SUMMARY
Cytochromes P450 (P450s) play a major role in the clearance of drugs, toxins, and environmental pollutants. Additionally, metabolism by P450s can result in toxic or carcinogenic products. The metabolism of pharmaceuticals by P450s is a major concern during the design of new drug candidates. Determining the interactions between P450s and compounds of very diverse structures is complicated by the variability in P450-ligand interactions. Understanding the protein structural elements and the chemical attributes of ligands that dictate their orientation in the P450 active site will aid in the development of effective and safe therapeutic agents. The goal of this review is to describe P450-ligand interactions from two perspectives. The first is the various structural elements that microsomal P450s have at their disposal to assume the different conformations observed in X-ray crystal structures. The second is P450-ligand dynamics analyzed by NMR relaxation studies.
Keywords: Cytochrome P450, drug metabolism, mammalian, microsomal, NMR, P450, plasticity, protein structure, xenobiotic, X-ray crystallography
1. Introduction
Microsomal P450 families 1–4 play a major role in the metabolism of the majority of pharmaceuticals and a large number of other xenobiotics. The diverse list of substrates encompasses a broad array of molecular shapes, volumes, geometries, and chemical properties. However, in many cases P450s are able to oxidize these compounds with exceptional regio- and stereospecificity. How these enzymes accomplish this impressive range of chemical modifications has been studied by numerous techniques including kinetic analysis [1], mutagenesis studies [2, 3], and computer modeling [4–7]. However, X-ray crystallography and NMR remain two of the most powerful tools to gain insight into P450 function and the ability to bind ligands. Structural information gained via these methods can aid in the design and development of pharmaceuticals with optimal metabolic properties. In addition, avoidance of adverse drug-drug interactions due to P450 inhibition is greatly assisted by detailed information on the active site (Section 2) and the conformations ligands are able to adopt when binding to these enzymes (Section 3).
2. X-ray crystallography of cytochromes P450
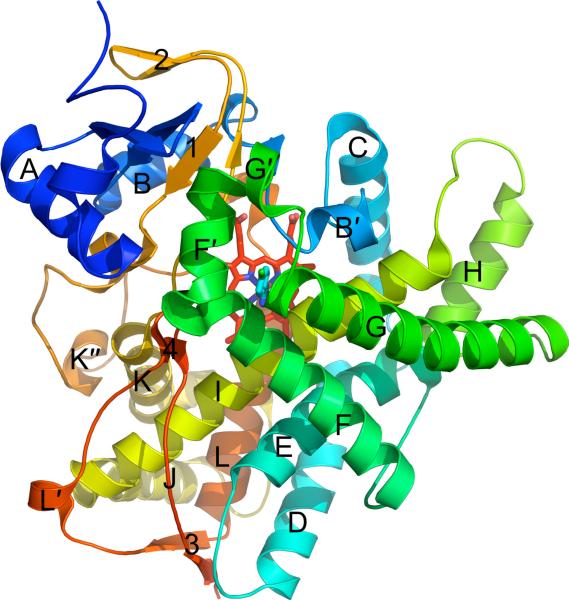
Numerous crystal structures have shown that all P450s share a common protein fold consisting of a large α-helical domain (with helices labeled A–L) and a smaller β-sheet domain (with sheets labeled 1–4) (Fig. 1). With such a high degree of structural similarity across enzyme families and subfamilies as well as across biological domains and kingdoms, it may not be readily apparent how P450s achieve substrate diversity. Numerous crystal structures of P450s have captured these enzymes in a variety of conformations, which highlight the ability of P450s to adapt their structures to accommodate their broad range of substrates. In particular, CYP2B4 has shown the largest degree of structural flexibility of any P450 to be crystallized. CYP2B4 has been observed in a similar closed, compact conformation when bound to four different small molecules [8, 9,PDB IDs 3KW4 & 3ME6], but has also been crystallized in three distinct open conformations in the presence of larger compounds [10, 11] or in the absence of ligand [12]. However, not all P450s are as plastic as CYP2B4. At the opposite end of the flexibility spectrum is CYP2A6, which has been crystallized in the presence of seven different ligands with varying sizes and shapes [13–16]. Despite the differences in compounds bound in its active site, CYP2A6 remains in a single closed conformation with little rearrangement of side chains. The evidence from X-ray crystal structures that some P450s adopt multiple conformations, whereas other P450s are observed in only one conformation, makes it is difficult to predict how P450s will behave with new compounds. Accordingly, significant obstacles remain in the rational design of pharmaceuticals that will maintain their efficacy and not cause adverse drug-drug interactions during combined therapeutic regimens. This section will discuss the various structural mechanisms that P450s utilize in order to bind ligands and adopt various conformations.
Figure 1.
A ribbon diagram of the CYP2B6–4-CPI complex (PDB ID 3IBD) shows the typical tertiary structure of a cytochrome P450. The twelve α-helices (A–L) and four β-sheets (1–4) are labeled, with minor helices denoted as 'prime' or 'double prime'. The protein is colored from blue (N-terminus) to red (C-terminus), while the heme and 4-CPI inhibitor are shown as red and cyan sticks, respectively.
2.1. CYP1A2
2.1.a. Substrate specificity
Family 1 enzymes are distinct from other P450s in their ability to metabolize polycyclic aromatic hydrocarbons (PAHs) [17], which also induce CYP1 expression [18]. Additionally, CYP1A2, which is the primary family 1 enzyme found in human liver, is known to N-oxidize arylamines to potent mutagens or carcinogens [19]. Differential expression and activity levels cause variations in drug clearance among individuals [20], but the cause is not well understood. In addition to PAHs and arylamines, CYP1A2 is also largely responsible for the metabolism of naturally occurring compounds such as caffeine and melatonin, as well as pharmaceuticals including flutamide, lidocaine, olanzapine, tacrine, theophylline, triamterene, and zolmitriptan [21]. The crystal structure of CYP1A2 shows the enzyme in complex with α-naphthoflavone (ANF) [20], which is a competitive inhibitor of family 1 P450s. In general, CYP1A2 substrates and inhibitors are relatively large molecules that contain aromatic, planar regions with various chemical side groups that may or may not be planar as well. The CYP1A2–ANF complex is the only family 1 structure found in the PDB.
2.1.b. Overall structure and active site
Like several other microsomal P450s, the CYP1A2–ANF complex contains additional, smaller helices (denoted as `prime' or `double prime') in addition to helices A–L. The most prominent of these small helices are B', F' and G'.
The active site cavity of CYP1A2 is relatively compact and closed with a volume of 390 Å3 (Table 1). The pocket is uniformly narrow and is lined by residues on the F and I helices, which define a mostly planar platform on either side of the cavity. The I helix bends across the heme, resulting in near coplanarity of the side chain of A317, the G316-A317 peptide bond, and the D320-T321 peptide bond. On the other side of the active site, F226 on the F helix creates a parallel binding surface. This size and shape of ANF fits well in the active site due to extensive van der Waals interactions with the hydrophobic side chains of the active site.
Table 1.
Active site cavity volumes of mammalian microsomal P450s.
| Enzyme | Ligand | PDB Code | Cavity Volumea (Å3) | Reference |
|---|---|---|---|---|
| CYP1A2 | ANF | 2HI4 | 390 | 20 |
| CYP2A6 | Coumarin | 1Z10 | 230 | 16 |
| CYP2A6 | D1G | 2FDU | 270 | 15 |
| CYP2A6 | D2G | 2FDV | 265 | 15 |
| CYP2A6 | D3G | 2FDW | 256 | 15 |
| CYP2A6 | 4,4'-Dipyridyl disulfide | 2FDY | 321 | 15 |
| CYP2A6 | Methoxsalen | 1Z11 | 243 | 16 |
| CYP2A6b | Phenacetin | 3EB6 | 300 | 13 |
| CYP2A6 N297Q | 2PG5 | 251 | 14 | |
| CYP2A6 L240C N297Q | 2PG6 | 245 | 14 | |
| CYP2A6 N297Q I300V | 2PG7 | 264 | 14 | |
| CYP2A13 | Indole | 2P85 | 307 | 26 |
| CYP2B4 | 1PO5 | NAc | 12 | |
| CYP2B4 | 1-CPI | 2Q6N | 421 | 9 |
| CYP2B4 | 4-CPI | 1SUO | 253 | 8 |
| CYP2B4 | 1-PBI | 3G5N | 391d | 10 |
| CYP2B4 | Bifonazole | 2BDM | 457d | 11 |
| CYP2B6 | 4-CPI | 3IBD | 582 | 37 |
| CYP2C5 | 1DT6 | 532d | 40 | |
| CYP2C5 | Diclofenac | 1NR6 | 406 | 41 |
| CYP2C5 | DMZ | 1N6B | 654 | 43 |
| CYP2C8 | Felodipine | 2NNJ | 1250 | 50 |
| CYP2C8 | Montelukast | 2NNI | 1403d | 50 |
| CYP2C8 | Palmitic Acide | 1PQ2 | 1580 | 51 |
| CYP2C8 | Retinoic Acid | 2NNH | 1290d | 50 |
| CYP2C8 | Troglitazone | 2VN0 | 951 | 50 |
| CYP2C9 | 1OG2 | 1135 | 55 | |
| CYP2C9 | Flurbiprofen | 1R9O | 978 | 54 |
| CYP2C9 | Warfarin | 1OG5 | 1271d | 55 |
| CYP2D6 | 2F9Q | 510 | 58 | |
| CYP2E1 | Indazole | 3E6I | 191 | 68 |
| CYP2E1 | 4-Methylpyrazole | 3E4E | 189 | 68 |
| CYP3A4 | 1W0E | 1173d,f | 78 | |
| CYP3A4 | 1TQN | 1226d,f | 79 | |
| CYP3A4 | Erythromycin | 2J0D | 2682d | 80 |
| CYP3A4 | Ketoconazole | 2V0M | 2017d | 80 |
| CYP3A4 | Metyrapone | 1W0G | 1332d | 78 |
| CYP3A4 | Progesteronee | 1W0F | 1193d,f | 78 |
Probe occupied cavities were calculated using a 1.4 Å probe radius in Voidoo [131].
Mutant with CYP2A13-like behavior.
Formation of dimer fills active site.
Water molecules were added to prevent cavity from extending into solvent.
Peripheral binding site.
Water molecules found inside cavity were removed for cavity volume calculation.
2.1.c. Conclusions and unusual features
A kink in the F helix occurs where the canonical hydrogen-bonding pattern of an α-helix is disrupted at V220 and K221. The space created is filled by two water molecules. T223 plays a crucial role in stabilizing this kink by interacting with one of these two water molecules, whereas residue 223 is Asn in CYP1A1 and CYP1B1. The overlaps in family 1 substrate specificities indicate that the active sites of CYP1A1 and CYP1B1 are not likely to be substantially different from CYP1A2. However, the lack of the F helix kink in other family 1 enzymes could result in less compact active sites than CYP1A2.
2.2. CYP2A6
2.2.a. Substrate specificity
CYP2A6 is the primary enzyme responsible for nicotine detoxification [22] and also activates numerous tobacco procarcinogens [16]. Inhibition of the enzyme by compounds such as methoxsalen may be useful in smoking cessation by regulating the plasma half-life of nicotine. CYP2A6 does not play an extensive role in drug metabolism, but is known oxidize fadrozole and losigamone [23] and activates tegafur [24]. The L240C and I300V mutants, which occur at the junction of the F, G, and I helices, display broader capacities to oxidize some substituted indoles, indicating that flexibility of the active site roof affects substrate specificity [14].
