Abstract
The cholera toxin genes of Vibrio cholerae are encoded by the filamentous phage, CTXΦ. Chromosomal CTXΦ prophage DNA is often found flanked by copies of a related genetic element designated RS1, and RS1 DNA can be packaged into filamentous phage particles (designated RS1Φ) by using the CTXΦ morphogenesis genes. RS1Φ is a satellite phage that further controls expression and dissemination of CTXΦ. Here we describe a CTXΦ-independent mechanism for production of RS1Φ. A nontoxigenic environmental V. cholerae strain (55V71) was identified that supports production of RS1Φ. However, newly infected CTX-negative strains did not produce RS1Φ, indicating that additional 55V71 genes were involved in production of RS1Φ. Analysis of nucleic acids from phage preparations of 55V71 revealed a 7.5-kb single-stranded DNA, whose corresponding replicative form was found in plasmid preparations. This DNA likely corresponds to the genome of a new filamentous phage, which we have designated KSF-1Φ. The replicative form DNA of KSF-1Φ was cloned into pUC18, and the resulting construct pKSF-1.1 supported the production of RS1Φ particles by CTX-negative V. cholerae strains. RS1Φ particles produced in this way infect recipient V. cholerae strains by a mechanism that is independent of the CTXΦ receptor, the toxin-coregulated pilus. Thus, KSF-1Φ is capable of facilitating the transfer of the RS1 element to strains that do not express toxin coregulated pilus. Given that RS1Φ can enhance coproduction of CTXΦ particles, KSF-1Φ-mediated dissemination of RS1 may indirectly promote the spread of toxin genes among V. cholerae strains. This study also shows that filamentous phages can package diverse DNA elements and thus may play a role in horizontal transfer of more genes than previously appreciated.
The Gram-negative bacterium Vibrio cholerae is the etiologic agent of cholera, a severe diarrheal disease that affects tens of thousands of people worldwide each year (1, 2). Cholera can occur as spreading epidemics, and pandemic clones exist that spread globally and, remarkably, displace local endemic strains (2). The properties that make one pathogenic clone of V. cholerae more evolutionarily fit than another are, for the most part, poorly understood but are thought to have their origin in the acquisition of novel genes or horizontally mobile genetic elements (1, 3–6).
The ability of pathogenic V. cholerae to cause disease depends primarily on the expression of two virulence factors: a potent enterotoxin (cholera toxin; CT) and a pilus colonization factor (toxin coregulated pilus; TCP) (1). The genes encoding cholera toxin (ctxAB) are carried in the genome of a filamentous bacteriophage designated CTXΦ (4). The genes required for TCP biogenesis and function also appear to be transferred as a cluster among V. cholerae strains, though the mechanism by which this occurs has not been clearly defined (6–8). Remarkably, the CTXΦ uses TCP as its receptor for infecting new strains (4), and thus these two horizontally mobile elements are linked evolutionarily. These and other observations have led to the theory that new pathogenic clones arise from the transfer of virulence-related gene clusters into environmental, nonpathogenic V. cholerae strains (1, 3). Thus, understanding the horizontal transfer of genes by bacteriophages, pathogenicity islands and other accessory genetic elements should provide insights into the emergence and evolution of bacterial pathogens.
Toxigenic V. cholerae strains are CTXΦ lysogens, and the CTXΦ prophage is inserted in a site called attRS1 near the terminus of DNA replication on the bacterium's large chromosome (9, 10). The CTXΦ genome consists of a core region that encodes CT as well as functions that are required for virion morphogenesis, and an RS2 region that encodes the regulation, replication, and integration functions of the CTXΦ genome (4, 11). The CTXΦ prophage is often flanked by a related genetic element known as the RS1 (10, 11). The RS1 element is very similar to the RS2 region of CTXΦ in genetic and functional aspects (11).
We recently reported that the RS1 element is in fact the genome of a satellite phage that utilizes CTXΦ morphogenesis genes to produce RS1Φ particles (12). RS1Φ particles formed with CTXΦ virion proteins efficiently infect only recipient V. cholerae strains that express the CTXΦ receptor, the TCP (12). Similar results have been reported by Davis et al. (13), who have further shown that RS1 controls production of CTXΦ phage particles. RS1 carries an additional ORF termed rstC that is not present in RS2 (11). RstC is an antirepressor that controls CTXΦ lysogeny, production of CTXΦ particles, and expression of cholera toxin (13). Together, these results suggest that the interplay between the RS1Φ and CTXΦ prophages promotes more efficient dissemination of the CT genes, while simultaneously enhancing the virulence and evolutionary fitness of at least those V. cholerae strains that carry TCP genes.
