12A (negative control). (B) An adjacent section hybridized to an anti-sense mHspA12A probe, revealing mHspA12A expression inside lesions, most likely in macrophages. (C) Magnified view of A. (D) Magnified view of B. (E) Lesion cross section hybridized to sense strand of mHspA12B (negative control). (F) An adjacent section hybridized to an anti-sense mHspA12B probe, revealing mHspA12B expression inside lesions, most likely in macrophages. (G) Magnified view of E. (H) Magnified view of F.
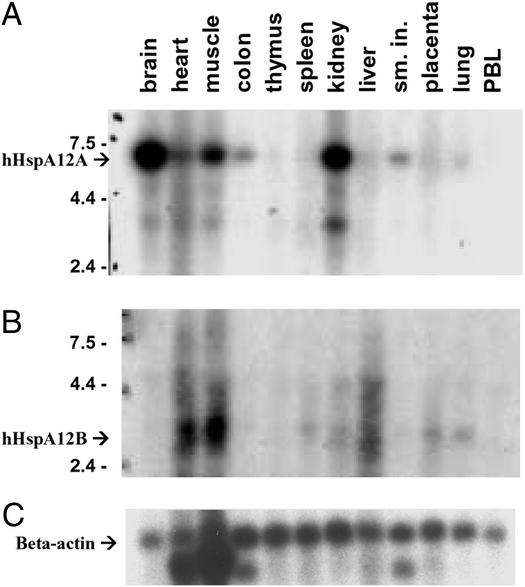
The tissue expression patterns of HspA12A and HspA12B are not confined to atherosclerotic aorta. As previously noted and shown in Fig. 1 B–D, mouse HspA12A is expressed in brain > kidney > heart and is low to absent in other tissues. In a similar analysis, mouse HspA12B is expressed most strongly in the heart and is low to absent in other tissues (data not shown). Multiple-tissue Northern blots were also done for human HspA12A and HspA12B and are shown in Fig. 5 A and B, respectively. Human HspA12A is expressed in brain > kidney > muscle > heart, whereas human HspA12B is expressed in muscle = heart > liver > kidney. For both genes, the most striking difference between human and mouse is muscle expression in the former but not in the latter. The major difference in the tissue-specific expression pattern of human HspA12A and HspA12B is brain expression in the former and liver expression in the latter. The differences in the tissue-specific expression patterns of these two closely related genes suggest they have distinct functions but does not help to explain why these genes are expressed in atherosclerotic lesions.
Figure 5.
Multiple tissue Northern blots of hHspA12A and -B. Hybridization was done with HspA12A (A), -B (B), and β-actin (C) probes.
Blasting of HspA12A and HspA12B against the National Center for Biotechnology Information database (nonredundant) revealed a hit with the Conserved Domain database for HSP70 (pfam00012.5, Hsp70). mHspA12A amino acids 58–244 align with the pfam00012.5, Hsp70 consensus sequence amino acids 1–175, showing 28% identity and 38% homology. The mHspA12A amino acids 312–542 align with the pfam00012.5, Hsp70 consensus sequence amino acids 189–398, showing 26% identity and 44% homology (Fig. 6A). The results for mHSPA12B are almost identical (Fig. 6B). Prototypic Hsp70s such as DnaK are 638 aa long, with amino acids 1–385 specifying the ATPase domain and 393–537, the substrate-binding domain. Thus HspA12A and HspA12B appear to have an atypical Hsp70 ATPase domain, which is in two parts separated by spacer amino acids 245–311.
Figure 6.
Schematic representation of mHSPA12A (A) and mHSPA12B (B) aligned with prototypic HSP70 sequences determined by the conserved domain database
In addition to the Conserved Domain, the blast search also revealed sequences in the database producing significant alignments. Blasting mHspA12A yielded 21 sequences that matched with E values ≤0.001. Eight matches, including the strongest seven, include mouse and human HspA12A and HspA12B full length and splice variants, and two of the matches were shorter forms of two of the proteins listed below. Aside from these, in descending order of alignment, as shown in Table 2, were a hypothetical protein from Trachhodesmium erythraeumforms, a hypothetical protein from Dictyostelium discoideum, Hsp70-related protein from Neurospora crassa, elongation factor-2 kinase isoform 1B related protein from D. discoideum, inducible Hsp70 from Leptinotarasa decemlineata (insect), Hsp68s from Drosophila mauritiana and Drosophila melanogaster, Hsp dnaK from Campylobacter jejuni, Hsp68 from Drosophila yakuba, Hsp cognate 2 from D. melanogaster, and Hsp68 from Drosophila auraria. All of these proteins, except for the two from Dictyostelium, match the Hsp70 consensus. Most of these proteins match HspA12A from amino acids 307–547, a region corresponding to the C-terminal one-half of the Hsp70 ATPase domain plus the linker region that connects to the substrate-binding domain. These alignments suggest that HspA12A and HspA12B may be distantly related Hsp70 family members and hence the names assigned by the Human Genome Organization Gene Nomenclature committee.