2.2.b. Overall structure and active site
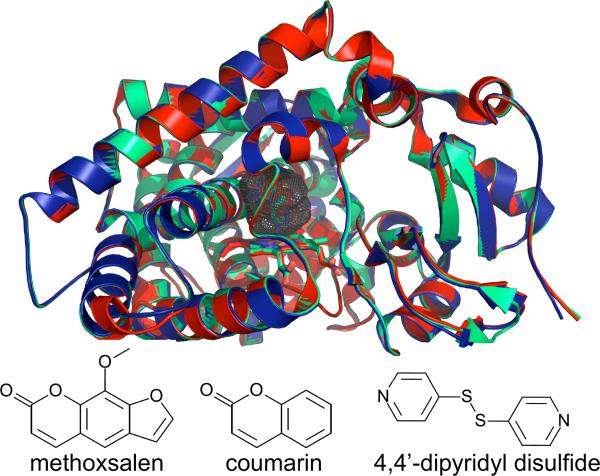
Despite the variety of substrates crystallized with CYP2A6, Cα traces of each structure are remarkably similar [13–16] (Fig. 2). The enzyme adopts a closed conformation with a compact active site that is lined by hydrophobic residues except for N297 and T305. With values ranging from 230–321 Å3, the cavity is less than 25% the size of CYP2C8, CYP2C9, and CYP3A4 (Table 1) due to the relatively low pitch of the F and G helices over the I helix. However, a ~20% increase in active site volume is observed in a CYP2A6 tetra-mutant (with CYP2A13-like phenacetin O-deethylase activity) without secondary structural rearrangement [13]. The shape of the active site favors small, nearly planar substrates that include a hydrogen-bonding partner for N297. The N297Q mutant is less able to bind/orient substrates, which could explain the lack of density for ligands in N297Q crystallization attempts [14]. The positions of ligand contact residues do not deviate among structures except for the 4,4'-dipyridyl disulfide and phenacetin complexes, where F209 swings out of the active site to accommodate the larger ligands [13, 15].
Figure. 2.
A Cα overlay of the methoxsalen (red, PDB ID 1Z11), coumarin (light green, PDB ID 1Z10), and 4, 4'-dipyridyl disulfide (dark blue, PDB ID 2FDY) complexes of CYP2A6. The enzyme requires very little structural rearrangement to bind these ligands. Gray mesh shows the active site cavity of the CYP2A6–coumarin complex and is representative of the cavity of other CYP2A6 structures. Stick diagrams (bottom) show the chemical structures of the ligands bound to each enzyme complex.
2.2.c. Conclusions and unusual features
Subtle deformation of the I helix in complexes with amine derivatives and 4,4'-dipyridyl disulfide [15] appears to be the only global difference observed when CYP2A6 conforms to ligands present during crystallization. Whether this is the only adaptable feature of the enzyme is unknown. However, the four mutations that push CYP2A6 to behave like CYP2A13 highlight how small changes in the active site can affect substrate binding and catalysis.
2.3. CYP2A13
2.3.a. Substrate specificity
CYP2A13 is primarily expressed in the respiratory tract and activates the most abundant nicotine-derived procarcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), into DNA-altering compounds [25] that can lead to lung cancer. CYP2A6 is also expressed in the lungs to a lesser extent, but has a much higher Km for NNK than CYP2A13 [26]. Reduction in lung cancer risk has been linked to a genetic polymorphism leading to a 2- to 3-fold reduction in catalytic efficiency of NNK activation [27, 28].
2.3.b. Overall structure and active site
The polypeptide backbone of CYP2A13 closely resembles that of CYP2A6, with an RMSD of 0.5 Å. The active site is small and highly hydrophobic with a cluster of six phenylalanines on the roof. All are found in CYP2A6 except for a F300I substitution. The cavity is planar and, with a volume of 307 Å3, is approximately the same size as CYP2A6 (Table 1). These two P450s have two of the smallest active site cavities that have been reported in X-ray crystal structures, with only CYP2E1 being smaller. As in CYP2A6, N297 appears to enable hydrogen bonding to ligands such as indole in the only structure available for CYP2A13 [26].
2.3.c. Conclusions and unusual features
Although indole was not added to crystallization setups, electron density was observed in each of the six monomers in the asymmetric unit, and the identity of the ligand present was determined by independent experiments [26]. The density in the active site of CYP2A13 allows for the modeling of two similar, but distinct conformations of indole. Both ligand binding modes place the phenyl ring closest to the heme with the flat plane of the molecule rotated approximately 28° relative to the length of the I helix. The difference in orientation occurs from rotation of the pyrrole ring toward the C helix. In one conformation, indole adopts a binding mode similar to that of coumarin in CYP2A6 [16]. The alternative orientation puts the pyrrole nitrogen in position to contact N297 through a water-mediated hydrogen bond. Along with their small, planar active site cavities, an Asn at residue 297 could be the defining characteristic of CYP2A enzymes and may control ligand orientation in this subfamily. Whether CYP2A13 shows as little conformational heterogeneity as CYP2A6 in the presence of other ligands is unknown, but is likely in light of the sequence identity of over 90%.
2.4. CYP2B4
2.4.a. Substrate specificity
Rabbit CYP2B4 is one of the most highly studied mammalian microsomal P450s and serves as a model for biochemical and biophysical P450 experiments. Substrates include 7-ethoxycoumarin, androstenedione [29], benzphetamine, cyclohexane, and 1-phenylethanol [30], and numerous imidazole compounds are potent inhibitors [31]. Despite the wealth of mechanistic data gathered on the enzyme, relatively little is known about products CYP2B forms with clinically used drugs.
2.4.b. Overall structure and active site
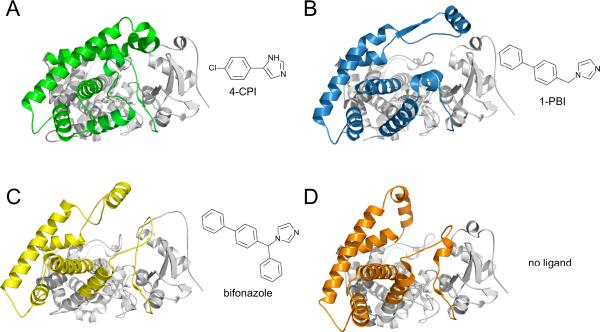
With seven entries deposited in the PDB, CYP2B4 is surpassed only by CYP2A6 in the number of structures determined. The CYP2B4 tertiary structure is the most plastic of any P450 studied by X-ray crystallography. Binding of the smaller molecules 1-(4-chlorophenyl)imidazole (1-CPI) [9], 4-(4-chlorophenyl)imidazole (4-CPI) [8], ticlopidine (PDB ID 3KW4), or clopidogrel (PDB ID 3ME6) results in a closed conformation not unlike that seen in other family 2 enzymes. Small changes in the positions of active site residues allow the enzyme to accommodate these different ligands. The flexibility of CYP2B4 is most evident in the absence of ligand [12] and in the complexes with the larger imidazole compounds 1-(phenylbenzyl)imidazole (1-PBI) [10] or bifonazole [11] (Fig. 3). These three structures all form interdigitated dimers that place portions of the F' helix in the active site of its partner. This causes the F-G region to stretch far above the plane of the heme and rearranges the B' helix and B/C loop. These changes cause significant movements of residues that line the active site of one structure but are shifted far away in another.
Figure 3.
Ribbon diagrams of the four markedly different conformations observed in CYP2B4 X-ray crystal structures. CYP2B4 forms a closed, compact structure when bound to the small inhibitor (A) 4-CPI (PDB ID 1SUO). The B', C, F, F', G', and H helices and the N-terminus of the I helix move to accomodate the larger inhibitors (B) 1-PBI (PDB ID 3G5N) and (C) bifonazole (PDB ID 2BDM) and (D) in the absence of ligand (PDB ID 1PO5). Stick diagrams show the chemical structures of inhibitors.
2.4.c. Conclusions and unusual features
The identification of a cluster of ten phenylalanine residues near the juncture of the F, G, and I helices [10] provides a possible explanation for how CYP2B4 achieves its remarkable flexibility. Eight of these residues are completely or partially conserved in other family 2 enzymes. The large, hydrophobic side chain of phenylalanine allows it to fill space and pack against other residues, as the plastic regions of the enzyme adopt their different conformations. The movement of these residues allows the F-G region of CYP2B4 to pivot over the I helix in direct response to the size of the ligand bound in the active site.
While this aromatic cluster allows for better packing in the interior of the enzyme, the movement of the F-G region also exposes other hydrophobic regions. The thermodynamic penalty incurred is compensated for by the packing of protein molecules in the crystals. However, it is likely that inserting portions of P450s into the membrane in vivo similarly compensates for the energetically unfavorable process of exposing the F-G region, portions of the N-terminus, and the B/C loop [11, 32]. It has yet to be determined what role the membrane plays in influencing the plasticity of CYP2B4 and other P450s.
2.5. CYP2B6
2.5.a. Substrate specificity
Despite its relatively low expression levels in the liver, CYP2B6 takes part in the metabolism of 3–12% of all drugs [33]. These include propofol, bupropion, clopidogrel, and sertraline [34]. The enzyme is also highly polymorphic, with 28 known alleles [34]. These genetic variations may lead to altered expression levels and drug metabolism. These changes can have serious consequences during treatment with drugs like efavirenz, which has a narrow therapeutic index and is primarily cleared by CYP2B6 [35]. Cyclophosphamide activation by CYP2B6 can also be compromised due to reduced enzyme activity [36, 37].
2.5.b. Overall structure and active site
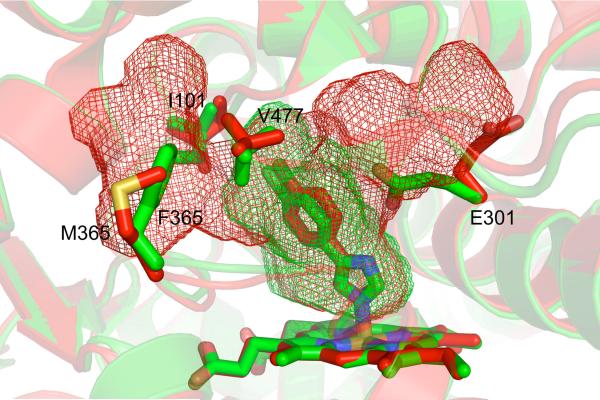
The closed, compact structure of CYP2B6 bound to the CYP2B inhibitor 4-CPI is very similar to the 2B4–4-CPI complex [38]. With an RMSD of 0.65 Å, these two structures are more related than the 1-CPI and 4-CPI complexes of CYP2B4 (0.76 Å RMSD), indicating that the same ligand may affect the conformation of a complex more than the primary sequence of the enzyme. The CYP2B6 active site closely matches that of CYP2B4 as well, with the only difference at residue 363, which is Leu in CYP2B6 and Ile in CYP2B4 (and Val in CYP2B1). In the case of 4-CPI, the identity of residue 363 does not affect ligand orientation. However, studies have shown that mutating this residue in CYP2B6 affects oxidation of 7-butoxycoumarin, pnitrophenol, and 7-ethoxy-4-trifluorocoumarin [39, 40], indicating variability at this location could be key to explaining differences in metabolism and product profiles among CYP2B enzymes, including CYP2B6. Despite their similarities, the 4-CPI complexes of CYP2B6 and CYP2B4 differ significantly in their active site cavity volumes, which are 582 Å3 and 253 Å3 respectively (Table 1). In one direction, the CYP2B6 active site cavity extends upward toward the region between the F and I helices (Fig. 4). This extension is enabled by the rotation of E301 out of the active site. In the other direction, the cavity reaches toward the A helix and β-sheet 1. Here, small rotations of I101 and V477 allow the inclusion of the extra volume. It is interesting to note in light of the observed global plasticity of CYP2B4, that the size and shape of CYP2B active sites can also change dramatically due to movement of a small number of side chains.
