Abstract
Although triglyceride-rich particles, such as very low-density lipoprotein (VLDL), contribute significantly to human atherogenesis, the molecular basis for lipoprotein-driven pathogenicity is poorly understood. We demonstrate that in macrophages, VLDL functions as a transcriptional regulator via the activation of the nuclear receptor peroxisome proliferator-activated receptor δ. The signaling components of native VLDL are its triglycerides, whose activity is enhanced by lipoprotein lipase. Generation of peroxisome proliferator-activated receptor δ null macrophages verifies the absolute requirement of this transcription factor in mediating the VLDL response. Thus, our data reveal a pathway through which dietary triglycerides and VLDL can directly regulate gene expression in atherosclerotic lesions.
Epidemiological studies indicate a strong link between the increased intake of dietary fats (cholesterol and fatty acids) and the dramatic rise in the incidence and prevalence of obesity, type II diabetes mellitus, and atherosclerosis in Western societies (1). Emerging from fatty streaks, atherosclerotic lesions are subject to chronic inflammation resulting from persistent injury to the vessel wall (2). Low-density lipoprotein (LDL) and its oxidation product, oxidized LDL (oxLDL), are key causative factors that result in injury to the vascular endothelium (3). Once circulating monocytes are recruited to these sites of injury, they differentiate into resident macrophages and internalize oxLDL via their scavenger receptors. We and others have previously identified two nuclear receptors, peroxisome proliferator-activated receptor (PPAR)γ and liver X receptors, that in macrophages act as sensors for oxidized lipids present in oxLDL (4–6). Indeed, activation of PPARγ results in the induction of two pathways that efficiently couple oxLDL uptake to cholesterol efflux in cultured macrophages, suggesting that PPARγ activation may serve this role in vivo (7–9).
Although LDL and oxLDL were previously considered to be the only lipid particles important in atherogenesis, recent studies in humans have established triglyceride-rich lipoproteins, such as the very low-density lipoprotein (VLDL), as an additional risk factor for development of atherosclerotic heart disease (10, 11). The detection of lipoprotein-derived triglycerides in human and rabbit atheromas supports a presumptive role in pathogenesis (12, 13). In addition, remnant particles (hydrolyzed chylomicra and VLDL particles) or triglycerides have been shown to predict the extent and progression of coronary artery disease in humans (14, 15). Consistent with this idea, treatment of hypertriglyceridemic patients with triglyceride-lowering therapy dramatically decreases the morbidity and mortality from ischemic heart disease (16). Despite these clinical advances, a molecular route through which VLDL or its component lipids promote the progression of macrovascular disease remains largely unexplained.
Here, we describe a transcriptional pathway, in the macrophage, driven by VLDL through the activation of PPARδ. The transcriptionally active components of native VLDL particle are its triglycerides, which can be released by the lipolytic action of lipoprotein lipase. Treatment of WT macrophages with VLDL results in triglyceride accumulation and the induction of adipose differentiation-related protein (ADRP). Remarkably, disruption of the PPARδ gene in macrophages completely abolishes this transcriptional response to native VLDL. Collectively, our work has identified a transcriptional pathway through which dietary triglycerides that are incorporated into VLDL can directly regulate gene expression in cells of the vessel wall.
Materials and Methods
Reporter and Expression Vector Constructs.
5′-RACE on the mouse ADRP gene was performed using RNA from 3T3-L1 cells, and the extended RACE product was used to screen a mouse genomic library (Stratagene). A 2-kb fragment containing the promoter and part of exon 1 was PCR amplified and cloned into the MluI/EcoRI sites of pGL3-basic vector to yield the −2,065 construct. StuI and PvuII digestions of −2,065 construct generated the deletion reporter constructs, and the mutations in the DR1 site were introduced by the QuikChange site-directed mutagenesis kit (Stratagene). Three copies of annealed oligos, containing the ADRP PPAR response element (PPRE), were cloned into the HindIII and BamHI sites of thymidine kinase (TK)-luciferase reporter vector. For the hemagglutinin (HA)-tagged ADRP expression plasmid, full-length ADRP cDNA was cloned into the EcoRI/XhoI sites of HA-tagged CMX expression vector. Full-length mouse PPARδ was cloned into the SnaBI site of pBabe-puro to generate the pBabe-puro-PPARδ retroviral expression plasmid (17).
Transient Transfections and Gel Mobility-Shift Assays.
A total of 5 × 105 CV-1 and 2 × 106 RAW cells were plated in 96- and 48-well plates, respectively, in DMEM supplemented with 10% FBS. Transfections were performed with Lipofectamine 2000 (Invitrogen) per manufacturer's instructions. Twenty-four hours after transfection, cells were treated with ligands (2 μM cPGI, 0.1 μM GW501516, 0.1 μM LG268), lipoproteins (Intracel, Frederick, MD), or triglycerides (50 μM, Sigma) for an additional 24 h. Triglyceride stocks were formulated in chloroform, and appropriate dilutions were made in 1% fatty acid-free BSA with vigorous agitation. All transfections were performed in triplicate and repeated at least three independent times. Gel-shift assays were performed as described using 32P-labeled oligos containing either the WT or mutant DR1 of the ADRP PPRE (WT oligo, 5′-TTGTAGGTGAAAGGGCAAAGA-3′; mutant oligo, 5′-TTGTAAGCTTAAGGGCAAAGA-3′).