Our earlier work showed that many clinical and environmental V. cholerae isolates that express TCP are susceptible to a kanamycin-resistance (KmR)-marked version of RS1Φ (12). However, RS1 can also be introduced into strains by electroporation with pRS1-Km, a KmR-marked plasmid form of the RS1 element (12). Transfer of RS1-Km into various clinical and environmental recipient usually resulted in the element either inserting into the chromosomal attRS site or being maintained as a plasmid, the latter particularly in strains that did not contain the attRS site (12). In CTX-negative strains, the RS1Φ genome did not produce RS1Φ particles. In the present study, we show that one nontoxigenic strain of V. cholerae produces RS1Φ even though this strain lacks the CTXΦ genome. Production of RS1Φ particles by this strain depends on functions encoded by a recently discovered filamentous phage that we have named KSF-1Φ. We also found that RS1Φ produced by means of this process are capable of infecting recipient strains in a TCP-independent manner. This finding constitutes another example of horizontal gene transfer mediated by alternative packaging of a different DNA element by a filamentous phage and has significant implications regarding the evolution of pathogenic V. cholerae strains.
Materials and Methods
Bacterial Strains, Plasmids, and Phages.
Bacterial strains, phages, and plasmids used in this study are described in Table 1. Clinical strains were originally isolated from patients who attended the treatment center of the International Centre for Diarrhoeal Disease Research, Bangladesh located in Dhaka. The environmental strains, including strain 55V71, were obtained from surface waters in Dhaka. Strains were stored either in lyophilized form or in sealed deep nutrient agar at room temperature. Before use, the identities of the V. cholerae cultures were confirmed by biochemical reaction and serology, and the presence or absence of the CTXΦ prophage, TCP pathogenicity island, RS1 element, as well as the CTXΦ attachment sequence attRS, was ascertained by using specific DNA probes or PCR assays as described below.
Table 1.
Characteristics of bacterial strains, plasmids, and phages used in the study
| Strains and plasmids | Relevant characteristics | Source or ref. |
|---|---|---|
| PRS1-Km | Extrachromosomal replicative form of the RS1 element carrying an intergenic KmR marker | 5 |
| RV-508 | V. cholerae O1 classical biotype strain that constitutively expresses CT, TCP, and other toxR-regulated gene products | 1 |
| 55V71 | Nontoxigenic (CTX-negative) environmental V. cholerae non-O1 non-O139 strain | This study |
| 55V71 (pRS1-Km) | Strain 55V71 carrying pRS1-Km | This study |
| KSF-1Φ | A filamentous phage isolated from strain 55V71 | This study |
| PKSF-1.1 | Replicative form of KSF-1Φ genome cloned into the XbaI site of pUC18 | This study |
| S-224 | Toxigenic V. cholerae O1 classical biotype strain | Collection |
| AF-1471, AK-31047 | Toxigenic V. cholerae O1 El Tor strains | Collection |
| AL-11089 | Toxigenic V. cholerae O139 strain | 5 |
| Env-99 | Environmental CTXΦ-negative V. cholerae O139 strain | 15 |
| Env-99 (RS1-Km) | Strain Env-99 lysogenized with RS1-KmΦ | This study |
| Env-99 (RS1-Km, pKSF1.1) | Env-99 (RS1-Km) electroporated with pKSF1.1 | This study |
| Env-002 | Environmental CTXΦ-negative V. cholerae O1 strain | 15 |
| Env-002 (RS1-Km) | Strain Env-002 lysogenized with RS1-KmΦ | This study |
| Env-002 (RS1-Km, pKSF-1.1) | Env-002 (RS1-Km) electroporated with pKSF1.1 | This study |
| SA-317, SA-406 | Clinical nontoxigenic V. cholerae O1 El Tor strains | 9 |
| SA-317 (RS1-Km) | Strain SA-317 lysogenized with RS1-KmΦ | This study |
| SA-406 (RS1-Km) | Strain SA-406 lysogenized with RS1-KmΦ | This study |
| AM-15714, AM-15729 | Clinical CTX-negative non-O1-non-0139 strains | Collection |
| AM-15714 (pRS1-Km) | Strain AM-15714 carrying pRS1-Km | This study |
| AM-15714 (pRS1-Km, pKSF1.1) | AM-15714(pRS1-Km) electroporated with pKSF1.1 | This study |
| AM-15729 (RS1-Km) | Strain AM-15729 lysogenized with RS1-KmΦ | This study |
| AM-15729 (RS1-Km, pKSF1.1) | AM-15729(RS1-Km) electroporated with pKSF1.1 | This study |
PCR and Hybridization Analysis.