Figure 4.
A comparison of the active site cavities of the 4-CPI complexes of CYP2B6 (red, PDB ID 3IBD) and 2B4 (green, PDB ID 1SUO). Despite their structural similarities (RMSD 0.65 Å), the two enzymes differ with respect to their active site cavity volumes (red and green mesh). The larger cavity in CYP2B6 is created by the movement of E301 out of the active site, the rotations of I101 and V477, and the the change from phenylalanine to methionine at position 365.
2.5.c. Conclusions and unusual features
The CYP2B6 structure contains the naturally occurring K262R mutation, which increases solubility for purification and crystallization. R262 is found to take part in a hydrogen-bonding network that also includes H252, T255, D263, and D266. Despite mobility of this region (G and H helices and their associated loop) in multiple CYP2B4 structures, these hydrogen bonds remain the same (and also in CYP2B6). In an alignment of 10 CYP2B enzymes (not shown), K262 in CYP2B6 was the only residue not conserved among these hydrogen-bonding partners. Disruption of this network by a Lys at position 262 could affect the G and I helices and alter the active site. This may explain how polymorphic mutations in P450s occurring far from the bound substrate affect ligand binding and catalysis.
2.6. CYP2C5
2.6.a. Substrate specificity
Rabbit CYP2C5, which belongs to the largest P450 subfamily (CYP2C), was the first mammalian microsomal P450 structure to be determined by X-ray crystallography [32]. Relatively few substrates for the enzyme are known, but CYP2C5 does catalyze the oxidation of the NSAID diclofenac [41]. Additionally, several sulfaphenazole derivatives are potent inhibitors [42], including 4-methyl-N-methyl-N-(2-phenyl-2H-pyrazol-3-yl)benzenesulfonamide (DMZ), which has been captured in a crystal structure [43]. Although there is no evidence that the enzyme is as plastic as CYP2B4, small shifts in the B/C loop and F and G helices and side chain movements in the CYP2C5 active site gave some of the earliest structural evidence of an induced fit model for P450 ligand binding [41, 43].
2.6.b. Overall structure and active site
All CYP2C5 structures align well to each other. The largest differences among the three are observed in the B', F', and G' helices. The B' helix is not present in the ligand free structure, but rather forms a loop [32]. No details are available for the F' and G' helices in this structure due to disorder in the electron density maps. Upon binding of either DMZ [43] or diclofenac [41], these regions adopt regular secondary structures. The placement of the B' helix in either ligand bound structure varies slightly, where the N-terminus of the helix points farther away from the heme. This flexibility is enabled by G×G motifs found at either end of the B' helix. These G×G motifs that flank the B' helix are highly conserved in family 2 enzymes. With only a single hydrogen bond (V106 main chain and K241 Nζ) holding the B' helix to the main body of the protein, this region could serve as a gateway to the active site for substrates.
2.6.c. Conclusions and unusual features
In the diclofenac complex, the position of the drug was clearly defined in electron density maps [41]. However, in another structure, fitting DMZ was more challenging. Fo-Fc maps did not result in a definitive ligand orientation. Using computer docking, two distinct, anti-parallel conformations that fit the electron density maps were determined and modeled with 50% occupancy each [43]. The CYP2C5–DMZ complex highlights how some P450 reactions result in multiple products. Based on these results, CYP2C5 is able to bind the same compound in two different conformations without changing the overall structure of the enzyme, but shifts in plastic regions also allow the enzyme to bind a separate compound with different geometry and chemical properties.
2.7. CYP2C8
2.7.a. Substrate specificity
Among CYP2C8 substrates are taxol, retinoic acid, arachidonic acid, troglitazone, and amiodarone [44]. Inhibition of CYP2C8 by gemfibrozil causes a significant, harmful drug-drug interaction with cerivastatin, causing rhabdomyolysis [45–47]. The enzyme is also inhibited by montelukast, felodipine, and tamoxifen [48]. Many CYP2C8 substrates are large, organic anions at physiological pH, which are oxidized approximately 13 Å away from the anionic group [49].
2.7.b. Overall structure and active site
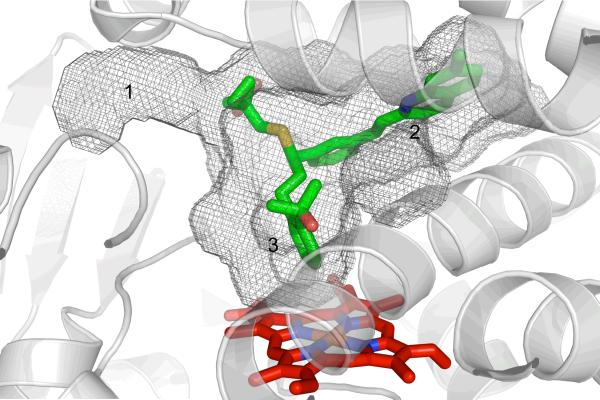
Despite being complexed with ligands of varying sizes and shapes, five crystal structures all display the enzyme in the same closed conformation [50, 51]. The active site, which is approximately two to three-fold larger than in CYP2C5 (Table 1), is consistent with the large substrates of CYP2C8. Part of this difference is caused by replacement of large residues in CYP2C5 with smaller residues in CYP2C8. The active site of CYP2C8 is T- or Y-shaped (Fig. 5), with branches of varying widths, lengths, and chemical properties. The bottom branch (#3 in Fig. 5) provides access to the heme, while branches 1 and 2 terminate in solvent access channels on either side of the B' helix. The cavity is largest at the junction of the three branches. A crystal structure with montelukast showed that the substrate occupies all three branches, whereas other compounds occupied only one or two [50]. For example, troglitazone occupies only the top portion of the active site. In the case of retinoic acid, two molecules fill the active site, where one is stacked upon the other.
Figure 5.
The T- or Y-shaped active site cavity (gray mesh) of the CYP2C8–montelukast complex (PDB ID 2NNI) is shown with three branches of varying lengths. The cavity is able to accommodate CYP2C8's various substrates by allowing them to bind in one or more of these branches. Montelukast (green sticks) fully occupies branches 2 and 3 and a portion of branch 1.
2.7.c. Conclusions and unusual features
The various binding modes of CYP2C8 substrates illustrate an important feature of P450-ligand interactions. In some cases, these compounds reside far away from the heme in positions that are not amenable for metabolism. This could indicate that binding of a second molecule (as in the retinoic acid complex) correctly orients the substrate for oxidation. Alternatively, perhaps binding of a redox partner causes a conformational change that brings the substrate closer to the heme.
The enzyme has also been shown to co-purify with E. coli fatty acids [51]. Two palmitic acid molecules, which were not added during crystallization, were found to stabilize a dimeric form of the enzyme by mediating contacts between the F-G regions at the interface. The binding of these fatty acids at a peripheral site could influence the shape of the active site through effects on the conformation and flexibility of the F-G region. Whether this is a process that occurs in vivo is not known, but given the role P450s play in fatty acid metabolism, it is plausible that these compounds could modulate P450 function in a similar manner as effectors of CYP3A4.
2.8. CYP2C9
2.8.a. Substrate specificity
Reports indicate that CYP2C9 prefers small, hydrophobic substrates with an acidic group, which include ibuprofen and other NSAIDs [7]. The enzyme may also be involved in the synthesis of arachidonic acid epoxides, which regulate blood pressure [52]. Polymorphism in CYP2C9, which affects metabolic activity, substrate binding, and enzymatic efficiency [53], can lead to adverse drug-drug interactions and toxicity. These implications are particularly important during treatment with CYP2C9 substrates with narrow, therapeutic windows, such as warfarin and phenytoin [7]. Structural studies of CYP2C9 may also yield insight into CYP2C19 function, given their 91% sequence identity. However, despite only one residue difference in their active sites, the enzymes display distinct substrate selectivities [54, 55].
2.8.b. Overall structure and active site
Distinct differences were found in the flurbiprofen complex [54] compared with the ligand free and warfarin bound enzyme [55]. While the latter two are nearly indistinguishable from one another (0.25 Å RMSD), the flurbiprofen complex contains an additional turn on the A helix, which displaces β-sheet 4 from its position in the other two structures. Additionally, the B' helix is unwound, and those residues form an extended loop. Furthermore, residues S209 to T229 form loops instead of the F' and G' helices, although portions of this region are not modeled due to disorder in electron density. Despite the implication that the CYP2C9 active site would include basic residues, none were found in the warfarin or ligand free complexes. However, based on the conformation of the B/C loop in the flurbiprofen complex, R108 enters into the active site.
2.8.c. Conclusions and unusual features
Warfarin was bound to CYP2C9 in the active site cavity far from the heme. This distal site could possibly be a `loading' site for molecules awaiting metabolism. The authors propose that an electron-transfer driven conformational change, perhaps due to reduction of the heme or through interaction with NADPH-cytochrome P450 reductase (CPR), would move the molecule closer for catalysis. It is also possible that this is a secondary binding site, while the area above the heme is the primary binding site, allowing for an allosteric or cooperative effect. This could explain the unusually large active site for an enzyme with a preference for smaller substrates. Further bolstering this possibility, dapsone was successfully docked in the active site alongside flurbiprofen [54]. The warfarin binding region is not part of the active site cavities of the ligand free structure or the flurbiprofen complex, but could easily be included by the rotation of F100 and F476 in a manner similar to how CYP2B6 increases its active site volume compared with CYP2B4.
2.9. CYP2D6
2.9.a. Substrate specificity
CYP2D6 takes part in the metabolism of at least 20% of all drugs [56]. Substrates generally contain a basic nitrogen atom and a planar aromatic ring [57]. Many of these features are found in drugs affecting the central nervous and cardiovascular systems. Several illicit drugs, including cocaine, hydrocodone, and amphetamine, are substrates or inhibitors of CYP2D6, and polymorphisms could play a role in determining substance abuse risk [57].