Generation of PPARδ Null Embryonic Stem (ES) Cells and ES Cell Differentiation into Macrophages.
Both the WT and PPARδ null ES cells were differentiated into macrophages as described (18).
Northern Analysis, Flow Cytometry, and Immunolocalization.
RNA was extracted from cells by using the Trizol reagent (Invitrogen), and Northern blot analysis for various genes was performed as described (18). For FACS analysis, cultured macrophages were removed from Petri dishes with ice-cold PBS supplemented with 5 mM EDTA. Samples were incubated for 15 min at 4°C with FITC-conjugated antibodies F4/80 (Serotec), Mac-1, Gr1, and 2.4G2 (PharMingen) and washed and analyzed on the Becton Dickinson FACScan using CELLQUEST software (BD Biosciences, Franklin Lakes, NJ). For immunolocalization studies, CV-1 cells were grown on chamber slides (Nalgene) and transfected with either 1 μg of pCMX-HA-ADRP or the CMX empty vector. Transfected cells were loaded with VLDL/lipoprotein lipase (LPL) for an additional 24 h. Subsequently, cells were fixed in 4% formaldehyde and stained with rabbit anti-HA antibody (Upstate Biotechnology, Lake Placid, NY) followed by FITC-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotechnology). Cellular images were obtained by using an Olympus IX-70 deconvolution microscope and deltavision software (Applied Precision, Issaquah, WA).
Stable Cell Lines.
Control and PPARδ expression plasmids were packaged into retroviruses by transient transfection of Phoenix A cells. NIH 3T3 cells were infected with viral supernatants at 50% confluence in 6-well plates and selected for stable integration with puromycin (2 μg/ml) over 7–10 days.
Results
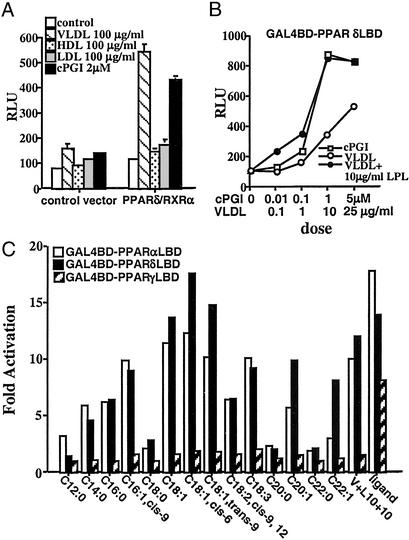
The identification of synthetic PPARδ agonists has revealed an important role for this receptor in lipid metabolism (19, 20). However, the endogenous signaling pathways that are key to its core biological function are poorly understood. Because we have previously shown that oxLDL can transcriptionally activate PPARγ, we explored the possibility that other native lipoprotein particles might constitute a comparable PPARδ activation pathway (4). Remarkably, VLDL emerged from this survey as an efficient activator of PPARδ in a reporter system (Fig. 1A). This activation was specific to VLDL, because neither LDL nor HDL particles produce PPARδ activation. Furthermore, a physiologic concentration of VLDL (100 μg/ml) closely approximated the maximal efficacy of synthetic PPARδ ligands such as carboprostacyclin (2 μM) and GW 501516 (0.1 μM) (Fig. 1B and data not shown). To determine whether the VLDL particle acts through the ligand-binding domain (LBD), we carried out transient transfection assays with a chimeric receptor, in which the GAL4 DNA binding domain (DBD) is fused in frame to the ligand binding domain of PPARδ. As expected, VLDL efficiently transactivated the GAL4-PPARδ fusion (Fig. 1B), suggesting that the VLDL particle may be an endogenous source of PPARδ ligands.
Figure 1.
PPARδ is activated by the VLDL particle. (A) VLDL particle transactivates full-length PPARδ. Transient transfection experiments were performed in triplicate in CV1 cells using the AOx-PPRE3 luciferase reporter construct (0.1 μg) and the CMX-PPARδ/CMX-RXRα expression vectors (10 ng each) as described in Materials and Methods. Transfected cells were cultured in lipoprotein-deficient FBS and treated with ligands or lipoproteins for 24 h before collection for reporter gene analysis. Luciferase activity was normalized for transfection efficiency to an internal β-galactosidase control. (B) VLDL particle activates via the ligand binding domain of PPARδ. CV-1 cells were cotransfected with the chimeric receptor, GAL4DBD-PPARδLBD, and the UAS3 luciferase reporter gene. Transfected cells were subsequently treated with varying concentrations of cPGI, VLDL, or VLDL coincubated with 10 μg/ml LPL. (C) Triglycerides present in VLDL transactivate PPARδ. Transient transfections were performed in CV-1 cells with GAL-PPAR (α, δ, and γ) and UAS3 luciferase reporter. Triglycerides were coincubated with LPL (10 μg/ml) in 1% BSA before their addition to the medium (50 μM final concentration).
