SUMMARY
The inherited neurodegenerative disease Friedreich’s ataxia (FRDA) is caused by GAA•TTC triplet repeat hyper-expansions within the first intron of the FXN gene, encoding the mitochondrial protein frataxin. Long GAA•TTC repeats causes heterochromatin-mediated gene silencing and loss of frataxin in affected individuals. We report the derivation of induced pluripotent stem cells (iPSCs) from FRDA patient fibroblasts by transcription factor reprogramming. FXN gene repression is maintained in the iPSCs, as are the global gene expression signatures reflecting the human disease. GAA•TTC repeats uniquely in FXN in the iPSCs exhibit repeat instability similar to patient families, where they expand and/or contract with discrete changes in length between generations. The mismatch repair enzyme MSH2, implicated in repeat instability in other triplet repeat diseases, is highly expressed in pluripotent cells, occupies FXN intron 1, and shRNA silencing of MSH2 impedes repeat expansion, providing a possible molecular explanation for repeat expansion in FRDA.
INTRODUCTION
Friedreich’s ataxia (FRDA), the most common inherited ataxia, is caused by heterochromatin-mediated silencing of the nuclear FXN gene, encoding the essential mitochondrial protein frataxin (Herman et al., 2006). The genetic mutation in FRDA is a GAA•TTC triplet-repeat expansion in the first intron of FXN (Campuzano et al., 1996), with unaffected alleles having 6–34 repeats in contrast to 66–1700 repeats in patient alleles. Longer repeats are associated with more severe gene repression, lower frataxin protein levels and earlier onset and increased disease severity (Bidichandani et al., 1998; Campuzano et al., 1996). Frataxin insufficiency leads to progressive spino-cerebellar neurodegeneration and associated movement disorders along with an increased risk for diabetes and cardiomyopathy, the latter being the most common cause of death in FRDA.
Unlike many triplet-repeat diseases (e.g., the polyglutamine expansion and the RNA toxicity diseases (Orr and Zoghbi, 2007)), GAA•TTC expansions in FXN are intronic and do not alter the frataxin protein sequence; thus, gene activation would be of therapeutic benefit (Gottesfeld, 2007; Herman et al., 2006). However, studies in FRDA pathogenesis and therapeutics are limited by poor cellular models, and available mouse models do not fully recapitulate gene silencing and frataxin protein levels (Al-Mahdawi et al., 2004; Miranda et al., 2002). Recent studies have shown that human fibroblasts can be reprogrammed to a pluripotent state by transduction of transcription factors (Takahashi et al., 2007), and importantly, the same has been demonstrated with fibroblasts from repeat-associated neurodegenerative disease patients such as Huntington’s disease (HD) and Fragile X syndrome (Park et al., 2008a; Urbach et al., 2010). We now report the derivation of FRDA iPSCs. We find that the FXN GAA•TTC repeats in FRDA iPSCs exhibit a repeat instability pattern similar to the human disease, where repeats expand and/or contract with discrete changes in length between generations (Campuzano et al., 1996; Pianese et al., 1997). We also provide evidence for the role of the mismatch repair (MMR) enzyme MSH2 in repeat instability. Our observations provide a cellular model system for mechanistic studies of repeat instability in FRDA and potentially in other triplet repeat diseases.
RESULTS
Derivation of iPSCs from FRDA Patient Fibroblasts
Primary fibroblasts from two FRDA patients (GM03816 and GM04078 from the NIGMS Coriell Cell Repository) were reprogrammed by transcription factor overexpression (Takahashi et al., 2007), and colonies with ES/iPS morphology were selected and expanded (Figure 1A). Analysis by qRT-PCR shows that our FRDA iPSC lines are indeed pluripotent (Figure 1B) and retain marked repression of FXN mRNA (Figure 1C). Further, expression of the integrated transgenic reprogramming factors is silenced in the iPSCs (Figure S1A, available online), a hallmark of full reprogramming (Lowry et al., 2008).