2.9.b. Overall structure and active site
The single ligand free structure of CYP2D6 displays a closed conformation [58], but the F-G region adopts a different orientation than other closed P450s. Instead of angling toward the A helix, the F helix is nearly perpendicular to the I helix. The F helix is also longer than in other P450s and kinks downward at K214 and E215, which compresses the roof of the active site. Furthermore, the N-terminus of the F helix displaces portions of the β-sheet domain, which shortens β-strands 1-1 and 1-2 and lengthens the loop between them. The F' helix is replaced by an extended loop region that terminates at the G' helix just above the B' helix. The G helix is also significantly shorter than in other P450s. The 510-Å3 active site of CYP2D6 (Table 1) is described as `foot-shaped' with a solvent access channel between the F helix and β-sheet 4. Two negatively charged active site residues, E216 and D301, are suggested to be substrate-binding partners. These two acidic residues presumably interact with basic groups on CYP2D6 substrates. Additionally, numerous aromatic residues in the active site, F120, F481, and F483, are available to control substrate orientation with respect to the heme.
2.9.c. Conclusions and unusual features
Given the unusual compacted conformation of the F-G region, the large active site volume of CYP2D6 is surprising. However, a majority of the active site cavity is found projecting in a V-shape toward either side of the F helix. Based on the wide variation of CYP2D6 substrates, alterations in the conformation of the active site roof could help accommodate different sized molecules. Modifications to the lengths and positions of the F, F', G, and G' helices are not uncommon in CYP2B4, CYP2C5, CYP2C9, and CYP3A4 and could be used by CYP2D6 to adopt a conformation more consistent with other P450s.
An alternative explanation for the irregular fold of the F-G region is an effect of internal mutations in the protein construct used during crystallization. Typically, microsomal P450s are modified for crystallization by removing the N-terminal transmembrane helix, mutating N-terminal residues to contain basic side chains, and adding a C-terminal histidine tag. The CYP2D6 enzyme that was crystallized contained additional internal mutations (L230D and L231R). The change from two hydrophobic side chains to hydrophilic aspartate and arginine in a region that adopts an atypical conformation has raised concern. In fact, ligand docking experiments using the CYP2D6 crystal structure were less successful than those using either a homology model of CYP2D6 [59] or molecular dynamics methods [60].
2.10. CYP2E1
2.10.a. Substrate specificity
CYP2E1 comprises over 50% of hepatic P450 mRNA [61] and 7% of hepatic P450 protein content [62]. The enzyme is also expressed in other tissues but at lower levels [63]. Substrates are generally small, neutral molecules [5], such as benzene, ethanol, and butadiene [7]. However, CYP2E1 is also known to metabolize endogenous compounds such as arachidonic acid [64] and epoxyeicosatrienoic acids [65]. Generation of toxic or carcinogenic products from such commonly used drugs as acetaminophen can occur often [66] and is influenced by factors, such as alcohol consumption [67].
2.10.b. Overall structure and active site
CYP2E1 has been crystallized in the presence of two small inhibitors, indazole or 4-methylpyrazole [68]. The enzyme binds both of these molecules with almost no structural rearrangement, similar to CYP2A6. Structurally, CYP2E1 is also similar to CYP2A6 and CYP2A13. Cα overlays with either CYP2A enzyme yield an RSMD of ~0.75 Å. Despite its structural similarities to these two enzymes, the active site cavity of CYP2E1 is smaller at 190 Å3 (Table 1). Although it does not interact with either indazole or 4-methylpyrazole, D295 is at an analogous position as N297 in CYP2A6 and CYP2A13. CYP2E1 may utilize this residue to orient ligands containing a polar group in the active site.
2.10.c. Conclusions and unusual features
A second 77-Å3 cavity that is unique to CYP2E1 was found adjacent to the active site and located between the B', F, G, and I helices. In CYP1A2 and CYP2A13, a small portion of this region is part of the active site, but it is filled by side chain atoms in CYP2A6. CYP2E1 side chains of F106 and F298 separate this cavity from the active site. It is possible that through movement of these residues, the active site cavity volume could be increased to include this region. While there is a solvent access channel on the opposite side of the B' helix where fatty acid substrates are proposed to bind, the additional flexibility provided by this secondary cavity could play a role in binding these larger substrates as well.
2.11. CYP3A4
2.11.a. Substrate specificity
CYP3A4 is the most highly expressed P450 in the liver, comprising up to 40% of hepatic P450 content [33]. The enzyme is also the most promiscuous P450 and contributes to the metabolism of approximately 50% of all pharmaceuticals on the market [33], including cyclosporin, bromocriptine, macrolide antibiotics [69], and statins [70]. Additionally, CYP3A4 is also the primary enzyme involved in food-drug interactions [71]. The enzyme provides striking examples of non-Michaelis-Menten kinetics, including homotropic cooperativity [72, 73]. Furthermore, studies suggest that effectors may act at a noncatalytic site within the binding pocket to modulate substrate oxidation [74, 75]. CYP3A4 is known to bind and metabolize multiple substrates simultaneously [76, 77].
2.11.b. Overall structure and active site
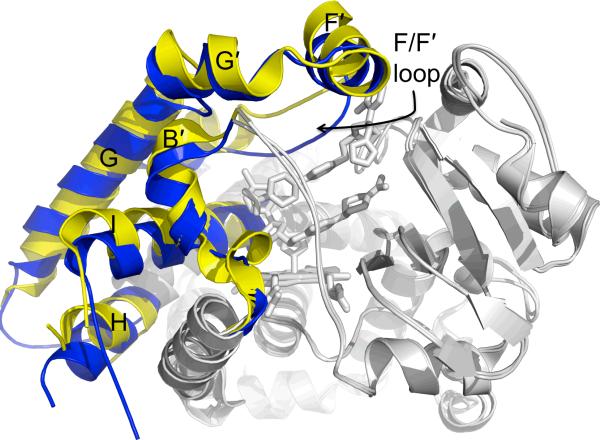
Despite its chemically diverse substrates, two ligand free structures and complexes of CYP3A4 with metyrapone or progesterone show little difference in the placement of secondary structural elements [78, 79]. The active site cavity volumes of these structures range from 1173 Å3 to 1332 Å3 (Table 1). However, binding of the larger ligands ketoconazole or erythromycin [80] increases the cavity volume to over 2000 Å3. Variation in the active site of CYP3A4 in the presence of these larger ligands is generated through plasticity in helices B', F, F', G, G', H, and the N-terminus of the I helix (Fig. 6). The most noticeable changes occur where an extended F/F' loop, stretching from K209 to D217, sags downward into the top of the active site in both ligand free structures and the progesterone and metyrapone complexes. In the erythromycin and ketoconazole complexes, the F helix is lengthened to F213, and portions of the ligands occupy space where the extended F/F' loop is observed in the other structures [80]. This loop is disordered in the erythromycin complex, but the ketoconazole complex shows it pushed upward, away from the heme. A majority of the additional cavity volume seen in these two complexes is created by movement of this loop. Additionally, numerous phenylalanine residues (F213, F215, F219, and F220) are found along this loop. Along with F108, F241, and F304, these residues comprise a unique cluster of aromatic side chains near the top of the CYP3A4 active site. Modifications to the length of the F helix and positions of residues on loops at the roof of the active site most likely allow CYP3A4 to adapt the size of its binding pocket to accommodate its long list of substrates.
Figure 6.
Cα overlay of ligand free CYP3A4 (blue, PDB ID 1TQN) and the CYP3A4–ketoconazole complex (yellow, PDB ID 2V0M). This ligand free structure is similar to a ligand free structure (PDB ID 1W0E) from another study and the CYP3A4 complexes of progesterone (PDB ID 1W0F) and metyrapone (PDB ID 1W0G). In this overlay, static portions of the enzyme (gray) do not change, but the B', F, F', G', and H helices and the N-terminus of the I helix alter their structures in response to the binding of two ketoconazole molecules. In particular, the F/F' loop changes position by ~5.5 Å.
2.11.c. Conclusions and unusual features
Binding of metyrapone to CYP3A4 fills only 230 Å3 [78] of the 1332 Å3 cavity (Table 1). This leaves a significant volume for the binding of an effector molecule, multiple metyrapone molecules, or even other substrates. Despite its larger volume, erythromycin also leaves additional room in the active site for the binding of other ligands [80]. Considering that erythromycin binds in a conformation that is not consistent with metabolism, the binding of an effector molecule could help orient the substrate properly. Further evidence for this model is provided by the binding of two ketoconazole molecules in the CYP3A4 active site [80]. However the biological relevance of this is unknown, since the stoichiometry of ketoconazole binding to CYP3A4 has not been reported.
3. NMR of drugs and cytochromes P450
NMR has lagged behind X-ray crystallography in providing molecular detail of P450-ligand interactions. However, whereas X-ray crystallography yields snapshots of the P450-ligand complex, NMR has the ability to analyze dynamic behavior of proteins and ligands under a wider range of conditions. Most P450 NMR studies have probed the orientation of ligands in the active sites [81] by using the Solomon-Bloembergen equation [82] to correlate the NMR relaxation of ligand protons to distances from the heme iron. This method has provided insight into the structural mechanism of metabolism, drug-drug interactions, and the influence of protein cofactors on drug metabolism. Although progress in directly examining P450 structure by NMR has been hindered by the inherent insolubility and instability of the proteins, recent NMR studies using solid-state NMR [83] and isotopically-labeled probes [84] show promise in overcoming these obstacles.
3.1. Regioselectivity and substrate specificity
Heme-induced relaxation of drug protons has been used to study the structural basis of regioselectivity in P450s. In many cases, the calculated distances place the sites of metabolism close to the heme iron [e.g. 85], which is consistent with experimental results. However, some studies show that orientation [81] and protein cofactors [86] play a significant role in dictating the sites of oxidation by these enzymes.
3.1.a. CYP1A1, CYP2B1, and CYP2B2
Phenobarbital-induced CYP2B1 and CYP2B2 are the major CYP2B enzymes from rat and metabolize a wide range of compounds including coumarins [87], steroids [88], and amphetamines [89]. The β-naphthoflavone induced CYP1A1 is associated with metabolism of polycyclic hydrocarbons such as benzo(a)pyrene [90] but metabolizes many of the same compounds as CYP2B1 and CYP2B2. The enzymes share < 40% sequence identity and often produce different products from the same substrate.
The analgesic acetaminophen (APAP) is metabolized by these P450s into the toxic metabolite n-acetyl-p-benzoquinone (NAPQI), which conjugates to proteins, or the non-toxic catechol, 3-hydroxyacetominophen. CYP1A1 shows a higher rate of APAP metabolism and NAPQI formation than CYP2B1 [91, 92]. The orientations of APAP determined in two NMR studies with unlabeled and isotopically-labeled APAP (i.e. 15N, or 13C) showed that APAP adopted different orientations in the active sites of CYP1A1 and CYP2B1 [91, 92]. In particular, APAP adopted parallel and perpendicular orientations to the heme in CYP1A1 and CYP2B1, respectively. The parallel orientation places the n-acetyl group of APAP closer to the heme, where it is readily oxidized to NAPQI.
Another substrate, aniline, is metabolized by CYP2B1 and CYP1A1 via two competing reactions: N-hydroxylation and p-hydroxylation [93–95]. An NMR study was performed with structural isomers of methylfluoroaniline and these P450s to study regioselectivity and the mechanism of aniline metabolism [96]. The calculated distances from the NMR relaxation were within ~1 Å for all the structural isomers, leading the authors to hypothesize that the metabolism was not strongly influenced by the position of functional groups in the structures [96]. However, for aniline, which is N- and p-hydroxylated at opposite ends of the molecule, the molecule likely adopts multiple binding orientations within the P450 active site. The lack of distance dispersion may result from averaging between different binding modes, and the small difference in the observed distances may reflect a preference toward one of the orientations.