VLDL is a large lipoprotein particle that is composed of triglycerides, free and esterified cholesterol, phospholipids, and apolipoproteins (21). Being a triglyceride-rich particle, VLDL transports liver-synthesized triglycerides to the peripheral tissues. In peripheral tissues, triglycerides present in VLDL are hydrolyzed by LPL into fatty acids, thereby allowing their entry into cells (22). We therefore hypothesized that if the activating component of the VLDL particle were its triglycerides, coincubation with LPL should enhance the ability of the particle to transactivate PPARδ. Indeed, as shown in Fig. 1B, coincubation with LPL greatly augmented the activity of VLDL on GAL4-PPARδ, with maximal stimulation attained at a 10-fold lower dose of VLDL (10 μg/ml). Furthermore, to determine whether any of the triglycerides that are normally found in native VLDL can activate PPARs, we next conducted a systematic screen with various triglycerides on all three PPAR subtypes, using the GAL4-PPAR(α,δ,γ) chimeric receptors. Remarkably, coincubation of triglycerides of varying chain lengths with LPL revealed that both mono- or polyunsaturated triglycerides are efficient activators of both GAL4-PPARδ and GAL4-PPARα, but not of GAL4-PPARγ (Fig. 1C). Lastly, although treatment of cells with triglycerides and LPL recapitulates the transcriptional response of VLDL/LPL on PPARδ, we cannot exclude the possibility that triglyceride-rich remnant lipoproteins, the residual product of VLDL lipolysis, might also serve to activate this receptor.
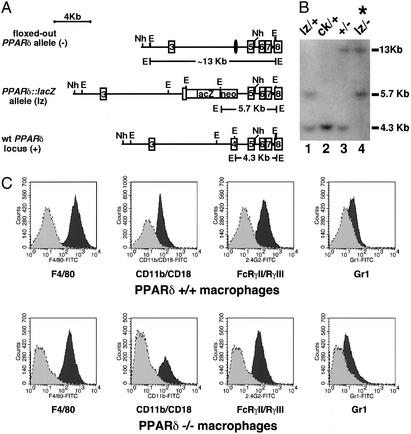
The role of PPARδ in mediating VLDL signaling in macrophages was genetically dissected using ES cells. We generated PPARδ-null ES cells by retargeting the remaining WT allele in the previously established PPARδ heterozygous ES cell line (clone IIIA4) (23). The retargeting construct harbored a knock-in of the lacZ gene upstream of the first zinc finger of the DNA-binding domain, generating an in-frame fusion between lacZ and a short N-terminal fragment of PPARδ (Fig. 2A). Homologous recombination of the second allele of PPARδ was confirmed by Southern blot analysis (Fig. 2B). To study the regulatory role of PPARδ in macrophage gene expression, we used an in vitro ES cell differentiation assay to generate both WT and PPARδ null macrophages (18). As shown in Fig. 2C, both PPARδ+/+ and PPARδ−/− ES cells were equally capable of differentiating into macrophages, as judged by cellular morphology and expression of myeloid cell-surface markers, such as F4/80, FcγRII/FcγRIII, and CD11b/CD18 (24).
Figure 2.
Generation of PPARδ-deficient ES cells and macrophages. (A) Targeting scheme for generation of PPARδ null ES cells. The G418-sensitive ES cell clone IIIA4 (23) consists of a mixture of cells heterozygous for the WT PPARδ allele (+, Bottom) and either a null “floxed-out” allele (−, Top) or a conditional “floxed” allele (ck, not shown). This clone was retargeted with a construct containing a lacZ gene knocked-in, which disrupts PPARδ upstream of the first zinc finger DNA-binding motif (lz, Middle). Patterned bars below each allele represent the corresponding predictions of EcoRI-flanked DNA fragments when probed with a 3′-external probe. Restriction sites: E, EcoRI; Nh, NheI. (B) Southern blot analysis of PPARδ null ES cells. The individual daughter clones generated by retargeting the IIIA4 cells contain different biallelic combinations of PPARδ: cells heterozygous for the WT and the newly introduced lacZ knock-in allele (+/lz; lane 1); two G418-resistant clones carrying the original IIIA4 allelic combinations ck/+ and +/− (lanes 2 and 3, respectively) and a likely nonhomologous integration of the neoR gene; and the PPARδ null ES clone analyzed functionally in this study (lz/−, lane 4; *), in which the lz allele integrated in place of the remaining WT allele of clone IIIA4. Allelic identity was further confirmed using a 5′ external probe (not shown). (C) FACS analysis of PPARδ+/+ and PPARδ−/− ES cell-derived macrophages. Macrophages were generated from ES cells as described in Materials and Methods. WT or PPARδ null macrophages were stained with the indicated antibodies (dark gray) or isotype control (light gray). Macrophage cell-surface markers include F4/80, Mac-1 (CD11b/CD18), and 2.4G2 (FcγRII/FcγRIII). Gr.1 (Ly-6G), a granulocyte lineage-specific marker, is used here as a negative control.