The RS1Φ genome was detected as previously described by positive hybridization with rstR and rstC gene probes and by negative reactions with probes corresponding to the CTXΦ core region (12). CTXΦ gene probes were described previously and included ctxA (14), zot, ace, and part of orfU (4, 15), or a 5.9-kb XbaI fragment of pCTX-Km representing almost the entire CTXΦ genome, except ctxA (4). Strand-specific oligonucleotide probes (5′-TTACAGTGATGGATCAGTCAAT and 5′-ATGAGTTTGAAACCATACACTTT, corresponding to the (+) and (−) strand respectively of the RS1 element) were also used to detect the single-stranded RS1Φ genome, and to distinguish between the phage genome and its replicative form (RF) DNA (12). A probe derived from the single-stranded KSF-1Φ genome by random-primed labeling was used to detect the RF of the phage genome (pKSF-1) in plasmid preparations. Conversely, relevant fragments of pKSF-1 were used to detect the KSF-1Φ genome whenever appropriate, including assessing the distribution of KSF-1Φ among naturally occurring V. cholerae strains.
Colony or Southern blots were prepared by using nylon filters (Hybond, Amersham Pharmacia), and processed by standard methods (16, 17). The polynucleotide probes were labeled by random priming (18) using a random primer DNA labeling kit (Invitrogen) and [α-32P]-deoxycytidine triphosphate (3,000 Ci/mmol, Amersham Pharmacia; 1 Ci = 37 GBq), and oligonucleotide probes were labeled by 3′tailing by using terminal deoxynucleotide transferase and [α-32P]-dCTP. Blots were hybridized with the probes and autoradiographed as described (12, 15).
PCR Assays.
All oligonucleotides used as probes or PCR primers were obtained from Oswel DNA Service (University of Edinburgh, Edinburgh), and PCR reagents were purchased from Perkin–Elmer. Presence of tcpA genes specific for the classical and El Tor biotypes was detected by using a multiplex PCR assay (19). Presence of the tcpI and acfB genes was also detected by PCR assays as described (15).
Preparation of Phage.
Overnight cultures of V. cholerae were diluted 100-fold in fresh LB medium and grown for 6 h at 30°C with shaking. Supernatants were sterilized by filtration through 0.22-μm filters (Millipore). To confirm that the filtrates did not contain bacterial cells, aliquots of the filtrates were streaked on Luria agar plates and incubated overnight at 37°C. For phage assays, aliquots of the sterile supernatants were incubated with the recipient strains and plated on appropriate antibiotic plates.
For isolation and analysis of phage nucleic acids, the filtrates were mixed with one-fourth volume of a solution containing 20% polyethylene glycol (PEG-6000) and 10% NaCl, and centrifuged at 12,000 × g to precipitate the phage particles. The precipitate was dissolved in a solution containing 20 mM Tris⋅Cl (pH 7.5), 60 mM Kcl, 10 mM MgCl, 10 mM NaCl, and digested with pancreatic DNaseI (100 units/ml) and RNase A (50 μg/ml) at 37°C for 2 h to remove possible nucleic acids carried over from lysed bacterial cells. The solution was extracted with phenolchloroform, and the total nucleic acids were precipitated with ethanol. The nucleic acids were analyzed by enzymatic digestion and by Southern blot hybridization using appropriate probes to detect the presence of relevant phage genomes.
Preparation and Analysis of pKSF-1.1.
All in vitro DNA manipulations were accomplished according to standard techniques (16). Plasmid DNA was prepared from strain 55V71 by standard methods and purified by using microcentrifuge filter units (Ultrafree-Probind; Sigma). Southern blots of the plasmid preparations were hybridized by using a probe derived from the single-stranded KSF-1Φ genome to identify the RF of the phage genome (pKSF-1). Southern blot analysis of restriction endonuclease cleavage sites within pKSF-1 was also performed (17). pKSF-1 was electroeluted from an agarose gel, and cloned into the XbaI site of pUC18 to construct pKSF-1.1.
Transduction Assays.