3.1.b. CYP1A2
The major product of caffeine metabolism by CYP1A2 is paraxanthine, which is formed by demethylation at position 3 of the drug [97, 98]. An NMR study of the enzyme with caffeine suggested that the molecule was relatively parallel to the heme with distances that only differed by 0.2 Å and the N-3 methyl slightly closer to the heme [99]. On the assumption that the distance from the heme iron dictates the rate of metabolism, the metabolites should be formed at the same rate in this parallel orientation, which contradicts the observation that metabolism primarily occurs at the 3 position [99, 100]. Because caffeine is metabolized into multiple products, the molecule might have multiple binding modes within the CYP1A2 active site as suggested by our analysis of methylfluoroaniline and CYP1A1. One possible explanation is that caffeine has two binding modes. In one orientation where the molecule spends a majority of time, caffeine is perpendicular to the heme with the methyl at position 3 close to the heme. The molecule then binds in another orientation, placing the methyl groups at the 1 and 7 positions close to the heme and the methyl at position 3 farther away. The averaging of the distances between these two binding modes would result in a seemingly parallel orientation. This model helps explain how 80% of caffeine metabolism yields paraxanthine along with minor products [99].
3.1.c. CYP2C9
The regioselectivity of substrate oxidation by CYP2C9 has been probed by NMR using three ligands [101]: tienilic acid, lauric acid, and diclofenac. During this study, three forms of tienilic acid were studied: tienilic acid (TA), a tienilic acid isomer (TAI), where the sulfur of the of the thiophene ring is shifted by one bond from the position in TA, and the tienilic acid phenol (TAP), which is a phenol formed when TA loses an acetate group. TA is either hydroxylated or forms an inactive conjugate with the P450 at position-5 of the thiophene ring [102]. In contrast, TAI is metabolized to form thiophene sulfoxide [102]. The orientations of TA and TAI, as determined by NMR, were similar. The thiophene and the phenyl functional groups face toward and away from the heme iron, respectively, which is consistent with metabolism occurring on the thiophene ring [101, 102]. In contrast, the NMR distances showed TAP in a different orientation than TA and TAI [101], suggesting that the acetic acid functional group is important for positioning TA in the CYP2C9 active site.
Another CYP2C9 substrate, lauric acid, which is metabolized primarily via ω-1 hydroxylation [103], was also examined during this study. Surprisingly, the calculated distances of protons in the middle section of lauric acid were closer on average to the heme than those of either the carboxylic acid or ω-1 methyl ends [101]. Because metabolism occurs at the ω-1 methyl position, this group might be expected to be closest to the heme. The chemically equivalent protons of the ω-1 methyl group were treated as if their distances from the heme were equal. However, by using newly developed methods that do not make this assumption [81, 104], the ω-1 proton does become the closest proton to the heme with a distance of 5.5 Å calculated from a relaxation time of 175 μsec (i.e. 524 μsec/3 methyl protons = 175 μsec). Therefore, chemical equivalence of protons should be carefully considered when determining ligand orientation based on NMR calculated distances.
The third substrate from this study, diclofenac, is hydroxylated at the 4'-postion by CYP2C9 [105]. The NMR relaxation of diclofenac in the presence of CYP2C9 showed that the phenyl acetic group was closer to the heme than the dichlorophenyl group. In this case, the orientation of the molecule seemed to play a stronger role in dictating the site of metabolism, since the 3' and 5' protons were closer to the heme than the proton at the 4' position [105].
Analysis of these three drugs allowed identification of some of the structural characteristics that may be important for CYP2C9 substrate selectivity [101]. Drugs possessing an anionic group and a hydrophobic region that lies between the hydroxylation site and the anionic group were proposed as good CYP2C9 substrates [101]. However, this model may not extend to other CYP2C9 substrates, since CYP2C9 can metabolize compounds lacking an anionic group, such as PAHs [106].
3.1.d. CYP2D6
The opiate codeine and the anti-nausea drug ondansetron are metabolized by CYP2D6 to morphine by O-demethylation of codeine [107] and by hydroxylation at position 16 of the phenyl ring of ondansetron [108]. NMR relaxation experiments with these drugs in the presence of CYP2D6 showed that the drugs were oriented with the sites of metabolism closest to the heme [85, 109].
The effect of the F120A mutation on drug orientation was investigated with the psychoactive drug 3,4-methylene dioxy-N-methylamphetamine (MDMA, a.k.a `ecstasy') [110]. The mutant N-hydroxylates MDMA to 3,4-methylenedioxy-N-hydroxy-N-alkylamphetamine in addition to forming the same product as wild type CYP2D6, dihydroxy-N-alklyamphetamine [110]. The calculated distance from NMR relaxation experiments of mutant and wild type enzyme showed that both oriented the methylene group of MDMA, which is the main site of metabolism, closer to the heme [110]. However, molecular dynamics showed that mutant CYP2D6 could orient MDMA in positions to explain either product [110].
3.2. Drug-drug interactions
Metabolism of compounds by mammalian microsomal P450s is often complex. The complexity arises from cooperativity of ligand binding, different conformational states of the enzyme, or interactions between P450s [90]. This section addressed studies that investigated homotropic and heterotropic cooperativity of ligand binding/metabolism.
3.2.a. CYP2C9
Flurbiprofen is metabolized by CYP2C9 and its allelic variants CYP2C9*2, CYP2C9*3 and CYP2C9*5 through 4'-hydroxylation of one of the phenyl rings [111]. The rate of 4'-hydroxylation of flurbiprofen is activated by the anti-leprosy drug dapsone [112]. In contrast, benzobromarone, which is used in the treatment of gout, inhibits flurbiprofen metabolism in wild type CYP2C9, but activates metabolism in CYP2C9*3 [113]. Distances calculated from NMR simply showed that dapsone shifted the proton at the 4' position closer to the heme [104, 114], while benzobromarone did not significantly affect the orientation [113]. Since both absolute distance and orientation did not seem to play a significant role in the effect of benzobromarone on flurbiprofen metabolism, the authors proposed that the kinetics of binding and release of the inhibitor and substrate were the driving force behind the activation and inhibition of flurbiprofen metabolism [113].
3.2.b. CYP3A4
The metabolism of midazolam by CYP3A4 leads to 1'-hydroxymidazolam and 4-hydroxymidazolam with very distinct kinetics [115]. Addition of testosterone decreases the ratio of 1'-hydroxymidazolam : 4-hydroxymidazolam produced, while ANF increases this ratio [116]. NMR calculated distances showed that the CYP3A4 effectors ANF and testosterone reoriented the 1' and 4 protons of midazolam into positions that were consistent with their effect on the product ratios.
In another study, caffeine was shown to activate the metabolism of APAP to NAPQI [81]. NMR relaxation of caffeine protons showed that it was positioned farther from the CYP3A4 heme than APAP [81]. Caffeine also disrupted the orientation of the APAP molecule [81]. Because there is no clear evidence from this study that the amide proton of APAP is closer to the heme, the results suggest that orientation of APAP plays a stronger role in NAPQI production than the absolute distance of the amide proton from the heme iron.
3.3. Effect of Protein Cofactors on Ligand Orientation
Catalytic turnover of P450s is complex and involves oxidation and reduction of the P450 heme iron in addition to the binding of two protein cofactors, cytochrome b5 and CPR [90]. The effect of CPR on the orientation of the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) in the active site of CYP2D6 was determined by NMR [86]. MPTP is metabolized by CYP2D6 at both ends to either 4-phenyl-1,2,3,6-tetrahydropyridine or 1-methyl-4-(4'-hydroxyphenyl)-1,2,3,6-tetrahydropyridine [86]. In the absence of the CPR, the NMR calculated distances place the pyridine ring closest to the heme [86]. In the presence of CPR, both ends of the molecules appear to be closest to heme, which would create a contorted orientation [86]. To explain both the formation of two products from a single substrate and the contorted orientation, the authors proposed that the calculated distances represent an average of two binding modes of the molecule [86].
3.4. Probing the Structure of Cytochrome P450s
Applying the NMR techniques to mammalian P450s that have been used with bacterial P450s [117–121] has been stymied by the general insolubility and instability of the mammalian enzymes. Some of these difficulties have been partially overcome through the use of unique methods. Solid state NMR has been used to probe the structure of isotopically-labeled CYP3A4 enzyme embedded in nanodiscs [83]. While solution NMR can be used to determine the entire structure of small proteins, this study was only able to identify a small number of CYP3A4 residues both in the absence and presence of ketoconazole [83]. Although the method might overcome the poor solubility of microsomal P450s, new issues arise, including control of the redox state of the enzyme in the solid state and correlating the results to those determined in solution. Another interesting method adds the compound 13C-methyl isocyanide to unlabeled CYP2B4 to probe the structure of the enzyme [84]. Using the NMR technique 1H-13C heteronuclear single quantum correlation (HSQC) spectroscopy, the authors observed a change in peak position in response to different buffer conditions that may reflect different protein conformations [84]. However, the structural information obtained by this method is limited, since only a single nucleus provides a signal.
4. Expert commentary
Despite the availability of a number of microsomal P450 X-ray crystal structures and ligand binding data generated by NMR studies, using these methods individually or in combination to design drugs with optimal metabolic properties remains a daunting task. Major impediments include the difficulty in predicting P450 structural response to ligand binding, as well as substrate binding mode. These issues may be overcome through the use of computational methods in conjunction with P450 crystal structures or NMR data. Examples include prediction of the site of metabolism for CYP2C9 and CYP2D6 substrates [110, 122], distinguishing type I and II interactions of CYP2C9 or CYP3A4 with ligands [123], differentiating competitive inhibitors and non-inhibitors of CYP2C9 [124], and improving the metabolic properties of a known CYP2C9 substrate [125]. Not surprisingly, other studies have shown that ligand orientations provided by computational methods were dependent on which P450 structure was used [126]. This is a particular problem with P450s that are known to alter their secondary structure significantly in response to ligand identity, such as CYP2B4, CYP2C5, and CYP3A4. However, when provided with known P450 metabolites of potential therapeutic agents, it is easier to make computational assessments of related compounds [126].
Future Perspective.
The rate of P450 structure determination by X-ray crystallography is increasing. Through advances in construct selection, purification, and crystallization, more P450-drug complexes should be reported in the future. These improvements were found to be useful in determining the structure of a particularly challenging P450, namely CYP2B6 [38]. With only two human drug-metabolizing P450 structures yet to be reported (CYP2C19 and CYP3A5), an emphasis in the future will likely be placed on crystallizing P450-ligand complexes that will aid in designing pharmaceutical candidates with optimal metabolic properties. Structures of P450 genetic variants may also help in the design of therapeutics that are beneficial to the widest segments of the population.