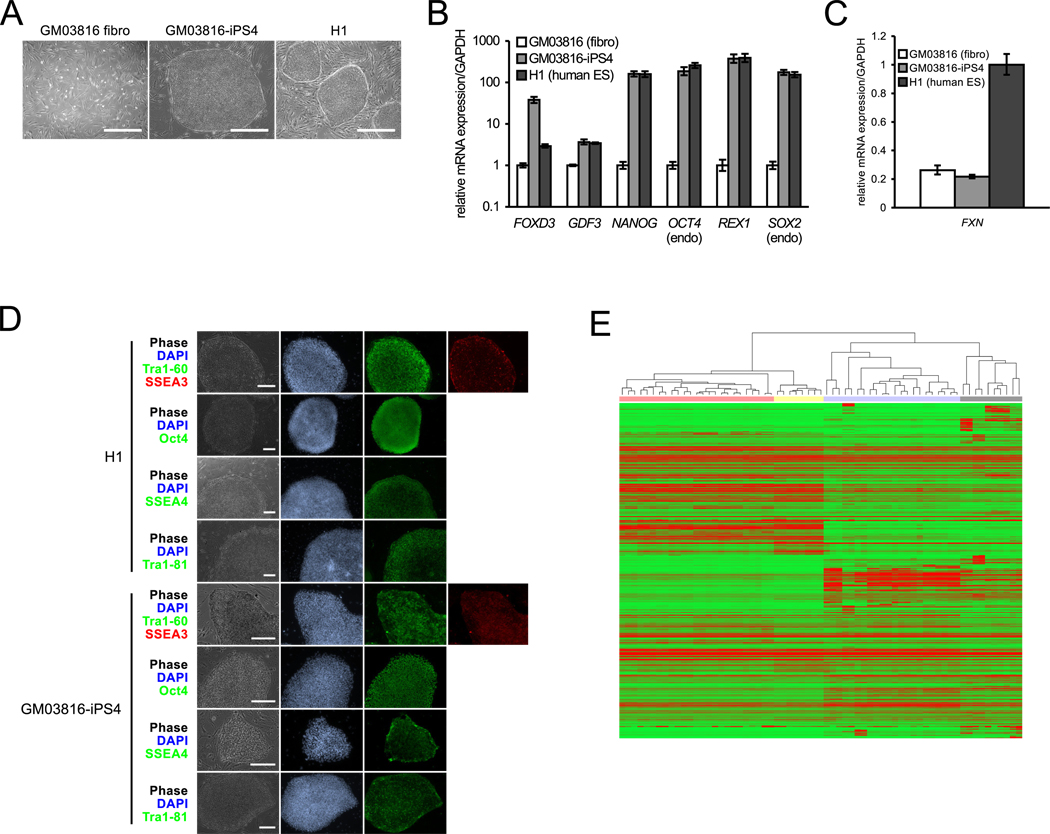
Figure 1. Characterization of FRDA iPSCs.
(A) Images of GM03816 FRDA fibroblasts (left), GM03816 iPSCs (middle), and H1 hESCs (right). Scale bars = 0.5 mm.
(B) GM03816-iPS4 shows similar expression of pluripotency mRNAs as H1 hESCs. GM03816 fibroblasts, white bars; GM03816 iPSCs, light grey; H1 hESCs, dark grey. mRNA levels are normalized to GAPDH. Error bars = SEM of duplicate measurements.
(C) Hallmark repression of FXN mRNA in GM03816 fibroblasts and the GM03816-iPS4 line as compared to the unaffected H1 line. Error bars = SEM of duplicate measurements.
(D) GM03816-iPS4 and H1 hES staining (contrast enhanced) of pluripotency markers. Phase contrast (gray); nuclear staining (blue); pluripotency markers staining (green and red) is as denoted by the colored text labels. Tra1–60 and Tra1–81, surface markers; SSEA-3 and -4, stage-specific embryonic antigens; Oct4, transcription factor. Scale bars = 0.25 mm.
(E) Microarray analysis of FRDA iPSCs versus unaffected iPSC/ESCs, human tissues, and cell lines. Red represents up-regulation, green represents down-regulation. Unaffected iPS/ESCs, red highlighting; FRDA iPSCs, yellow; unaffected human tissue, blue; human cell lines, gray. See also Figure S1 and Tables S1–S3.
Immunostaining of FRDA iPSCs for pluripotent markers (SSEA3 and SSEA4; Oct4; and Tra1–60 and Tra1–81) was also found to be comparable to that of H1 ESCs (Figure 1D). Genotyping of the FXN gene GAA•TTC repeats and cytogenetic analysis demonstrated that the iPSCs indeed originated from FRDA fibroblasts and are karyotypically normal (Figures 2A and S1B), and ChIP experiments confirm heterochromatin histone marks near the FXN GAA•TTC repeats (Al-Mahdawi et al., 2008; Herman et al., 2006; Rai et al., 2008) (Figure S1C to E). Finally, teratoma analysis shows full in vivo differentiation capacity (Figure S1F), providing additional evidence of the pluripotent nature of the FRDA iPSCs.