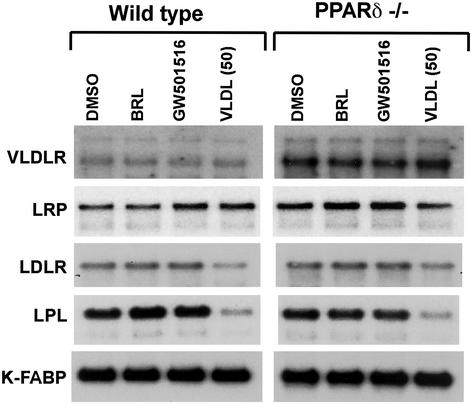
We have previously described a lipid cycle in macrophages, in which components of oxLDL activate PPARγ and induce the expression of the scavenger receptor CD36 and the oxysterol receptor liver X receptor α (LXRα) (7, 25). Because PPARγ and PPARδ share a high degree of homology in their DNA-binding domains (26), we expected PPARδ to also transcriptionally regulate these genes. However, to our surprise, gene-expression analysis of WT and PPARδ null macrophages revealed that, unlike PPARγ, PPARδ activation does not significantly induce CD36 or LXRα (data not shown). Therefore, we next sought to determine whether PPARδ participates in the regulation of a subset of genes relevant to VLDL metabolism. Various cellular proteins, including cell-surface receptors and LPL, mediate the uptake and clearance of VLDL (27). Although WT ES cell-derived macrophages express low levels of the VLDL receptor, no induction of this gene is observed on treatment with either PPARδ or PPARγ ligands. In contrast, the expression of VLDL receptor gene is dramatically induced in PPARδ null macrophages (Fig. 3), suggesting that this gene is either directly or indirectly suppressed by PPARδ. Furthermore, the induction of VLDL receptor gene was specific, as the expression of both the chylomicron remnant receptor (LRP1) and LDL receptor (LDLR) was unchanged in both the WT and PPARδ−/− macrophages. Finally, although LPL is abundantly expressed in macrophages of both genotypes, lipid loading of macrophages with VLDL down-regulates its expression in a PPARδ-independent manner (Fig. 3), suggesting a potential regulatory involvement with the SREBPs (28, 29). Collectively, these data suggest that by sensing different components in lipoprotein particles, PPARγ and PPARδ orchestrate distinct transcriptional responses to regulate macrophage lipid metabolism.
Figure 3.
VLDL receptor is dramatically induced in PPARδ null macrophages. WT or PPARδ−/− ES-derived macrophages were treated with vehicle, rosiglitazone (1 μM), GW 501516 (0.1 μM), or VLDL (50 μg/ml) for 24 h. Total RNA (5 μg) was analyzed by Northern blotting using 32P-labeled cDNA probes. An equivalent amount of intact RNA was run present in each lane as accessed by hybridization to K-FABP cDNA probe.
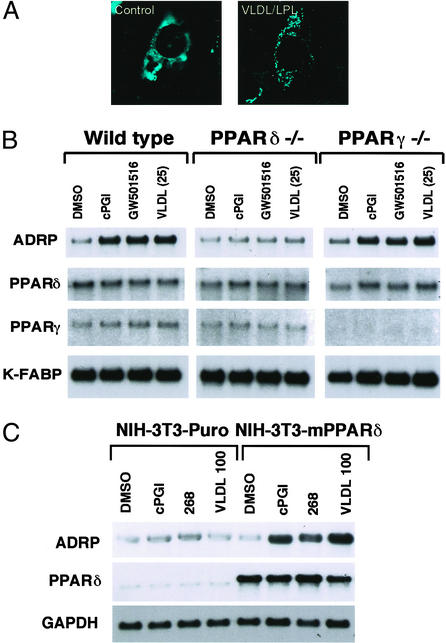
Treatment of differentiated macrophages or various other cell types with the VLDL/LPL combination results in accumulation of cytoplasmic lipid droplets that are composed of triglycerides and esterified cholesterol (data not shown). Because ADRP has been shown to be associated with small lipid droplets in various cell types (30), we next evaluated whether loading of cells with a VLDL/LPL combination results in recruitment of ADRP to the lipid droplet. For these studies, CV-1 cells were transiently transfected with HA-ADRP, and the cytoplasmic distribution of HA-ADRP was followed after VLDL/LPL loading. As shown in Fig. 4A, HA-tagged ADRP has a nonspecific staining pattern in untreated CV-1 cells. However, VLDL/LPL treatment of transfected CV-1 cells results in redistribution of HA-tagged ADRP onto discrete oil red O-positive droplets (Fig. 4A and data not shown). We next sought to determine whether the activation of PPARδ by VLDL leads to the induction of the ADRP gene in differentiated macrophages. Indeed, treatment of WT or PPARγ null macrophages with both the synthetic and natural PPARδ ligands resulted in dramatic induction of the ADRP mRNA (Fig. 4B). In contrast, no change in ADRP expression was observed with ligand treatment of PPARδ null macrophages, indicating unequivocally that the endogenous PPARδ gene in macrophages mediates transcriptional response to the VLDL particle (Fig. 4B).