NMR techniques that have been used to study bacterial P450s are now being applied to a growing number of mammalian P450 systems [117–121]. Difficulties arising from low solubility and stability of these enzymes have been partially overcome by using solid-state NMR techniques and isotopically-labeled ligands. These techniques have been used to probe the structure of CYP3A4 embedded in nanodiscs [83] and of CYP2B4 [84]. However, neither technique is developed enough for common use. Other NMR techniques such as saturation transfer difference [127] and water ligand observed via gradient spectroscopy (WaterLOGSY) [128–130] can be used to probe the interaction of ligands within the active site of CO-reduced P450s. In the future, dramatic improvements in the interpretation of NMR data are anticipated by integrating NMR-generated restraints into computational studies, which should facilitate prediction of the sites of metabolism of new drugs. However, the combination of NMR, X-ray crystallography, and computational methods may ultimately provide a more complete picture of the molecular basis of drug metabolism.
Executive Summary.
X-ray crystal structures of P450s yield detailed descriptions of the active sites and can aid in determining the flexibility of a given P450 in response to ligand binding.
Various P450s have different means of accommodating their wide range of substrates. Rotations of active site residues and movement of the B', C, F, F', G', G, and H helices and the N-terminus of the I helix are the most common ways P450s adapt to bind ligands.
When predicting metabolism based on P450 structural information, caution must be used to determine which X-ray crystal structure is most relevant to the compound in question.
The main advantage of NMR over X-ray crystallography is the ability to analyze dynamic behavior of proteins and ligands under a wider range of conditions.
A number of NMR studies have examined the orientation of drugs in mammalian microsomal P450 active sites using the Solomon-Bloembergen equation to correlate the NMR relaxation of ligand protons to distances from the heme iron.
Probing the structure of drug metabolizing P450s directly by NMR has been stymied by the inherent insolubility and instability of the enzymes, although recent work with solid-state NMR and using isotopically labeled probes show promise.
Acknowledgments
Studies in the authors' laboratory described in this paper are supported by National Institute of Health grants ES003619 and ES006676 to James R. Halpert.
References
- 1.Kramer MA, Tracy TS. Studying cytochrome P450 kinetics in drug metabolism. Expert Opin Drug Metab Toxicol. 2008;4(5):591–603. doi: 10.1517/17425255.4.5.591. [DOI] [PubMed] [Google Scholar]
- 2.Domanski TL, Halpert JR. Analysis of mammalian cytochrome P450 structure and function by site-directed mutagenesis. Curr Drug Metab. 2001;2(2):117–137. doi: 10.2174/1389200013338612. [DOI] [PubMed] [Google Scholar]
- 3.Guengerich FP, Parikh A, Yun C-H, et al. What makes P450s work? Searches for answers with known and new P450s. Drug Metab Rev. 2000;32(3–4):267–281. doi: 10.1081/dmr-100102334. [DOI] [PubMed] [Google Scholar]
- 4.de Graaf C, Vermeulen NP, Feenstra KA. Cytochrome P450 in silico: an integrative modeling approach. J Med Chem. 2005;48(8):2725–2755. doi: 10.1021/jm040180d. [DOI] [PubMed] [Google Scholar]
- 5.Lewis DF. Human P450s involved in drug metabolism and the use of structural modelling for understanding substrate selectivity and binding affinity. Xenobiotica. 2009;39(8):625–635. doi: 10.1080/00498250903000255. [DOI] [PubMed] [Google Scholar]
- 6.Lewis DF, Ito Y, Goldfarb PS. Structural modelling of the human drug-metabolizing cytochromes P450. Curr Med Chem. 2006;13(22):2645–2652. doi: 10.2174/092986706778201567. [DOI] [PubMed] [Google Scholar]
- 7.Lewis DF, Modi S, Dickins M. Structure-activity relationship for human cytochrome P450 substrates and inhibitors. Drug Metab Rev. 2002;34(1–2):69–82. doi: 10.1081/dmr-120001391. [DOI] [PubMed] [Google Scholar]
- 8.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 A resolution: Insight into the range of P450 conformations and coordination of redox partner binding. J Biol Chem. 2004;279(26):27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Sun L, Muralidhara BK, et al. Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4. Biochemistry. 2007;46(41):11559–11567. doi: 10.1021/bi7011614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay SC, Sun L, Maekawa K, Halpert JR, Stout CD. Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: Ligand induced structural response through α-helical repositioning. Biochemistry. 2009;48(22):4762–4771. doi: 10.1021/bi9003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281(9):5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 12.Scott EE, He YA, Wester MR, et al. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci USA. 2003;100(23):13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeVore NM, Smith BD, Urban MJ, Scott EE. Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes. Drug Metab Disp. 2008;36(12):2582–2590. doi: 10.1124/dmd.108.023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sansen S, Hsu M-H, Stout CD, Johnson EF. Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants. Arch Biochem Biophys. 2007;464(1):197–206. doi: 10.1016/j.abb.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yano JK, Denton TT, Cerny MA, Zhang X, Johnson EF, Cashman JR. Synthetic inhibitors of cytochrome P-450 2A6: Inhibitory activity, difference spectra, mechanism of inhibition, and protein cocrystallization. J Med Chem. 2006;49(24):6987–7001. doi: 10.1021/jm060519r. [DOI] [PubMed] [Google Scholar]
- 16.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12(9):822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 17.Kim YD, Todoroki H, Oyama T, et al. Identification of cytochrome P450 isoforms involved in 1-hydroxylation of pyrene. Environ Res. 2004;94(3):262–266. doi: 10.1016/S0013-9351(03)00134-8. [DOI] [PubMed] [Google Scholar]
- 18.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279(23):23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 19.Kim D, Guengerich FP. Cytochrome P450 activations of arylamines and heterocyclic amines. Annu Rev Pharmacol Toxicol. 2005;45(1):27–49. doi: 10.1146/annurev.pharmtox.45.120403.100010. [DOI] [PubMed] [Google Scholar]
- 20.Sansen S, Yano JK, Reynald RL, et al. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem. 2007;282(19):14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 21.Agundez JA. Cytochrome P450 gene polymorphism and cancer. Curr Drug Metab. 2004;5(3):211–214. doi: 10.2174/1389200043335621. [DOI] [PubMed] [Google Scholar]
- 22.Cashman JR, Park SB, Yang ZC, Wrighton SA, Jacob P, III, Benowitz NL. Metabolism of nicotine by human liver microsomes: stereoselective formation of trans-nicotine N'-oxide. Chem Res Toxicol. 1992;5(5):639–646. doi: 10.1021/tx00029a008. [DOI] [PubMed] [Google Scholar]
- 23.Torchin CD, McNeilly PJ, Kapetanovic IM, Strong JM, Kupferberg HJ. Stereoselective metabolism of a new anticonvulsant drug candidate, losigamone, by human liver microsomes. Drug Metab Disp. 1996;24(9):1002–1008. [PubMed] [Google Scholar]
- 24.Ikeda K, Yoshisue K, Matsushima E, et al. Bioactivation of tegafur to 5-fluorouracil is catalyzed by cytochrome P-450 2A6 in human liver microsomes in vitro. Clin Cancer Res. 2000;6(11):4406–4415. [PubMed] [Google Scholar]
- 25.Wong HL, Zhang X, Zhang Q-Y, et al. Metabolic activation of the tobacco carcinogen 4-(methylnitrosamino)-(3-pyridyl)-1-butanone by cytochrome P450 2A13 in human fetal nasal microsomes. Chem Res Toxicol. 2005;18(6):913–918. doi: 10.1021/tx0500777. [DOI] [PubMed] [Google Scholar]
- 26.Smith BD, Sanders JL, Porubsky PR, Lushington GH, Stout CD, Scott EE. Structure of the human lung cytocrhome P450 2A13. J Biol Chem. 2007;282(23):17306–17313. doi: 10.1074/jbc.M702361200. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Tan W, Hao B, et al. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63(22):8057–8061. [PubMed] [Google Scholar]
- 28.Zhang X, Su T, Zhang QY, et al. Genetic polymorphisms of the human CYP2A13 gene: identification of single-nucleotide polymorphisms and functional characterization of an Arg257Cys variant. J Pharmacol Exp Ther. 2002;302(2):416–423. doi: 10.1124/jpet.302.2.416. [DOI] [PubMed] [Google Scholar]
- 29.He YQ, Szklarz GD, Halpert JR. Interconversion of the androstenedione hydroxylase specificities of cytochromes P450 2B4 and 2B5 upon simultaneous site-directed mutagenesis of four key substrate recognition residues. Arch Biochem Biophys. 1996;335(1):152–160. doi: 10.1006/abbi.1996.0493. [DOI] [PubMed] [Google Scholar]
- 30.Coon MJ, Vaz AD, McGinnity DF, Peng HM. Multiple activated oxygen species in P450 catalysis: Contributions To specificity in drug metabolism. Drug Metabolism and Disposition. 1998;26(12):1190–1193. [PubMed] [Google Scholar]
- 31.Muralidhara BK, Negi S, Chin CC, Braun W, Halpert JR. Conformational flexibility of mammalian cytochrome P450 2B4 in binding imidazole inhibitors with different ring chemistry and side chains. Solution thermodynamics and molecular modeling. J Biol Chem. 2006;281(12):8051–8061. doi: 10.1074/jbc.M509696200. [DOI] [PubMed] [Google Scholar]
- 32.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5(1):121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Tompkins LM. CYP2B6: New insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab. 2008;9(7):598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. Polymorphic CYP2B6: Molecular mechanisms and emerging clinical significance. Pharmacogenetics. 2007;8(7):743–759. doi: 10.2217/14622416.8.7.743. [DOI] [PubMed] [Google Scholar]
- 35.Ward BA, Gorski CJ, Jones DR, Hall SD, Flockhart DA, Desta Z. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: Implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther. 2003;306(1):287–300. doi: 10.1124/jpet.103.049601. [DOI] [PubMed] [Google Scholar]
- 36.de Jonge ME, Huitema AD, Rodenhuis S, Beijnen JH. Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet. 2005;44(11):1135–1164. doi: 10.2165/00003088-200544110-00003. [DOI] [PubMed] [Google Scholar]
- 37.Xie HJ, Yasar U, Lundgren S, et al. Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J. 2003;3(1):53–61. doi: 10.1038/sj.tpj.6500157. [DOI] [PubMed] [Google Scholar]