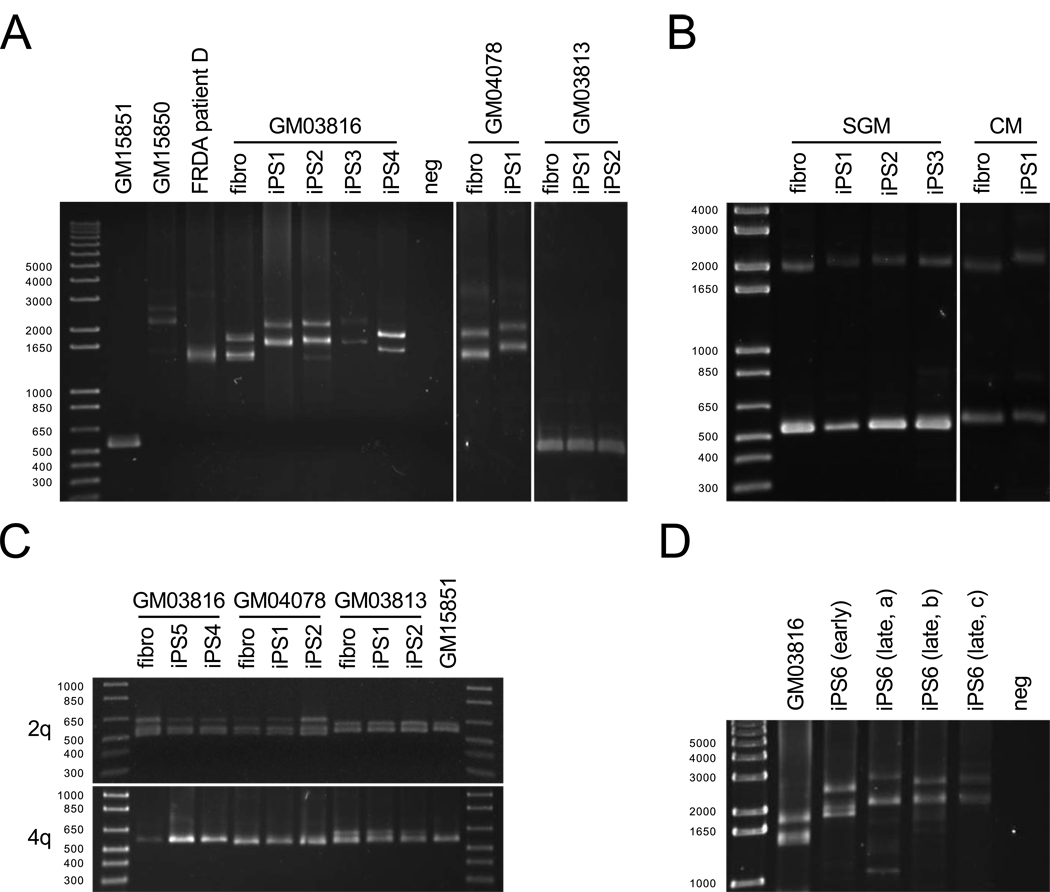
Figure 2. Analysis of GAA•TTC repeat lengths.
(A) PCR analysis of FXN GAA•TTC repeat length in cell lines, patient lymphocytes, and FRDA iPSCs. DNA length markers are shown in bp. Lanes are as follows (left to right): Left panel: GM15851 unaffected and GM15850 FRDA lymphoblasts; FRDA patient D lymphocytes; GM03816 FRDA fibroblasts; GM03816-derived iPS lines 1–4; no DNA control (neg). Middle panel: GM04078 FRDA fibroblasts; GM04078-derived iPSCs (GM0478-iPS1). Right panel: GM03813 SMA fibroblasts and two corresponding iPSC lines (GM03813-iPS1 and -iPS-2).
(B) PCR products of the FXN GAA•TTC repeat region in carrier parents of a FRDA patient. Left panel (left to right): maternal fibroblast and three corresponding iPSC lines. Right panel (left to right): paternal fibroblast and one corresponding iPSC line.
(C) GAA•TTC repeats at two unrelated genetic loci in FRDA (GM03816- and GM04078-derived) and SMA (GM03813-derived) fibroblasts and two iPSC lines. PCR products from GM15851 FRDA lymphoblasts are also shown.
(D) FXN GAA•TTC repeat length PCR measurements over time. Left to right: FRDA fibroblasts (GM03816), early passage FRDA iPSC (iPS6 early), and three FRDA iPSC single colony samples at passage 15 (iPS6 late a–c).
Global mRNA Expression Profiles of FRDA iPSCs
Hierarchical clustering of global gene expression profiles of four FRDA iPSC lines (two from GM03816, two from GM04078) with a set of unaffected iPSCs, hESCs, and various human tissues and cell lines show that our iPSC lines group to the same cluster as other iPS/ESCs, though in a separate, distinct subset (Figure 1E). We attribute this distinction to a 5–7% global expression difference between our FRDA iPSCs and other iPS/ESCs, whereas an internal variation of only 2–3% was observed among other iPS/ESCs, a difference that likely reflects the diseased nature of the FRDA genetic background. Moreover, functional annotation clustering using the Database for Annotation, Visualization and Integrated Discovery (DAVID) of the top differentially expressed genes in FRDA iPSCs identified gene groups related to mitochondrial function, DNA repair, and DNA damage response (Table S1). DNA repair also appeared as a top GO category significantly enriched in our dataset (Table S2), consistent with recent studies on FRDA patients (Haugen et al., 2010). Additional significant GO categories were related to cell cycle, protein modification/ubiquitination, lipid metabolism and carbohydrate biosynthesis, all of which have been previously associated with altered function in FRDA patients (Coppola et al., 2006; Haugen et al., 2010). Global microRNA profiling also shows that the FRDA iPSCs express many miRNAs associated with pluripotency, but distinct differences, again presumably due to FRDA pathogenesis, were also noted (Figure S1G–H and Table S3).