Figure 4.
VLDL and PPARδ regulate ADRP expression. (A) Lipid loading with VLDL/LPL results in redistribution of cytoplasmic ADRP. Expression vector containing HA-tagged mouse ADRP was transfected into CV-1 cells grown on chamber slides. After a 24-h treatment with VLDL/LPL, cells were stained with anti-HA antibody and visualized with FITC-conjugated secondary antibody. (B) PPARδ and VLDL regulate ADRP expression in macrophages. WT, PPARδ, or γ null macrophages were treated with cPGI (2 μM), GW 501516 (0.1 μM), or VLDL (25 μg/ml) for 24 h. (C) Ectopic expression of PPARδ confers VLDL responsiveness onto cells. NIH vector and NIH-PPARδ cells were treated with vehicle, cPGI (2 μM), LG 268 (0.1 μM), or VLDL (100 μg/ml) for 24 h. Gene expression analyses were performed by Northern blots with 32P-labeled cDNA probes.
Having established that PPARδ is essential for mediating the transcriptional response of the VLDL particle in macrophages, we next sought to determine whether ectopic expression of PPARδ is sufficient for this process. For these experiments, a retroviral vector (Babe-PPARδ) was used to express PPARδ in NIH 3T3 cells. Treatment of control cells with the PPARδ ligand carboprostacylin (cPGI), the retinoid X receptor (RXR)-specific ligand LG268, or VLDL had no effect on the expression of ADRP (Fig. 4C). In contrast, treatment of NIH-PPARδ cells with cPGI, LG268, or VLDL resulted in a robust induction of ADRP (Fig. 4C), suggesting that forced expression of PPARδ is sufficient to confer transcriptional responsiveness to the VLDL particle.
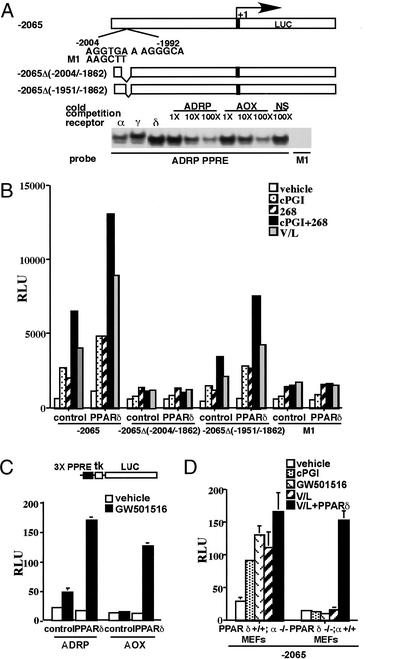
To determine whether the ADRP gene is a direct target for transcriptional regulation by PPARδ/RXR heterodimers, we performed transient transfection experiments with the mouse ADRP promoter. A reporter construct containing 2.1 kb of the proximal promoter of ADRP was efficiently activated by both synthetic and natural PPARδ ligands in RAW 267.4 cells (Fig. 5A). Sequence analysis of the responsive promoter fragment revealed a potential DR-1 element, the preferred binding site for PPAR/RXR heterodimers (31), between −2,004 and −1,992 bp (Fig. 5A). Consistent with the notion that this element may mediate ADRP induction by PPARδ, deletion of the region between −2,004 and −1,862 bp, but not of the adjacent region between −1,951 and −1,862 bp, abrogated the response of the promoter to liganded PPARδ/RXR heterodimers (Fig. 5 A and B). Gel mobility shift assays revealed that all three PPAR subtypes could bind the DR-1 PPRE of the ADRP promoter. This binding of ADRP PPRE by PPARγ/RXRα and PPARα/RXRα heterodimers is consistent with the observed regulation of this gene by PPARγ and PPARα ligands in other cell types (refs. 32–34 and data not shown). Competition with excess unlabeled probes further established the specificity of binding by PPAR/RXR heterodimers. In addition, mutations in the ADRP PPRE abolished both the binding and activation by liganded PPARδ/RXRα heterodimers (Fig. 5 A and B). Transient transfection experiments with a reporter gene containing three copies of the ADRP PPRE upstream of a heterologous promoter (ADRP PPRE3-TK-Luc) confirmed that the identified element was as efficiently activated as the control reporter (AOx PPRE3-TK-Luc) by PPARδ/RXR heterodimers in vivo (Fig. 5C). Finally, to evaluate if endogenous PPARδ and VLDL/LPL are sufficient to activate the ADRP promoter, we performed transient transfection assays with a −2,065-bp reporter construct in PPARα−/− or PPARδ−/− mouse embryo fibroblasts (MEFs). As expected, treatment of transfected PPARα−/− MEFs, which express PPARδ (data not shown), with both the synthetic PPARδ ligands and VLDL/LPL combination resulted in robust activation of the reporter gene (Fig. 5D). In contrast, neither the synthetic ligands nor the VLDL/LPL combination could transactivate the ADRP promoter in PPARδ null MEFs (Fig. 5D). This was easily rescued by exogenous expression of PPARδ, further verifying that the endogenous PPARδ is sufficient to mediate transcriptional effects of VLDL on the ADRP promoter.