Table 2.
List of closely homologous proteins to HspA12A
| Accession no. | Identity | E value | Protein vs. HSPA12A* | Protein vs. Hsp70 consensus** |
|---|---|---|---|---|
| NZ_AAAU01000064 | Hypothetical protein (T. erythraeum IMS101) | 8.00E-26 | 185–396 vs. 323–539 | 290–376 vs. 283–368 |
| AC116955 | Hypothetical protein (D. discoideum) | 2.00E-24 | 4–193 vs. 57–246 | |
| AL355932 | Hsp70 related protein (N. crassa) | 6.00E-22 | 1–588 vs. 16–629 | 57–414 vs. 1–334 |
| AC116955 | Elongation factor-2 kinase 1B (D. discoideum) | 4.00E-19 | 8–225 vs. 446–628 | |
| AF288978 | Inducible hsp70 protein (L. decemlineata) | 3.00E-04 | 100–326 vs. 307–547 | 1–520 vs. 86–599 |
| AF247555 | Hsp68 (D. mauritiana) | 4.00E-04 | 181–407 vs. 307–547 | 1–602 vs. 5–600 |
| NM_079750 | Hsp68 (D. melanogaster) | 6.00E-04 | 187–413 vs. 307–547 | 3–608 vs. 1–600 |
| NC_002163 | Hsp dnaK (C. jejuni) | 0.001 | 183–407 vs. 306–547 | 4–600 vs. 1–600 |
| AF277570 | Hsp68 (D. yakuba) | 0.001 | 181–407 vs. 307–547 | 1–602 vs. 5–600 |
| NM_079615 | Hsp cognate 2 (D. melanogaster) | 0.001 | 190–416 vs. 307–547 | 6–605 vs. 1–593 |
| AF247553 | Hsp68 (D. auraria) | 0.001 | 181–407 vs. 307–547 | 1–601 vs. 5–599 |
The list was generated by blasting the mHspA12A amino acid sequence, and repetative entries were excluded. The homologous regions were obtained by blasting the nonredundant database (*) and Conserved Domain database (**).
Discussion
Gene expression profiling revealed a Hsp70 family member, HspA12A, expressed at higher levels in elicited peritoneal macrophages of an atherosclerosis susceptible compared with an atherosclerosis-resistant mouse strain. This gene maps to an atherosclerosis lesion area quantitative trait locus (QTL) on chromosome 19 identified in an intercross between C57BL/6 and FVB/N apoE knockout mice. HspA12A has a closely related homologue, HspA12B, which maps to a different region of the genome. Both HspA12A and HspA12B are expressed in atherosclerotic lesions of apoE knockout mice. These two HSP family members may play a role in atherosclerotic lesion development.
In an intercross between C57BL/6 and FVB/N apoE knockout mice, QTL mapping revealed three atherosclerosis susceptibility loci as follows: chromosome 10, marker D10Mit213 [logarithm of odds (LOD) 7.8, highly significant]; chromosome 14, marker D14Mit60 (LOD 3.2, suggestive); and chromosome 19, marker D19Mit120 (LOD 3.2, suggestive) (10). The genotypic means for atherosclerotic lesion area of C57 homozygotes, heterozygotes, and FVB homozygotes at the marker D19Mit120 were 48,541, 72,927, and 72,356 μm2, respectively. The marker D19Mit 120 is located at 42 Mb (Ensembl database), whereas HspA12A is located at 59 Mb. The corresponding genotypic means for the closest tested markers flanking HspA12A, D19Mit105 (57 Mb) and D19Mit71 (60 Mb), were 53,700, 69,899, and 71,341 μm2, and 55,433, 68,333, 71,244 μm2, respectively. For marker D19Mit120, the genotypic means for homozygotes for C57 differ from both heterozygotes and homozygotes for FVB, whereas the significance for markers D19Mit105 and D19Mit71 is much lower to absent, but the same trends are present. Although HspA12A is located 19 Mb distal to the peak LOD marker, D19Mit120, it cannot be excluded as the gene underlying the chromosome 19 locus in this cross.