Susceptibility of recipient strains to RS1-KmΦ was assayed with phage prepared from culture supernatants of 55V71 (pRS1-Km) by using the 55V71 native strain as well as strain RV508 as positive control recipients. The assays were done both under in vitro laboratory conditions and in adult rabbit ileal loops by using described methods (20, 21). Briefly, phage particles were precipitated from 50-ml aliquots of filtered sterile supernatants, and the pellet was suspended in 100 μl of TES buffer (20 mM Tris⋅HCl, pH 7.5/10 mM NaCl/0.1 mM Na2EDTA). Recipient cells were grown in LB at 30°C, precipitated by centrifugation, and washed in fresh LB. Approximately 105 bacterial cells were mixed with 10 μl of the phage preparation in a final volume of 100 μl, inoculated into ileal loops of adult rabbits obtained from the Animal Resources Branch of International Centre for Diarrhoeal Disease Research, Bangladesh, and prepared as described (21). For each recipient strain, at least five loops were inoculated. After 16 h, the rabbits were killed, and the contents of the ileal loops were collected. The insides of the loops were washed out with 1 ml of 10 mM PBS (pH 7.4), and dilutions of the ileal loop washes were plated on Luria agar plates containing kanamycin (50 μg/ml), and on plates without kanamycin. The ratio of KmR-transduced colonies to the total number of colonies derived from the recipient strain was calculated and expressed as the percentage of recipient cells infected. For in vitro assays, mixtures of phage and recipient cells were prepared as described above. Each mixture was inoculated into 5 ml of LB and incubated for 16 h at 30°C, and aliquots of the culture were plated as described above to determine the percentage of recipient cells infected.
Representative infected colonies were grown in LB containing kanamycin (50 μg/ml), and analyzed for the production of fresh RS1-KmΦ particles in the culture supernatant. Integration of the phage genome into the chromosome of the recipient cells was studied by comparative Southern blot analysis of total DNA and plasmid preparations from the phage-infected and the corresponding native strains as described (20, 21).
Results
CTXΦ-Independent Production of RS1Φ.
Previous work showed that the RS1 element is a satellite phage that utilizes CTXΦ morphogenesis genes to produce RS1Φ particles, and that the CTXΦ genes are essential for the production of RS1Φ (12, 13). To further define the functions required for RS1Φ production, we introduced pRS1-Km by electroporation into a number of different strains, including some that were TCP-negative (unpublished data). To determine whether any of these strains were capable of producing extracellular phage particles, fresh culture supernatants of the strains were examined for the presence of KmR transducing particles. As expected, toxigenic strains carrying pRS1-KmΦ produced phage particles in the culture supernatant (Table 3), but surprisingly, one CTXΦ-negative strain (55V71) was also able to produce extracellular RS1-KmΦ. DNA hybridization and PCR assays showed that strain 55V71 was negative for all CTXΦ genes, as well as for the TCP pathogenicity island and the attachment sequence attRS (Table 2). Despite the absence of the CTXΦ genome in 55V71, cell-free culture supernatants from derivatives of 55V71 carrying pRS1-Km were able to transmit KmR to a number of recipient V. cholerae strains (Table 2), suggesting that KmR-transducing particles were produced when a genetically marked RS1Φ genome (pRS1-Km) was present in the same cells. The distribution of the CTXΦ, the TCP pathogenicity island, and the CTXΦ attachment sequence attRS, in the recipient strains is also shown in Table 2.
Table 3.
Production of RS1-Km phage particles by V. cholerae strains carrying an RS1-Km element
| Strain | Titer of RS1-KmΦ (particles per ml) using different host strains*
|
|
|---|---|---|
| RV508 | 55V71 | |
| 55V71 (pRS1-Km)† | 3.21 × 103 | 3.72 × 103 |
| AF1471 (pRS1-Km) | 0.19 × 102 | 0.15 × 102 |
| AL-11089 (pRS1-Km) | 0.18 × 102 | 0.16 × 102 |
| SA-317 (RS1-Km) | 0 | 0 |
| SA-317 (RS1-Km, pUC18) | 0 | 0 |
| SA-317(RS1-Km, pKSF-1.1) | 0.09 × 102 | 0.11 × 102 |
| Env-02 (pRS-Km) | 0 | 0 |
| Env-02 (pRS1-Km, pUC18) | 0 | 0 |
| Env-02 (pRS1-Km, pKSF-1.1) | 0.05 × 102 | 0.12 × 102 |
| Env-99 (pRS1-Km) | 0 | 0 |
| Env-99 (pRS1-Km, pUC18) | 0 | 0 |
| Env-99 (pRS1-Km, pKSF-1.1) | 0.21 × 102 | 0.11 × 102 |
| AOE-12/39 (RS1-Km) | 0 | 0 |
| AOE-12/39 (RS1-Km, pUC18) | 0 | 0 |
| AOE-12/39 (RS1-Km, pKSF-1.1) | 0.16 × 102 | 0.21 × 102 |
| Env-002 (pRS1-Km) | 0 | 0 |
| Env-002 (pRS1-Km, pUC18) | 0 | 0 |
| Env-002 (pRS1-Km, pKSF-1.1) | 2.33 × 102 | 2.96 × 103 |
| AM15729 (RS1-Km) | 0 | 0 |
| AM15729 (RS1-Km, pUC18) | 0 | 0 |
| AM15729 (RS1-Km, pKSF-1.1) | 0.31 × 102 | 0.26 × 102 |
| AM15714 (pRS1-Km) | 0 | 0 |
| AM15714 (pRS1-Km, pUC18) | 0 | 0 |
| AM15714 (pRS1-Km, pKSF-1.1) | 3.11 × 102 | 4.16 × 102 |
Values are averages of three independent assays.