GAA•TTC Repeat Expansion in FRDA iPSCs
PCR analysis of the FXN GAA•TTC repeats showed repeat instability in the iPSC lines (Figure 2A), a phenomenon not seen in donor fibroblasts (data not shown). PCR products from unaffected (GM15851) and FRDA (GM15850) lymphoblasts and an unrelated patient DNA are also shown for comparison. In all cases, an apparent expansion of both alleles of FRDA iPSCs was observed (with certain caveats addressed below). PCR analysis of iPSCs from a second patient (GM04078) similarly showed repeat expansion (Figure 2A, middle panel), confirming the general nature of this observation. In contrast, wild-type FXN alleles do not change in size in a non-FRDA iPSC control (Figure 2A; right panel, GM03813 spinal muscular atrophy iPSCs) and in non-disease unaffected iPSCs (data not shown). Due to the allele ambiguity of our PCR assay, shifts in PCR bands could represent expansion of both alleles or contraction of one and expansion of the other. Therefore, iPSCs were generated from carrier parents of a third FRDA patient and subjected to PCR analysis, showing repeat expansion in both parental pathogenic alleles (Figure 2B). The wild-type FXN allele, as expected, did not expand, suggesting a gender-neutral instability in pathogenic FXN alleles. As in somatic cells, GAA•TTC repeat expansions at two unrelated genetic loci (2q36, 16 repeats; 4q31.1, 30 repeats; (Rindler et al., 2006)) remain unchanged between our iPSCs and their three corresponding donor fibroblasts (Figure 2C), even though these loci are at Alu elements similar to FXN intron 1. Altogether, the data suggest that changes at the FXN gene are a consequence of its particular repeat expansion (and perhaps its length) and are not a general phenomenon throughout the human genome. Further, we find that GAA•TTC repeat lengths in FRDA iPSCs change over time in culture (Figure 2D).
Molecular Basis For Repeat Instability
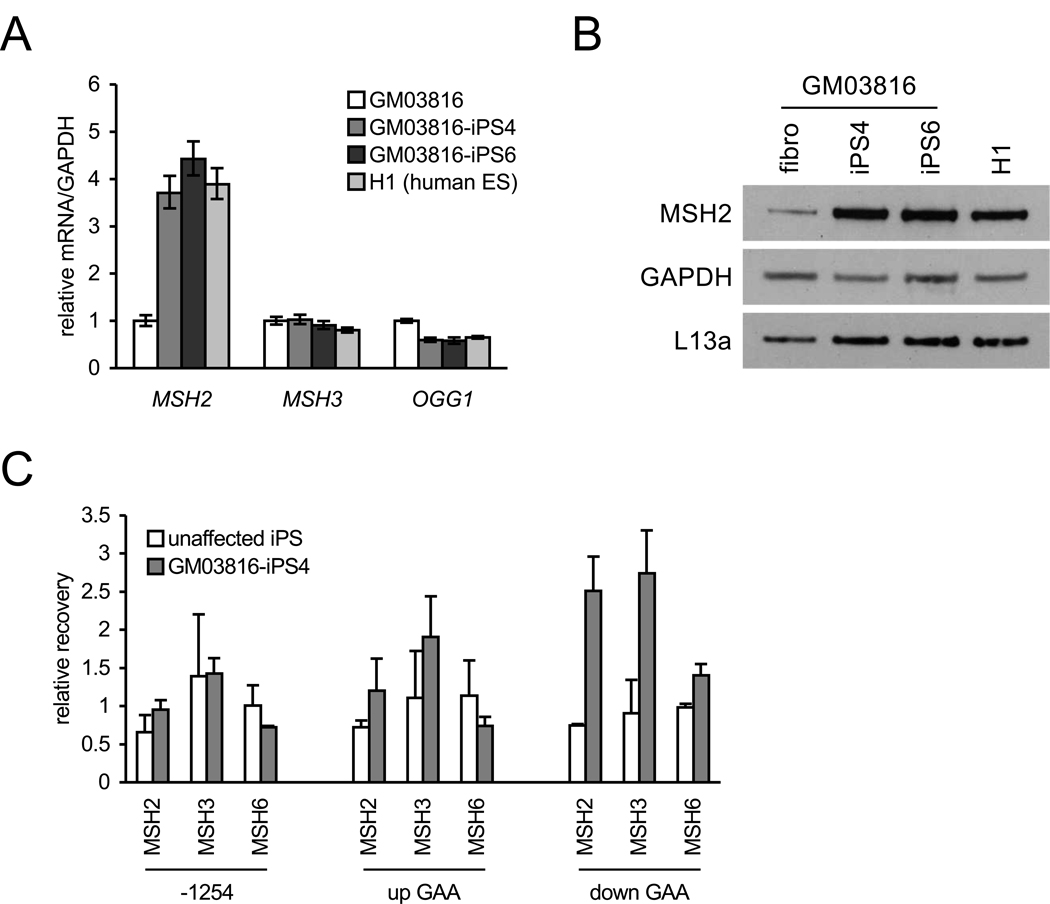
Previous reports have implicated the MMR enzymes MSH2, MSH3 and MSH6 in CAG•CTG and CTG•CAG somatic and intergenerational repeat instability in HD and DM1 (myotonic dystrophy type I) transgenic mice, respectively (reviewed in (Dion and Wilson, 2009)). Other studies have implicated the oxoguanine-DNA glycosylase OGG1 in somatic instability (Kovtun et al., 2007). In FRDA iPSCs, mRNA expression analysis shows large increases in MSH2 compared to donor fibroblasts (Figure 3A). No differences in MSH3 mRNA, however, were found, and a small decrease in OGG1 mRNA was noted in the iPS/ESCs. Western blotting further shows corresponding increases in MSH2 protein in FRDA iPSCs and H1 ESCs compared to FRDA fibroblasts (Figure 3B). ChIP assays, at a resolution of ~1 kb, show increased occupancy of MSH2 and MSH3 downstream of the GAA•TTC repeats in FRDA iPSCs compared to an unaffected iPSC line but, in contrast, not 1254 bp upstream of the FXN transcriptional start site nor directly upstream of the GAA•TTC repeats (Figure 3C). No differences in MSH6 occupancy were found at any of the regions probed.