Hsp70 family members consist of two distinct domains: a highly conserved 44-kDa N-terminal ATPase domain and a more divergent 25-kDa C-terminal substrate-binding domain (12). Hsp70 proteins recognize and bind to short stretches of exposed hydrophobic peptides that arise when proteins are denatured under stress or in the processes of synthesis, folding, assembly, and translocation to an appropriate subcellular compartment, and prevents them from irreversible aggregation (13). When released from Hsp70, the misfolded polypeptides have a chance to resume correct folding; however, repeated misfolding increases the likelihood of degradation by the ubiquitin–proteasome system (14). Mammalian Hsp70 family members are either induced or constitutively expressed and generally localized to the cytoplasm, but specific family members can localize to nuclei, mitochondria, and endoplasmic reticulum (15).
Anti-Hsp70 antibodies that may identify a number of family members localize Hsp70 mainly to macrophages in atherosclerotic lesions (16). There is some suggestion that Hsp70 expression is maximal in central portions of more thickened atherosclerotic lesions around sites of necrosis and lipid accumulation (17). In apoE knockout mice, Hsp70 was shown to be expressed in macrophages, smooth muscle cells, and endothelial cells in atherosclerotic lesions with the expression level reduced in advanced lesions (18). OxLDL, a crucial mediator of atherogenesis, induces Hsp70 expression in human endothelial (19) and smooth muscle cells (20). In the current study, HspA12A and HspA12B mRNA localized to similar regions of apoE knockout mouse atherosclerotic lesions.
Increased expression of Hsp70s protects against apoptosis induced by oxidative stress, toxins, and heat-shock and cellular damage after ischemia- or sepsis-induced injury (21–26). Given these functions of Hsp70s, it is not unreasonable to assume they would also protect against atherosclerosis. Therefore, the markedly increased expression of HspA12A and HspA12B in atherosclerotic lesions compared with normal aorta can be interpreted in two ways. If HspA12A is responsible for the chromosome 19 QTL, then its increased expression in C57BL/6 macrophages could be directly protective against atherosclerosis. This is compatible with the finding that the C57BL/6 allele acts as a recessive protective allele, based on the genotypic means for atherosclerotic lesion area at the chromosome 19 locus (see above). Alternatively, if HspA12A is not responsible for the chromosome 19 QTL, the increased expression of HspA12A in macrophages and HspA12A and HspA12B in lesions may simply be a reaction to proatherosclerotic stresses, which may be greater in C57BL/6 than FVB/N. Even if HspA12A is not playing a primary role in the difference in atherosclerosis susceptibility between the strains, HspA12A and HspA12B may still be important players in atherogenesis as modulators of inflammation and/or apoptosis.
Despite HspA12A and HspA12B localization to macrophages in lesions and their placement into the Hsp70 family by computer algorithms, we cannot be certain that they share any of the functions of Hsp70s. As shown, blast searching did not reveal homology to any of the mammalian Hsp70s. blast searching did reveal homology to the Hsp70 conserved domain ATPase-binding region but not to the substrate-binding region. Moreover, the HspA12A and HspA12B ATPase homology to the conserved domain consensus sequence is in two parts separated by about 70 amino acids. Finally, the most significant homologies with Hsp70s of lower organisms are only with the second half of the ATPase domain. Regardless of whether HspA12A or HspA12B act like mammalian Hsp70s, their increased expression in atherosclerotic lesions suggests they play a role in atherogenesis.
In summary, we have discovered two distantly related members of the Hsp70 family that are expressed in atherosclerotic lesions. The correct interpretation of the role of HspA12A and HspA12B in atherosclerosis will be provided only by over- or underexpressing these genes in transgenic and knockout mice as well as by the precise identification of the atherosclerosis susceptibility gene on chromosome 19.
Supplementary Material
Acknowledgments
We thank Dr. Guoqing Chang for technical help with in situ hybridization and Dr. Daniel Teupser for help in mouse aorta dissection. This work was supported by National Institutes of Health Grant 5 P01 HL54591 and the Glorney–Raisbeck Program of the New York Academy of Medicine.