Figure 5.
ADRP promoter is a target for direct regulation by VLDL and PPARδ/RXR heterodimers. (A) ADRP promoter contains a PPRE. Schematic outline of reporter constructs containing the WT, point mutations, and internal deletions of the ADRP promoter. The potential PPRE is located between −2,004 and −1,992 bp of the ADRP promoter. Gel mobility shift assays were performed using in vitro translated proteins and 32P-end-labeled ADRP and acyl CoA oxidase (AoX) PPRE oligonucleotides. PPARα/RXR, PPARδ/RXR, and PPARγ/RXR heterodimers can bind to the ADRP PPRE. Unlabeled PPRE oligonucleotides or nonspecific DNA were used at the indicated molar excess to perform competition assays. Point mutations in the 5′ half site of the ADRP PPRE (M1) abolished binding of the PPARδ/RXR heterodimers. (B) PPARδ/RXR heterodimers transactivate the ADRP promoter. RAW 264.7 cells were cotransfected with ADRP reporter constructs (0.1 μg) and CMX-PPARδ/CMX-RXR expression vectors (10 ng) as described in Materials and Methods. Transfected cells were treated with ligands (2 μM cPGI, 0.1 μM LG 268, 2 μM cPGI + 0.1 μM LG 268, and VLDL (25 mg/ml)/LPL (10 mg/ml) for 24 h before collection for reporter gene assays. (C) The ADRP PPRE is a functional PPARδ response element. Three copies of the ADRP or the AOx PPRE were cloned upstream of the TK-luciferase reporter gene. CV-1 cells were cotransfected with reporters and receptor expression vectors and analyzed for luciferase activity as stated in B. (D) Endogenous PPARδ and VLDL transactivate the ADRP promoter. Transient transfection experiments were performed in PPARα−/− and PPARδ−/− MEFs with the luciferase reporter construct containing the proximal promoter (2,065 bp) of the ADRP gene.
Discussion
Whereas VLDL and serum triglycerides are known additional risk factors for coronary artery disease, how they modulate macrophage response and lesion progression is largely unknown. The data presented here show that PPARδ mediates the transcriptional response to native VLDL in macrophages. Transcriptional activation of PPARδ by VLDL leads to the induction of the ADRP gene, which may potentially allow lesion macrophages to store excess triglycerides. Consistent with this, deletion of PPARδ gene in macrophages completely abolishes the induction of the ADRP gene by VLDL. Taken together with our previous studies, these data show that in contrast to PPARγ, which responds to the oxidized fatty acids present in oxLDL (4), PPARδ functions as a sensor for the triglycerides present in the VLDL particle.
VLDL is synthesized and secreted by the liver to transport triglycerides and cholesterol to peripheral tissues (22). The composition of the fatty acids present as the triglyceride moieties in VLDL particles closely reflects the hepatic pool of preformed fatty acids (35, 36). These preformed fatty acids are either proximally derived from dietary intake (37) or from a pool of fatty acids being mobilized from adipose tissue in states of insulin resistance, such as type II diabetes mellitus (38, 39). Because both PPARδ and PPARα are activated by the triglycerides present in the VLDL particle, their distinctive functions can be attributed to PPARα expression in the liver and PPARδ expression in peripheral tissues. Thus, our findings suggest that the coupled activation of PPARδ and PPARα by VLDL would allow for a coordinated cellular response to incoming fatty acids and triglycerides. For instance, the activation of PPARδ by VLDL might facilitate the “short-term” storage of triglycerides as ADRP-coated lipid droplets, whereas stimulation of PPARα would result in the induction genes important in fatty acid oxidation (40, 41). This coupled response to VLDL could efficiently target triglycerides present in cytoplasmic lipid droplets for oxidation in peroxisomes and mitochondria. Consistent with this notion, treatment of rats with etomoxir, an inhibitor of CPT-1 and mitochondrial fatty acid oxidation pathways, results in accumulation of cytoplasmic lipids and the induction of ADRP (42), further raising the possibility that lipid droplet-associated proteins participate in cytoplasmic trafficking of lipids. In this regard, we have recently identified perilipin, which plays a critical role in the “long-term” storage of lipids (43, 44), as a PPARγ target gene in adipocytes (data not shown, AC and RME).