Figure 3. MSH2 expression and localization in FRDA iPSCs.
(A) MSH2, MSH3, and OGG1 mRNA levels in GM03816 fibroblasts (white), two GM03816 iPSC lines (iPS4, medium grey; iPS6, dark grey) and H1 cells (light grey). Levels are normalized to GAPDH. Errors bars = SEM of duplicate measurements.
(B) Western blot of MSH2 protein. Left to right: GM03816 fibroblast, two GM03816 iPSC lines (4 and 6), and H1 hESCs. GAPDH and L13a are loading controls.
(C) MSH2, MSH3, and MSH6 occupancy on the FXN gene 1254 bp upstream of the transcriptional start site (−1254), upstream (up GAA) and downstream (down GAA) of the GAA•TTC repeat expansion (measured by ChIP). IP recovery is relative to FXN intron 2 (11482 bp downstream of the transcriptional start site). Error bars = SEM of independent experiments (MSH2, n = 4; MSH3 and MSH6, n = 2).
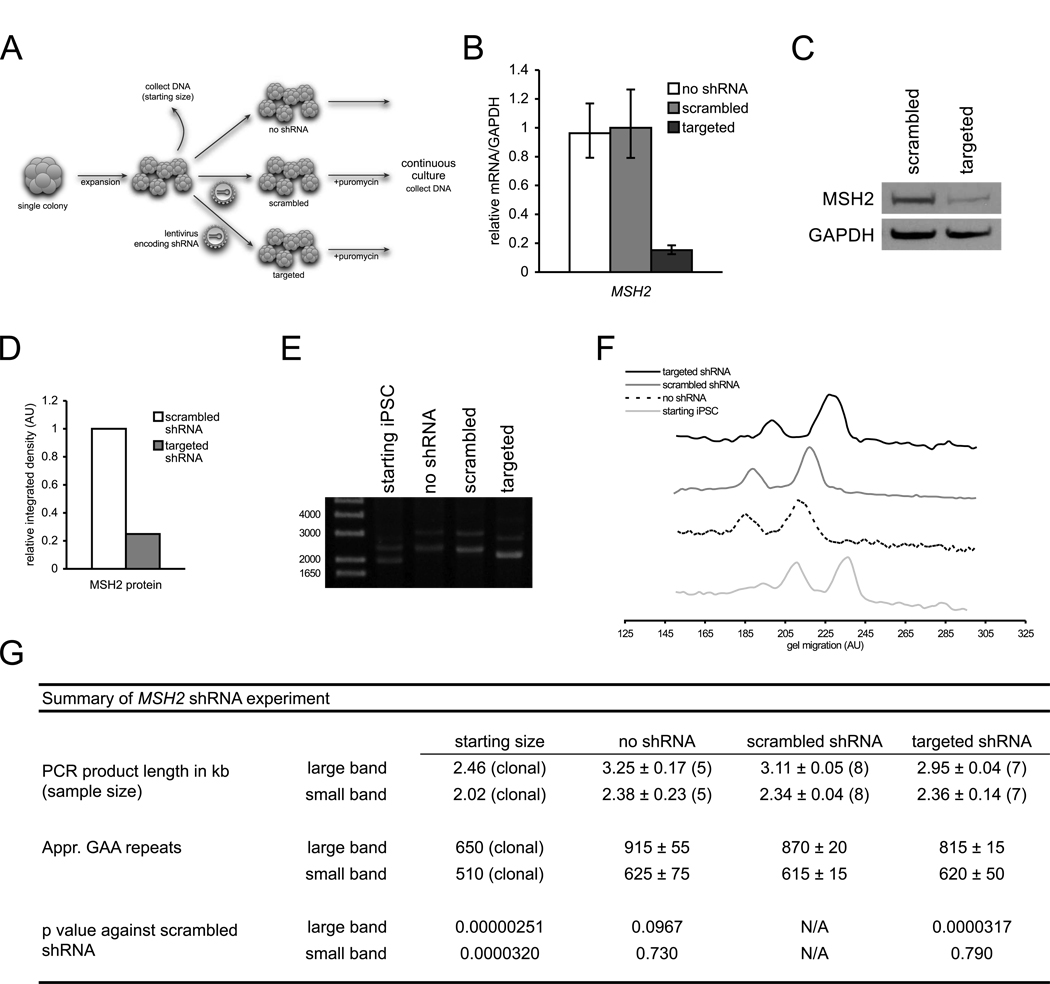
To further investigate the role of MSH2 in repeat instability, lentiviral shRNA constructs were integrated into single colony-expanded FRDA iPSCs (to limit repeat length heterogeneity) and assayed for GAA•TTC repeat length (summarized in Figure 4A). As shown in Figures 4B–D, stable expression of MSH2-targeted shRNA achieved relatively high levels of mRNA and protein knockdown compared to a scrambled shRNA. Additionally, we verify that MSH2 silencing does not affect pluripotency (Figure S2A). After eight passages, repeat length PCR analysis showed that MSH2 knockdown results in a significantly smaller large allele compared to a scrambled shRNA (Figure 4E–G). No statistical significance, however, was observed for the smaller allele based on pooled data, contrary to the single-point data shown in Figure 4E–F. shRNA silencing of MSH2 in FRDA fibroblasts followed by reprogramming also yielded similar results (Figure S2). Collectively, these data implicate the involvement of MSH2 in GAA•TTC repeat instability.