- 38.Gay SC, Shah MB, Talakad JC, et al. Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0 A resolution. Mol Pharmacol. 2010;77(4):529–538. doi: 10.1124/mol.109.062570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domanski TL, Schultz KM, Roussel F, Stevens JC, Halpert JR. Structure-function analysis of human cytochrome P-450 2B6 using a novel substrate, site-directed mutagenesis, and molecular modeling. J Pharmacol Exp Ther. 1999;290(3):1141–1147. [PubMed] [Google Scholar]
- 40.Spatzenegger M, Liu H, Wang Q, Debarber A, Koop DR, Halpert JR. Analysis of differential substrate selectivities of CYP2B6 and CYP2E1 by site-directed mutagenesis and molecular modeling. J Pharmacol Exp Ther. 2003;304(1):477–487. doi: 10.1124/jpet.102.043323. [DOI] [PubMed] [Google Scholar]
- 41.Wester MR, Johnson EF, Marques-Soares C, et al. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 A resolution: evidence for an induced fit model of substrate binding. Biochemistry. 2003;42(31):9335–9345. doi: 10.1021/bi034556l. [DOI] [PubMed] [Google Scholar]
- 42.Marques-Soares C, Dijols S, Macherey AC, et al. Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: identification of a new high affinity substrate common to those enzymes. Biochemistry. 2003;42(21):6363–6369. doi: 10.1021/bi027391+. [DOI] [PubMed] [Google Scholar]
- 43.Wester MR, Johnson EF, Marques-Soares C, Dansette PM, Mansuy D, Stout CD. Structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3 A resolution: Evidence for multiple substrate binding modes. Biochemistry. 2003;42(21):6370–6379. doi: 10.1021/bi0273922. [DOI] [PubMed] [Google Scholar]
- 44.Totah RA, Rettie AE. Cytochrome P450 2C8: substrates, inhibitors, pharmacogenetics, and clinical relevance. Clin Pharmacol Ther. 2005;77(5):341–352. doi: 10.1016/j.clpt.2004.12.267. [DOI] [PubMed] [Google Scholar]
- 45.Prueksaritanont T, Tang C, Qiu Y, Mu L, Subramanian R, Lin JH. Effects of fibrates on metabolism of statins in human hepatocytes. Drug Metab Disp. 2002;30(11):1280–1287. doi: 10.1124/dmd.30.11.1280. [DOI] [PubMed] [Google Scholar]
- 46.Wang JS, Neuvonen M, Wen X, Backman JT, Neuvonen PJ. Gemfibrozil inhibits CYP2C8-mediated cerivastatin metabolism in human liver microsomes. Drug Metab Disp. 2002;30(12):1352–1356. doi: 10.1124/dmd.30.12.1352. [DOI] [PubMed] [Google Scholar]
- 47.Williams D, Feely J. Pharmacokinetic-pharmacodynamic drug interactions with HMG-CoA reductase inhibitors. Clin Pharmacokinet. 2002;41(5):343–370. doi: 10.2165/00003088-200241050-00003. [DOI] [PubMed] [Google Scholar]
- 48.Walsky RL, Gaman EA, Obach RS. Examination of 209 drugs for inhibition of cytochrome P450 2C8. J Clin Pharmacol. 2005;45(1):68–78. doi: 10.1177/0091270004270642. [DOI] [PubMed] [Google Scholar]
- 49.Melet A, Marques-Soares C, Schoch GA, et al. Analysis of human cytochrome P450 2C8 substrate specificity using a substrate pharmacophore and site-directed mutants. Biochemistry. 2004;43(49):15379–15392. doi: 10.1021/bi0489309. [DOI] [PubMed] [Google Scholar]
- 50.Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J Biol Chem. 2008;283(25):17227–17237. doi: 10.1074/jbc.M802180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8: Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279(10):9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 52.Fleming I. Cytochrome P450 epoxygenases as EDHF synthase(s) Pharmacol Res. 2004;49(6):525–533. doi: 10.1016/j.phrs.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 53.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12(3):251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 54.Wester MR, Yano JK, Schoch GA, et al. The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. J Biol Chem. 2004;279(34):35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 55.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424(6947):464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 56.Bertz RJ, Granneman GR. Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokinet. 1997;32(3):210–258. doi: 10.2165/00003088-199732030-00004. [DOI] [PubMed] [Google Scholar]
- 57.Yu A-I, Idle JR, Gonzalez FJ. Polymorphic cytochrome P450 2D6: Humanized mouse model and endogenous substrates. Drug Metab Rev. 2004;36(2):243–277. doi: 10.1081/dmr-120034000. [DOI] [PubMed] [Google Scholar]
- 58.Rowland P, Blaney FE, Smyth MG, et al. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281(11):7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- 59.Unwalla RJ, Cross JB, Salaniwal S, et al. Using a homology model of cytochrome P450 2D6 to predict substrate site of metabolism. J Comput Aided Mol Des. 2010;24(3):237–256. doi: 10.1007/s10822-010-9336-6. [DOI] [PubMed] [Google Scholar]
- 60.Hritz J, de Ruiter A, Oostenbrink C. Impact of plasticity and flexibility on docking results for cytochrome P450 2D6: a combined approach of molecular dynamics and ligand docking. J Med Chem. 2008;51(23):7469–7477. doi: 10.1021/jm801005m. [DOI] [PubMed] [Google Scholar]
- 61.Bièche I, Narjoz C, Asselah T, et al. Reverse transcriptase-PCR quantification of mRNA levels from cytochrome (CYP)1, CYP2 and CYP3 families in 22 different human tissues. Pharmacogenet Genomics. 2007;17(9):731–742. doi: 10.1097/FPC.0b013e32810f2e58. [DOI] [PubMed] [Google Scholar]
- 62.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270(1):414–423. [PubMed] [Google Scholar]
- 63.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43(1):149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 64.Laethem RM, Balazy M, Falck JR, Laethem CL, Koop DR. Formation of 19(S)-, 19(R)-, and 18(R)-hydroxyeicosatetraenoic acids by alcohol-inducible cytochrome P450 2E1. J Biol Chem. 1993;268(17):12912–12918. [PubMed] [Google Scholar]
- 65.Roy U, Joshua R, Stark RL, Balazy M. Cytochrome P450/NADPH-dependent biosynthesis of 5, 6-trans-epoxyeicosatrienoic acid from 5, 6-trans-arachidonic acid. Biochem J. 2005;390(Pt 3):719–727. doi: 10.1042/BJ20050681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ioannides C, Lewis DF. Cytochromes P450 in the bioactivation of chemicals. Curr Top Med Chem. 2004;4(16):1767–1788. doi: 10.2174/1568026043387188. [DOI] [PubMed] [Google Scholar]
- 67.Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radical Med Chem. 2008;44(5):723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porubsky PR, Meneely KM, Scott EE. Structures of human cytochrome P450 2E1: Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J Biol Chem. 2008;283(48):33698–33707. doi: 10.1074/jbc.M805999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rendic S, DiCarlo FJ. Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab Rev. 1997;29(1&2):413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 70.Molden E. Variability in cytochrome P450-mediated metabolism of cardiovascular drugs: Clinical implications and practical attempts to avoid potential problems. Heart Drug. 2004;4(2):55–79. [Google Scholar]
- 71.Fujita K. Food-drug interactions via human cytochrome P450 3A (CYP3A) Drug Metabol Drug Interact. 2004;20(4):195–217. doi: 10.1515/dmdi.2004.20.4.195. [DOI] [PubMed] [Google Scholar]
- 72.Guengerich FP. Cytochrome P-450 3A4: Regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39(1):1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 73.Hutzler JM, Tracy TS. Atypical kinetic profiles in drug metabolism reactions. Drug Metab Disp. 2002;30(4):355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 74.Harlow GR, Halpert JR. Analysis of human cytochrome P450 3A4 cooperativity: Construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc Natl Acad Sci USA. 1998;95(12):6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson EF, Schwab GE, Vickery LE. Positive effectors of the binding of active site-directed amino acid steroid to rabbit cytochrome P450 3c. J Biol Chem. 1988;263(33):17672–17677. [PubMed] [Google Scholar]
- 76.Korzekwa KR, Krishnamachary N, Shou M, et al. Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry. 1998;37(12):4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- 77.Shou M, Grogan J, Mancewica JA, et al. Activation of CYP 3A4: Evidence for the simultaneous binding of two substrates in a cytochrome P450 active site. Biochemistry. 1994;33(21):6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 78.Williams PA, Cosme J, Vinkovic DM, et al. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305(5684):683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 79.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279(37):38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 80.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA. 2006;103(37):13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cameron MD, Wen B, Roberts AG, Atkins WM, Campbell AP, Nelson SD. Cooperative binding of acetaminophen and caffeine within the P450 3A4 active site. Chem Res Toxicol. 2007;20(10):1434–1441. doi: 10.1021/tx7000702. [DOI] [PubMed] [Google Scholar]
- 82.Dwek RA. Nuclear magnetic resonance (N.M.R.) in biochemistry: Applications to enzyme systems. Oxford University Press; London: 1973. [Google Scholar]
- 83.Kijac AZ, Li Y, Sligar SG, Rienstra CM. Magic-angle spinning solid-state NMR spectroscopy of nanodisc-embedded human CYP3A4. Biochemistry. 2007;46(48):13696–13703. doi: 10.1021/bi701411g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCullough CR, Pullela PK, Im SC, Waskell L, Sem DS. 13C-Methyl isocyanide as an NMR probe for cytochrome P450 active sites. J Biolmol NMR. 2009;43(3):171–178. doi: 10.1007/s10858-009-9300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lewis DF, Ito Y, Eddershaw PJ, Dickins M, Goldfarb PS. An evaluation of ondansetron binding interactions with human cytochrome P450 enzymes CYP3A4 and CYP2D6. Drug Metab Lett. 2010;4(1):25–30. doi: 10.2174/187231210790980408. [DOI] [PubMed] [Google Scholar]
- 86.Modi S, Gilham DE, Sutcliffe MJ, et al. 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine as a substrate of cytochrome P450 2D6: allosteric effects of NADPH-cytochrome P450 reductase. Biochemistry. 1997;350(15):4461–4470. doi: 10.1021/bi962633p. [DOI] [PubMed] [Google Scholar]
- 87.Kobayashi Y, Fang X, Szklarz GD, Halpert JR. Probing the active site of cytochrome P450 2B1: metabolism of 7-alkoxycoumarins by the wild type and five site-directed mutants. Biochemistry. 1998;37(19):6679–6688. doi: 10.1021/bi9731450. [DOI] [PubMed] [Google Scholar]
- 88.Lin HL, Zhang H, Waskell L, Hollenberg PF. Threonine-205 in the F helix of P450 2B1 contributes to androgen 16 beta-hydroxylation activity and mechanism-based inactivation. J Pharmacol Exp Ther. 2003;306(2):744–751. doi: 10.1124/jpet.103.050260. [DOI] [PubMed] [Google Scholar]
- 89.Yamamoto T, Takano R, Egashira T, Yamanaka Y. Metabolism of methamphetamine, amphetamine and p-hydroxymethamphetamine by rat-liver microsomal preparations in vitro. Xenobiotica. 1984;14(11):867–875. doi: 10.3109/00498258409151485. [DOI] [PubMed] [Google Scholar]