In addition to human epidemiological studies, recent experiments in mouse models of atherosclerosis have found an increased susceptibility to atherosclerosis with high VLDL levels. For instance, when LDLR−/− mice are challenged with diets of different compositions, a linear correlation was observed between atheroslcerotic plaque burden and diets that raise serum VLDL levels (45). Because this tight correlation is apparently independent of other lipid parameters, such as serum LDL and HDL, it suggests that activation of PPARδ by the VLDL particle might increase the lipid content of atheroslcerotic plaques. However, the true contribution of PPARδ to the atherogenic process is difficult to predict. On the one hand, impaired clearance of VLDL and triglyceride particles by peripheral tissues, as in patients with type II diabetes, may result in pathologic activation of this receptor in lesion macrophages and exacerbation of atherosclerosis. On the other hand, pharmacologic induction of genes, such as UCP3 and PDK4 (Y. Wang and R.M.E., unpublished data) by PPARδ ligands could enhance the uptake and clearance of the triglyceride-rich lipoprotein particles in peripheral tissues and rob the growing lesion of its atherogenic substrate. Therefore, future studies in mouse models of atherosclerosis with either PPARδ activators or with PPARδ-deficient bone marrow will be needed to clarify the role of this receptor in vascular disease.
Acknowledgments
We thank Elaine Stevens for administrative assistance and members of the Evans laboratory for critical comments. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. A.C. was supported by the Physician Postdoctoral Fellowship of the Howard Hughes Medical Institute. This work was supported by the Howard Hughes Medical Institute and the Hilblom Foundation.
Abbreviations
- LDL
low-density lipoprotein
- VLDL
very low-density lipoprotein
- oxLDL
oxidized LDL
- ADRP
adipose differentiation-related protein
- TK
thymidine kinase
- HA
hemagglutinin
- ES
embryonic stem
- LPL
lipoprotein lipase
- RXR
retinoid X receptor
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR response element
- MEFs
mouse embryo fibroblasts
References
- 1.Spiegelman B M, Flier J S. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 2.Ross R. Am Heart J. 1999;138:419–420. doi: 10.1016/s0002-8703(99)70266-8. [DOI] [PubMed] [Google Scholar]
- 3.Glass C K, Witztum J L. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 4.Nagy L, Tontonoz P, Alvarez J G, Chen H, Evans R M. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 5.Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 6.Chawla A, Repa J J, Evans R M, Mangelsdorf D J. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 7.Chawla A, Boisvert W A, Lee C, Laffitte B A, Barak Y, Joseph S B, Liao D, Nagy L, Edwards P A, Curtiss L K, et al. Mol Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 8.Chinetti G, Lestavel S, Bocher V, Remaley A T, Neve B, Torra I P, Teissier E, Minnich A, Jaye M, Duverger N, et al. Nat Med. 2001;7:53–58. doi: 10.1038/83348. [DOI] [PubMed] [Google Scholar]
- 9.Lazar M A. Nat Med. 2001;7:23–24. doi: 10.1038/83301. [DOI] [PubMed] [Google Scholar]
- 10.Malloy M J, Kane J P. Adv Intern Med. 2001;47:111–136. [PubMed] [Google Scholar]
- 11.Forrester J S. Curr Opin Cardiol. 2001;16:261–264. doi: 10.1097/00001573-200107000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Rapp J H, Lespine A, Hamilton R L, Colyvas N, Chaumeton A H, Tweedie-Hardman J, Kotite L, Kunitake S T, Havel R J, Kane J P. Arterioscler Thromb. 1994;14:1767–1774. doi: 10.1161/01.atv.14.11.1767. [DOI] [PubMed] [Google Scholar]
- 13.Mamo J C, Wheeler J R. Coron Artery Dis. 1994;5:695–705. doi: 10.1097/00019501-199408000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Drexel H, Amann F W, Beran J, Rentsch K, Candinas R, Muntwyler J, Luethy A, Gasser T, Follath F. Circulation. 1994;90:2230–2235. doi: 10.1161/01.cir.90.5.2230. [DOI] [PubMed] [Google Scholar]
- 15.Phillips N R, Waters D, Havel R J. Circulation. 1993;88:2762–2770. doi: 10.1161/01.cir.88.6.2762. [DOI] [PubMed] [Google Scholar]