Figure 4. shRNA silencing of MSH2 in FRDA iPSCs.
(A) Diagram of silencing experiment.
(B) MSH2 mRNA levels of targeted and scrambled shRNA-expressing FRDA iPSCs. mRNA levels are normalized to GAPDH. Errors bars = SEM of duplicate measurements.
(C) Western blot of MSH2 protein in FRDA iPSCs with scrambled shRNA and targeted shRNA. GAPDH, loading control.
(D) Densitometric analysis of MSH2 protein normalized to GAPDH of previous panel.
(E) PCR of GAA•TTC repeat lengths pre-silencing (starting iPSC) and eight passages after transduction with no shRNA (no shRNA), scrambled shRNA (scrambled), and shRNA against MSH2 (targeted). Note: data are from single point data and do not represent pooled data (panel G).
(F) Signal intensity traces of ethidium-stained agarose gel from panel E. Higher gel migration represents smaller PCR bands.
(G) Summary of pooled GAA•TTC repeat length data. See Supplemental Experimental Procedures for quantitation of repeat lengths.
See also Figure S2.
DISCUSSION
To our knowledge, this is the first report of triplet repeat instability occurring in patient-specific iPSCs. Previous studies have analyzed triplet expansions in disease-specific iPSCs but either did not compare donor fibroblast repeats or the iPSCs did not show any changes after reprogramming (Park et al., 2008a; Urbach et al., 2010). In our case, we observe repeat expansion, perhaps resembling intergenerational instability as in FRDA families with one exception. In patient families, only the maternal pathogenic allele is reported to undergo intergenerational expansion, whereas the paternal allele usually remains the same length or can contract (Campuzano et al., 1996; Pianese et al., 1997). However, we find expansion of both parental pathogenic alleles in iPSCs. One interpretation for this difference is that both alleles undergo the same cellular changes during in vitro reprogramming as opposed to differential gametogenesis.
Our finding of MSH2 and possibly MSH3 as components involved in repeat expansion is supported by extensive prior studies in various triplet repeat disorders (Dion and Wilson, 2009). Early work in Fragile X syndrome models pointed towards MSH2 as a component responsible for repeat instability (Kramer et al., 1996). Other studies have shown that MSH2 has a role in both intergenerational and somatic instability in various HD models (Dragileva et al., 2009), and similar findings have been presented in DM1 studies as well (Savouret et al., 2003). Along similar lines, our present results implicate MSH2 as one of the proteins responsible for GAA•TTC repeat expansion. Although we have no direct evidence for an MSH2–MSH3 complex (MutSβ), we believe that such a complex is responsible in our case, as not only do both MSH2 and MSH3 localize near pathogenic FXN alleles, but other studies also point to the involvement of MutSβ as well (Dragileva et al., 2009; Kim et al., 2008).
Mechanistically, there is also debate over GAA•TTC repeat instability, and our results are consistent with several models of FRDA pathogenesis (Ditch et al., 2009; Dragileva et al., 2009; Iyer and Wells, 1999; Shishkin et al., 2009). One model proposes the formation of an exposed single-stranded DNA hairpin (resembling mismatched DNA) originating from a triple-stranded structure formed by long GAA•TTC repeats, which recruits MMR machinery (Wells, 2008). This recruitment then stabilizes the slipped-stranded intermediates, leading to repeat expansion (reviewed in (Mirkin, 2007)). Alternatively, sense/antisense transcription at the FXN locus (de Biase et al., 2009) could allow for transcription-coupled DNA repair and GAA•TTC repeat tract expansion (Ditch et al., 2009). Interestingly, transcription-coupled repair has been shown to interact with the MutSβ complex (Zhao et al., 2009), further supporting our observations.
There currently exist few model systems in which one can study GAA•TTC expansions (Al-Mahdawi et al., 2004; Ditch et al., 2009; Iyer and Wells, 1999; Shishkin et al., 2009). Despite the fact that a recent report revealed cellular physiological differences between a human disorder and its iPSC model and urged caution when making associations between the two (Urbach et al., 2010), these key differences are likely to be highly context-dependent. In our case, we expect that FRDA iPSCs will provide a valuable, more accessible resource to study repeat instability mechanisms as well as for differentiation into cell types affected in this human disease (sensory neurons and cardiomyocytes). Such cellular models will be useful to dissect disease mechanisms and to screen potential therapeutic agents.
EXPERIMENTAL PROCEDURES
Cell culture
Fibroblasts were grown at 37°C and 5% CO2 with 10% FBS (Lonza) in MEM, 2 mM glutamine, 1% NEAA, 20 mM HEPES, and 1% antibiotic-antimycotic (all from Invitrogen). ES/iPSCs were grown at 37°C and 5% CO2 on γ-irradiated MEFs (GlobalStem) in D-MEM/F12 with 20% Knockout Serum Replacement, 1 mM glutamine, 1% NEAA, 15 mM HEPES, 0.1 mM β-mercaptoethanol (all from Invitrogen), 20 ng/mL basic FGF (Stemgent) and were passaged manually every five to seven days. Phoenix cells were grown with 10% FBS (Lonza) in DMEM, 2 mM glutamine, 20 mM HEPES, and 1% NEAA (all from Invitrogen).