- 90.Ortiz de Montellano PR. Cytochrome P450: Structure, mechanism, and biochemistry. 3rd Plenum Publishing; New York: 2004. [Google Scholar]
- 91.Myers TG, Thummel KE, Kalhorn TF, Nelson SD. Preferred orientations in the binding of 4'-hydroxyacetanilide (acetaminophen) to cytochrome P450 1A1 and 2B1 isoforms as determined by 13C- and 15N-NMR relaxation studies. J Med Chem. 1994;37(6):860–867. doi: 10.1021/jm00032a021. [DOI] [PubMed] [Google Scholar]
- 92.van de Straat R, de Vries J, de Boer HJ, Vromans RM, Vermeulen NP. Relationship between paracetamol binding to and its oxidation by two cytochromes P-450 isozymes--a proton nuclear magnetic resonance and spectrophotometric study. Xenobiotica. 1987;17(1):1–9. doi: 10.3109/00498258709047169. [DOI] [PubMed] [Google Scholar]
- 93.Blaauboer BJ, Van Holsteijn CW. Formation and disposition of N-hydroxylated metabolites of aniline and nitrobenzene by isolated rat hepatocytes. Xenobiotica. 1983;13(5):295–302. doi: 10.3109/00498258309052267. [DOI] [PubMed] [Google Scholar]
- 94.Vainio H, Hanninen O. Enhancement of aniline p-hydroxylation by acetone in rat liver microsomes. Xenobiotica. 1972;2(3):259–267. doi: 10.3109/00498257209111056. [DOI] [PubMed] [Google Scholar]
- 95.Wisniewska-Knypl JM, Jablonska JK. The rate of aniline metabolism in vivo in rats exposed to aniline and drugs. Xenobiotica. 1975;5(8):511–519. doi: 10.3109/00498257509056121. [DOI] [PubMed] [Google Scholar]
- 96.Koerts J, Rietjens IM, Boersma MG, Vervoort J. 1H NMR T1 relaxation rate study on substrate orientation of fluoromethylanilines in the active sites of microsomal and purified cytochromes P450 1A1 and 2B1. FEBS Lett. 1995;368(2):279–284. doi: 10.1016/0014-5793(95)00672-v. [DOI] [PubMed] [Google Scholar]
- 97.Tassaneeyakul W, Birkett DJ, McManus ME, et al. Caffeine metabolism by human hepatic cytochromes P450: Contributions of 1A2, 2E1 and 3A isoforms. Biochem Pharmacol. 1994;47(10):1767–1776. doi: 10.1016/0006-2952(94)90304-2. [DOI] [PubMed] [Google Scholar]
- 98.Welfare MR, Aitkin M, Bassendine MF, Daly AK. Detailed modelling of caffeine metabolism and examination of the CYP1A2 gene: lack of a polymorphism in CYP1A2 in Caucasians. Pharmacogenetics. 1999;9(3):367–375. doi: 10.1097/00008571-199906000-00012. [DOI] [PubMed] [Google Scholar]
- 99.Regal KA, Nelson SD. Orientation of caffeine within the active site of human cytochrome P450 1A2 based on NMR longitudinal (T1) relaxation measurements. Arch Biochem Biophys. 2000;384(1):47–58. doi: 10.1006/abbi.2000.2073. [DOI] [PubMed] [Google Scholar]
- 100.Berthou F, Guillois B, Riche C, Dreano Y, Jacqz-Aigrain E, Beaune PH. Interspecies variations in caffeine metabolism related to cytochrome P4501A enzymes. Xenobiotica. 1992;22(6):671–680. doi: 10.3109/00498259209053129. [DOI] [PubMed] [Google Scholar]
- 101.Poli-Scaife S, Attias R, Dansette PM, Mansuy D. The substrate binding site of human liver cytochrome P450 2C9: an NMR study. Biochemistry. 1997;36(42):12672–12682. doi: 10.1021/bi970527x. [DOI] [PubMed] [Google Scholar]
- 102.Lopez-Garcia MP, Dansette PM, Mansuy D. Thiophene derivatives as new mechanism-based inhibitors of cytochromes P-450: inactivation of yeast-expressed human liver cytochrome P-450 2C9 by tienilic acid. Biochemistry. 1994;33(1):166–175. doi: 10.1021/bi00167a022. [DOI] [PubMed] [Google Scholar]
- 103.Clarke SE, Baldwin SJ, Bloomer JC, Ayrton AD, Sozio RS, Chenery RJ. Lauric acid as a model substrate for the simultaneous determination of cytochrome P450 2E1 and 4A in hepatic microsomes. Chem Res Toxicol. 1994;7(6):836–842. doi: 10.1021/tx00042a018. [DOI] [PubMed] [Google Scholar]
- 104.Hummel MA, Gannett PM, Aguilar J, Tracy TS. Substrate proton to heme distances in CYP2C9 allelic variants and alterations by the heterotropic activator, dapsone. Arch Biochem Biophys. 2008;475(2):175–183. doi: 10.1016/j.abb.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang W, Stearns RA, Wang RW, Chiu SH, Baillie TA. Roles of human hepatic cytochrome P450s 2C9 and 3A4 in the metabolic activation of diclofenac. Chem Res Toxicol. 1999;12(2):192–199. doi: 10.1021/tx9802217. [DOI] [PubMed] [Google Scholar]
- 106.Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45(6):525–538. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Armstrong SC, Cozza KL. Pharmacokinetic drug interactions of morphine, codeine, and their derivatives: theory and clinical reality, part I. Psychosomatics. 2003;44(2):167–171. doi: 10.1176/appi.psy.44.2.167. [DOI] [PubMed] [Google Scholar]
- 108.Dixon CM, Colthup PV, Serabjit-Singh CJ, et al. Multiple forms of cytochrome P450 are involved in the metabolism of ondansetron in humans. Drug Metab Disp. 1995;23(11):1225–1230. [PubMed] [Google Scholar]
- 109.Modi S, Paine MJ, Sutcliffe MJ, et al. A model for human cytochrome P450 2D6 based on homology modeling and NMR studies of substrate binding. Biochemistry. 1996;35(14):4540–4550. doi: 10.1021/bi952742o. [DOI] [PubMed] [Google Scholar]
- 110.Keizers PH, de Graaf C, de Kanter FJ, et al. Metabolic regio- and stereoselectivity of cytochrome P450 2D6 towards 3, 4-methylenedioxy-N-alkylamphetamines: In silico predictions and experimental validation. J Med Chem. 2005;48(19):6117–6127. doi: 10.1021/jm050338+. [DOI] [PubMed] [Google Scholar]
- 111.Tracy TS, Rosenbluth BW, Wrighton SA, Gonzalez FJ, Korzekwa KR. Role of cytochrome P450 2C9 and an allelic variant in the 4'-hydroxylation of (R)- and (S)-flurbiprofen. Biochem Pharmacol. 1995;49(9):1269–1275. doi: 10.1016/0006-2952(95)00048-5. [DOI] [PubMed] [Google Scholar]
- 112.Hutzler JM, Wienkers LC, Wahlstrom JL, Carlson TJ, Tracy TS. Activation of cytochrome P450 2C9-mediated metabolism: Mechanistic evidence in support of kinetic observations. Arch Biochem Biophys. 2003;410(1):16–24. doi: 10.1016/s0003-9861(02)00665-3. [DOI] [PubMed] [Google Scholar]
- 113.Hummel MA, Locuson CW, Gannett PM, et al. CYP2C9 genotype-dependent effects on in vitro drug-drug interactions: Switching of benzbromarone effect from inhibition to activation in the CYP2C9.3 variant. Mol Pharmacol. 2005;68(3):644–651. doi: 10.1124/mol.105.013763. [DOI] [PubMed] [Google Scholar]
- 114.Hummel MA, Gannett PM, Aguilar JS, Tracy TS. Effector-mediated alteration of substrate orientation in cytochrome P450 2C9. Biochemistry. 2004;43(22):7207–7214. doi: 10.1021/bi036158o. [DOI] [PubMed] [Google Scholar]
- 115.Williams JA, Ring BJ, Cantrell VE, et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Disp. 2002;30(8):883–891. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- 116.Cameron MD, Wen B, Allen KE, et al. Cooperative binding of midazolam with testosterone and alpha-naphthoflavone within the CYP3A4 active site: A NMR T1 paramagnetic relaxation study. Biochemistry. 2005;44(43):14143–14151. doi: 10.1021/bi051689t. [DOI] [PubMed] [Google Scholar]
- 117.Lampe JN, Brandman R, Sivaramakrishnan S, Ortiz de, Montellano PR. Two-dimensional NMR and all-atom molecular dynamics of cytochrome P450 CYP119 reveal hidden conformational substates. J Biol Chem. 2010;285(13):9594–9603. doi: 10.1074/jbc.M109.087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lampe JN, Floor SN, Gross JD, et al. Ligand-induced conformational heterogeneity of cytochrome P450 CYP119 identified by 2D NMR spectroscopy with the unnatural amino acid (13)C-p-methoxyphenylalanine. J Am Chem Soc. 2008;130(48):16168–16169. doi: 10.1021/ja8071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pochapsky SS, Pochapsky TC, Wei JW. A model for effector activity in a highly specific biological electron transfer complex: the cytochrome P450(cam)-putidaredoxin couple. Biochemistry. 2003;42(19):5649–5656. doi: 10.1021/bi034263s. [DOI] [PubMed] [Google Scholar]
- 120.Roberts AG, Diaz MD, Lampe JN, et al. NMR studies of ligand binding to P450(eryF) provides insight into the mechanism of cooperativity. Biochemistry. 2006;45(6):1673–1684. doi: 10.1021/bi0518895. [DOI] [PubMed] [Google Scholar]
- 121.Zhang W, Pochapsky SS, Pochapsky TC, Jain NU. Solution NMR structure of putidaredoxin-cytochrome P450cam complex via a combined residual dipolar coupling-spin labeling approach suggests a role for Trp106 of putidaredoxin in complex formation. J Mol Biol. 2008;384(2):349–363. doi: 10.1016/j.jmb.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zamora I, Afzelius L, Cruciani G. Predicting drug metabolism: A site of metabolism prediction tool applied to the cytochrome P450 2C9. J Med Chem. 2003;46(12):2313–2324. doi: 10.1021/jm021104i. [DOI] [PubMed] [Google Scholar]
- 123.Ahlström MM, Zamora I. Characterization of type II ligands in CYP2C9 and CYP3A4. J Med Chem. 2008;51(6):1755–1763. doi: 10.1021/jm701121y. [DOI] [PubMed] [Google Scholar]
- 124.Afzelius L, Masimirembwa CM, Karlén A, Andersson TB, Zamora I. Discriminant and quantitative PLS analysis of competitive CYP2C9 inhibitors versus non-inhibitors using alignment independent GRIND descriptors. J Comput Aided Mol Des. 2002;16(7):443–458. doi: 10.1023/a:1021281008423. [DOI] [PubMed] [Google Scholar]
- 125.Ahlström MM, Ridderström M, Zamora I, Luthman K. CYP2C9 structure-metabolism relationships: Optimizing the metabolic stability of COX-2 inhibitors. J Med Chem. 2007;50(18):4444–4452. doi: 10.1021/jm0705096. [DOI] [PubMed] [Google Scholar]
- 126.Ahlström MM, Ridderström M, Zamora I. CYP2C9 structure-metabolism relationships: Substrates, inhibitors, and metabolites. J Med Chem. 2007;50(22):5382–5391. doi: 10.1021/jm070745g. [DOI] [PubMed] [Google Scholar]
- 127.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123(25):6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 128.Gossert AD, Henry C, Blommers MJ, Jahnke W, Fernandez C. Time efficient detection of protein-ligand interactions with the polarization optimized PO-WaterLOGSY NMR experiment. J Biolmol NMR. 2009;43(4):211–217. doi: 10.1007/s10858-009-9303-5. [DOI] [PubMed] [Google Scholar]
- 129.Dalvit C, Pevarello P, Tato M, Veronesi M, Vulpetti A, Sundstrom M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J Biolmol NMR. 2000;18(1):65–68. doi: 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- 130.Shimotakahara S, Furihata K, Tashiro M. Application of NMR screening techniques for observing ligand binding with a protein receptor. Magn Reson Chem. 2004;43(1):69–72. doi: 10.1002/mrc.1492. [DOI] [PubMed] [Google Scholar]
- 131.Kleywegt GJ, Jones TA. Detection delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. 1994;D50(Pt 2):178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]