- 16.Rubins H B, Robins S J, Collins D, Fye C L, Anderson J W, Elam M B, Faas F H, Linares E, Schaefer E J, Schectman G, et al. N Engl J Med. 1999;341:410–418. doi: 10.1056/NEJM199908053410604. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 18.Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans R M. Nat Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 19.Leibowitz M D, Fievet C, Hennuyer N, Peinado-Onsurbe J, Duez H, Bergera J, Cullinan C A, Sparrow C P, Baffic J, Berger G D, et al. FEBS Lett. 2000;473:333–336. doi: 10.1016/s0014-5793(00)01554-4. [DOI] [PubMed] [Google Scholar]
- 20.Oliver W R, Jr, Shenk J L, Snaith M R, Russell C S, Plunket K D, Bodkin N L, Lewis M C, Winegar D A, Sznaidman M L, Lambert M H, et al. Proc Natl Acad Sci USA. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shelness G S, Sellers J A. Curr Opin Lipidol. 2001;12:151–157. doi: 10.1097/00041433-200104000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Ginsberg H N. Endocrinol Metab Clin North Am. 1998;27:503–519. doi: 10.1016/s0889-8529(05)70023-2. [DOI] [PubMed] [Google Scholar]
- 23.Barak Y, Liao D, He W, Ong E S, Nelson M C, Olefsky J M, Boland R, Evans R M. Proc Natl Acad Sci USA. 2002;99:303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKnight A J, Gordon S. Adv Immunol. 1998;68:271–314. doi: 10.1016/s0065-2776(08)60562-3. [DOI] [PubMed] [Google Scholar]
- 25.Tontonoz P, Nagy L, Alvarez J G, Thomazy V A, Evans R M. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 26.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain M M, Strickland D K, Bakillah A. Annu Rev Nutr. 1999;19:141–172. doi: 10.1146/annurev.nutr.19.1.141. [DOI] [PubMed] [Google Scholar]
- 28.Hannah V C, Ou J, Luong A, Goldstein J L, Brown M S. J Biol Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 29.Horton J D, Goldstein J L, Brown M S. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Londos C, Brasaemle D L, Schultz C J, Segrest J P, Kimmel A R. Semin Cell Dev Biol. 1999;10:51–58. doi: 10.1006/scdb.1998.0275. [DOI] [PubMed] [Google Scholar]
- 31.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 32.Vosper H, Patel L, Graham T L, Khoudoli G A, Hill A, Macphee C H, Pinto I, Smith S A, Suckling K E, et al. J Biol Chem. 2001;276:44258–44265. doi: 10.1074/jbc.M108482200. [DOI] [PubMed] [Google Scholar]
- 33.Buechler C, Ritter M, Duong C Q, Orso E, Kapinsky M, Schmitz G. Biochim Biophys Acta. 2001;1532:97–104. doi: 10.1016/s1388-1981(01)00121-4. [DOI] [PubMed] [Google Scholar]
- 34.Gupta R A, Brockman J A, Sarraf P, Willson T M, DuBois R N. J Biol Chem. 2001;276:29681–29687. doi: 10.1074/jbc.M103779200. [DOI] [PubMed] [Google Scholar]
- 35.Parks E J, Krauss R M, Christiansen M P, Neese R A, Hellerstein M K. J Clin Invest. 1999;104:1087–1096. doi: 10.1172/JCI6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aarsland A, Chinkes D, Wolfe R R. J Clin Invest. 1996;98:2008–2017. doi: 10.1172/JCI119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aarsland A, Wolfe R R. J Lipid Res. 1998;39:1280–1286. [PubMed] [Google Scholar]
- 38. Ginsberg, H. N. Diabetes 45, Suppl. 3, S27–S30, 1996. [DOI] [PubMed]
- 39.Ginsberg H N. J Clin Invest. 2000;106:453–458. doi: 10.1172/JCI10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djouadi F, Weinheimer C J, Kelly D P. Adv Exp Med Biol. 1999;466:211–220. [PubMed] [Google Scholar]
- 41.Gonzalez F J. Biochimie. 1997;79:139–144. doi: 10.1016/s0300-9084(97)81506-4. [DOI] [PubMed] [Google Scholar]
- 42.Steiner S, Wahl D, Mangold B L, Robison R, Raymackers J, Meheus L, Anderson N L, Cordier A. Biochem Biophys Res Commun. 1996;218:777–782. doi: 10.1006/bbrc.1996.0138. [DOI] [PubMed] [Google Scholar]
- 43.Tansey J T, Sztalryd C, Gruia-Gray J, Roush D L, Zee J V, Gavrilova O, Reitman M L, Deng C X, Li C, Kimmel A R, Londos C. Proc Natl Acad Sci USA. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Botas J, Anderson J B, Tessier D, Lapillonne A, Chang B H, Quast M J, Gorenstein D, Chen K H, Chan L. Nat Genet. 2000;26:474–479. doi: 10.1038/82630. [DOI] [PubMed] [Google Scholar]
- 45.Merkel M, Velez-Carrasco W, Hudgins L C, Breslow J L. Proc Natl Acad Sci USA. 2001;98:13294–13299. doi: 10.1073/pnas.231490498. [DOI] [PMC free article] [PubMed] [Google Scholar]