Isolation of primary fibroblasts
Dermal explant cultures were established from dispase-treated skin biopsies on fibronectin underneath a glass coverslip with fibroblast media after 5–7 days. After establishment, primary dermal fibroblasts were cultured as described above. Biopsies were performed at the University of California, Los Angeles, under an approved Human Subjects Protocol.
Generation of viruses
Retroviruses were packaged using Phoenix cells (a gift from the laboratory of W. Balch) and Fugene6 (Roche). The four reprogramming vectors ((Takahashi et al., 2007); www.addgene.org) were packaged individually and pseudotyped with VSV-G. Lentiviruses were generated by co-transfecting into 293T cells shRNA constructs with psPAX2 and pMD2.G. Virus-containing ES media supernatant was collected throughout 48 hours after transfection (See Supplemental Experimental Procedures).
Derivation of iPSCs
Previous methods were followed with minor deviations (Park et al., 2008b). Donor fibroblasts were transduced daily for three consecutive days, and four to six days after the last transduction, cells were replated onto MEFs. Beginning one day following, cells were given ES media daily. Colonies were picked between 21 and 28 days after transduction.
Lentiviral shRNA transduction
FRDA iPSCs were subjected to two lentiviral transductions with lentivirus overnight at 37°C with 5 µg/mL polybrene. Cells were then expanded and subjected to 6 days of puromycin selection (0.4 µg/mL) on DR4 drug-resistant MEFs (GlobalStem).
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 (all in PBS). Primary antibodies were incubated at 4°C overnight, and secondary antibodies were incubated at room temperature for one hour followed by nuclear staining with DAPI (See Supplemental Experimental Procedures for antibodies used).
Nucleic Acid Purification
Total RNA was purified using the RNeasy Plus Mini kit (Qiagen) according to the manufacturer. Genomic DNA was purified by phenol/chloroform extraction followed by isopropanol precipitation of cell lysates prepared in total cell lysis buffer (100 mM Tris, 5 mM EDTA, 0.2% SDS, 0.2 M NaCl, 200 µg/mL proteinase K, pH 8).
PCR and Quantitative RT-PCR
For GAA•TTC repeat length PCRs, Phusion polymerase (New England Biolabs) was used according to the manufacturer. Quantitative RT-PCR analysis was done using the qScript One-Step SYBRGreen qRT-PCR kit (Quanta Biosciences) according to the manufacturer. All primers for pluripotent markers were as described (Park et al., 2008b). FXN, MSH2, MSH3, OGG1, and GAPDH primers are described in Supplemental Experimental Procedures. Analysis of relative qRT data was performed using the ΔΔCT method (Livak and Schmittgen, 2001). Detection of retroviral transgenes was done by absolute qRT-PCR using the same kit as above with previously described retrovirus transcript-specific primers (Takahashi et al., 2007). (See Supplemental Experimental Procedures.)
Western analysis
Whole cell extracts (in 50 mM Tris pH 7.4, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100, protease inhibitor; Roche) were electrophoresed in polyacrylamide gels and transferred onto nitrocellulose membranes. Primary antibodies were incubated overnight, and secondary antibodies were incubated one hour at room temperature. Signals were detected using HRP-conjugated secondary antibodies and enhanced chemiluminescence (SuperSignal West Pico, Thermo Scientific). (See Supplemental Experimental Procedures for antibodies used.)
Chromatin immunoprecipitation
Cells were crosslinked first with 1.5 mM dithiobis-succinimidyl propionate (DSP) followed by 1% formaldehyde. Subsequent ChIP procedures were as described (Herman et al., 2006) with MSH2 antibody (Santa Cruz). Analysis by qPCR using primers for the FXN promoter, the region upstream of the GAA•TTC repeats, and the region downstream of the repeats were as described (Herman et al., 2006). Additional primers are listed in Supplemental Experimental Procedures.
Microarray analysis
RNA purification with the MirVana RNA extraction kit (Ambion), labeling with the TotalPrep kit (Ambion), and hybridization to Illumina HT12 arrays were as according to the manufacturers. Data were then filtered, normalized, and hierarchically clustered (Eisen et al., 1998). Differentially expressed genes compared to a set of unaffected iPSCs were detected using a false discovery rate-corrected Student’s t-test at a significance level of αFDR < 0.01. Genes with expression level changes greater than 1.75 were then subjected to functional annotation analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; (Dennis et al., 2003; Huang et al., 2009)) at a significance level of αFDR < 0.01.
HIGHLIGHTS.