Abbreviation
- apo
apolipoprotein
Footnotes
References
- 1.Plump A S, Smith J D, Hayek T, Aalto-Setala K, Walsh A, Verstuyft J G, Rubin E M, Breslow J L. Cell. 1992;71:343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- 2.Zhang S H, Reddick R L, Piedrahita J A, Maeda N. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 3.Smith J D, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Proc Natl Acad Sci USA. 1995;92:8264–8268. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boring L, Gosling J, Cleary M, Charo I F. Nature. 1998;394:894–897. doi: 10.1038/29788. [DOI] [PubMed] [Google Scholar]
- 5.Dawson T C, Kuziel W A, Osahar T A, Maeda N. Atherosclerosis. 1999;143:205–211. doi: 10.1016/s0021-9150(98)00318-9. [DOI] [PubMed] [Google Scholar]
- 6.Cybulsky M I, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos J C, Connelly P W, Milstone D S. J Clin Invest. 2001;107:1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dansky H M, Barlow C B, Lominska C, Sikes J L, Kao C, Weinsaft J, Cybulsky M I, Smith J D. Arterioscler Thromb Vasc Biol. 2001;21:1662–1667. doi: 10.1161/hq1001.096625. [DOI] [PubMed] [Google Scholar]
- 8.Ravagnan L, Gurbuxani S, Susin S A, Maisse C, Daugas E, Zamzami N, Mak T, Jaattela M, Penninger J M, Garrido C, Kroemer G. Nat Cell Biol. 2001;3:839–843. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- 9.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Atherosclerosis. 1985;57:65–73. doi: 10.1016/0021-9150(85)90138-8. [DOI] [PubMed] [Google Scholar]
- 10.Dansky H M, Shu P, Donavan M, Montagno J, Nagle D L, Smutko J S, Roy N, Whiteing S, Barrios J, McBride T J, et al. Genetics. 2002;160:1599–1608. doi: 10.1093/genetics/160.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung V G, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Nat Genet. 1999;21:15–19. doi: 10.1038/4439. [DOI] [PubMed] [Google Scholar]
- 12.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 13.Hartl F U, Hayer-Hartl M. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 14.Meacham G C, Patterson C, Zhang W, Younger J M, Cyr D M. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 15.Gething M-J. Guidebook to Molecular Chaperones and Protein-Folding Catalysts. Oxford: Oxford Univ. Press; 1997. [Google Scholar]
- 16.Berberian P A, Myers W, Tytell M, Challa V, Bond M G. Am J Pathol. 1990;136:71–80. [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson A D, Berberian P A, Tytell M, Bond M G. Arterioscler Thromb Vasc Biol. 1995;15:27–36. doi: 10.1161/01.atv.15.1.27. [DOI] [PubMed] [Google Scholar]
- 18.Kanwar R K, Kanwar J R, Wang D, Ormrod D J, Krissansen G W. Arterioscler Thromb Vasc Biol. 2001;21:1991–1997. doi: 10.1161/hq1201.100263. [DOI] [PubMed] [Google Scholar]
- 19.Zhu W, Roma P, Pellegatta F, Catapano A L. Biochem Biophys Res Commun. 1994;200:389–394. doi: 10.1006/bbrc.1994.1461. [DOI] [PubMed] [Google Scholar]
- 20.Zhu W M, Roma P, Pirillo A, Pellegatta F, Catapano A L. FEBS Lett. 1995;372:1–5. doi: 10.1016/0014-5793(95)00834-v. [DOI] [PubMed] [Google Scholar]
- 21.Buzzard K A, Giaccia A J, Killender M, Anderson R L. J Biol Chem. 1998;273:17147–53. doi: 10.1074/jbc.273.27.17147. [DOI] [PubMed] [Google Scholar]
- 22.Garrido C, Gurbuxani S, Ravagnan L, Kroemer G. Biochem Biophys Res Commun. 2001;286:433–442. doi: 10.1006/bbrc.2001.5427. [DOI] [PubMed] [Google Scholar]
- 23.Jaattela M, Wissing D, Kokholm K, Kallunki T, Egeblad M. EMBO J. 1998;17:6124–6134. doi: 10.1093/emboj/17.21.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marber M S, Mestril R, Chi S H, Sayen M R, Yellon D M, Dillmann W H. J Clin Invest. 1995;95:1446–1456. doi: 10.1172/JCI117815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J H, Redmond H P, Watson R W, Condron C, Bouchier-Hayes D. Arch Surg. 1995;130:1260–1265. doi: 10.1001/archsurg.1995.01430120014002. [DOI] [PubMed] [Google Scholar]
- 26.Yaglom J A, Gabai V L, Meriin A B, Mosser D D, Sherman M Y. J Biol Chem. 1999;274:20223–20228. doi: 10.1074/jbc.274.29.20223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.