FRDA iPSCs retain key molecular features characteristic of the human disease
GAA•TTC repeat instability is observed in iPSCs in the disease-relevant FXN gene
Repeat instability is dependent on the mismatch repair enzyme MSH2
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Perlman (UCLA) for providing skin biopsies and I. Singec, J. Clark, and C. Desponts for valuable advice and guidance. Additionally, we thank V. Lukiyanchuk for virus expertise, K. Clingerman for veterinary assistance, and C. Lynch for microarray analysis. This work was supported by the National Institutes of Neurological Disorders and Stroke (NIH), The Friedreich’s Ataxia Research Alliance (FARA), GoFAR, Ataxia UK, Friedreich’s Ataxia Society Ireland, and Repligen Corporation (Waltham, MA) to J.M.G. and by a fellowship from Families of Spinal Muscular Atrophy (to S.K.). L.C.L. is supported by NIH WRHR K12 Career Development Award and the Hartwell Foundation, and G.A. and J.F.L. are supported by CIRM (CL1-00502, RT1-01108, TR1-01250), NIH (R21MH087925), the Millipore Foundation, and the Esther O’Keefe Foundation. M.N. is supported by the National Ataxia Foundation and the FARA "Kyle Bryant Translational Research Award."
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Microarray data are deposited in the Gene Expression Omnibus as Accession No. GSE22651.
SUPPLEMENTAL INFORMATION
Supplemental Information includes two figures, three tables, Supplemental Experimental Procedures, and can be found with this article online at http://www.cell.com/cell-stem-cell/supplemental/etc.
REFERENCES
- Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- Al-Mahdawi S, Pinto RM, Ruddle P, Carroll C, Webster Z, Pook M. GAA repeat instability in Friedreich ataxia YAC transgenic mice. Genomics. 2004;84:301–310. doi: 10.1016/j.ygeno.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Coppola G, Choi SH, Santos MM, Miranda CJ, Tentler D, Wexler EM, Pandolfo M, Geschwind DH. Gene expression profiling in frataxin deficient mice: microarray evidence for significant expression changes without detectable neurodegeneration. Neurobiol Dis. 2006;22:302–311. doi: 10.1016/j.nbd.2005.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Biase I, Chutake YK, Rindler PM, Bidichandani SI. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One. 2009;4:e7914–e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack Da, Yang J, Gao W, Lane HC, Lempicki Ra. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3–P3. [PubMed] [Google Scholar]
- Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. 2009:297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA.TTC repeat expansion in human cell lines. PLoS genetics. 2009;5:e1000704–e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, Kucherlapati RS, Edelmann W, Lunetta KL, MacDonald ME, et al. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld JM. Small molecules affecting transcription in Friedreich ataxia. Pharmacol Ther. 2007;116:236–248. doi: 10.1016/j.pharmthera.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen AC, Di Prospero Na, Parker JS, Fannin RD, Chou J, Meyer JN, Halweg C, Collins JB, Durr A, Fischbeck K, et al. Altered gene expression and DNA damage in peripheral blood cells from Friedreich's ataxia patients: cellular model of pathology. PLoS genetics. 2010;6:e1000812. doi: 10.1371/journal.pgen.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nature Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki Ra. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iyer RR, Wells RD. Expansion and Deletion of Triplet Repeat Sequences in Escherichia coli Occur on the Leading Strand of DNA Replication. Journal of Biological Chemistry. 1999;274:3865–3865. doi: 10.1074/jbc.274.6.3865. [DOI] [PubMed] [Google Scholar]
- Kim HM, Narayanan V, Mieczkowski PA, Petes TD, Krasilnikova MM, Mirkin SM, Lobachev KS. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. Embo J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer PR, Pearson CE, Sinden RR. Stability of triplet repeats of myotonic dystrophy and fragile X loci in human mutator mismatch repair cell lines. Hum Genet. 1996;98:151–157. doi: 10.1007/s004390050179. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda CJ, Santos MM, Ohshima K, Smith J, Li L, Bunting M, Cossee M, Koenig M, Sequeiros J, Kaplan J, et al. Frataxin knockin mouse. FEBS Lett. 2002;512:291–297. doi: 10.1016/s0014-5793(02)02251-2. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Ann Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Park I-H, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I-H, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008b;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pianese L, Cavalcanti F, De Michele G, Filla A, Campanella G, Calabrese O, Castaldo I, Monticelli A, Cocozza S. The effect of parental gender on the GAA dynamic mutation in the FRDA gene. Am J Hum Genet. 1997;60:460–463. [PMC free article] [PubMed] [Google Scholar]
- Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Coppola G, Geschwind DH, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PloS one. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rindler PM, Clark RM, Pollard LM, De Biase I, Bidichandani SI. Replication in mammalian cells recapitulates the locus-specific differences in somatic instability of genomic GAA triplet-repeats. Nucleic Acids Res. 2006;34:6352–6361. doi: 10.1093/nar/gkl846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. The EMBO journal. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishkin Aa, Voineagu I, Matera R, Cherng N, Chernet BT, Krasilnikova MM, Narayanan V, Lobachev KS, Mirkin SM. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Molecular cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential Modeling of Fragile X Syndrome by Human Embryonic Stem Cells and Induced Pluripotent Stem Cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RD. DNA triplexes and Friedreich ataxia. Faseb J. 2008;22:1625–1634. doi: 10.1096/fj.07-097857. [DOI] [PubMed] [Google Scholar]
- Zhao J, Jain A, Iyer RR, Modrich PL, Vasquez KM. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Research. 2009;37:4420–4429. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.