Strain 55V71 is a naturally occurring environmental strain that carries KSF-1 phage.
Table 2.
Presence of CTX- and TCP-specific genes among clinical and environmental V. cholerae O1 and non-O1 strains, and susceptibility of the strains to RS1-KmΦ derived from strain 55V71(pRS1-Km)
| Strain | Description | Presence of*
|
Susceptibility to RS1-Kmֆ
|
Integration of phage genome‡ | |||
|---|---|---|---|---|---|---|---|
| CTX | TCP | attRS | In vitro | In rabbit | |||
| AK-31047 | O1, El Tor | + | + | + | 1.1 × 10−5 | 7.3 × 10−6 | − |
| AF-1471 | O1, El Tor | + | + | + | 4.1 × 10−6 | 2.8 × 10−6 | − |
| SA-317 | O1, El Tor | − | + | + | 1.8 × 10−4 | 2.1 × 10−4 | + |
| SA-406 | O1, El Tor | − | + | + | 7.8 × 10−4 | 1.1 × 10−3 | + |
| RV508 | O1, classical | + | + | + | 2.5 × 10−3 | 1.9 × 10−3 | − |
| S-224 | O1, classical | + | + | + | 1.2 × 10−4 | 2.7 × 10−3 | − |
| AL-11089 | O139 | + | + | + | 1.6 × 10−5 | 2.1 × 10−5 | − |
| Env-99 | O139 | − | + | + | 7.5 × 10−4 | 5.2 × 10−4 | + |
| Env-002 | O1, El Tor | − | + | + | 6.3 × 10−4 | 1.9 × 10−3 | + |
| 55V71 | Non-O1, non-O139 | − | − | − | 2.4 × 10−3 | 9.5 × 10−4 | − |
| AM-15714 | Non-O1, non-O139 | − | − | − | 5.9 × 10−4 | 7.5 × 10−4 | − |
| AM15729 | Non-O1, non-O139 | − | − | + | 1.8 × 10−4 | 2.2 × 10−4 | + |
Presence of different genes was detected by using DNA probes and PCR assays.
Susceptibility to the phage was determined by using a genetically marked phage. See text for details.
Integration of the phage genome into the host chromosome was determined by Southern blot analysis of genomic and plasmid DNA isolated from native and infected strains.
V. cholerae Strain 55V71 Possesses a Novel Filamentous Phage, KSF-1Φ.
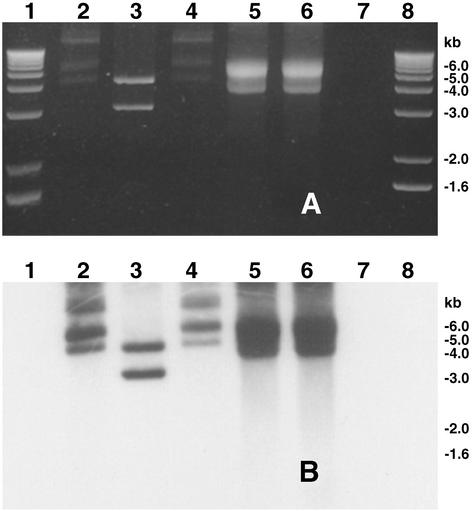
Because it was known that the RS1 element itself does not encode all of the functions required for morphogenesis of an RS1Φ (12, 13), the finding that 55V71 produced RS1-Km transducing particles raised the question of whether CTXΦ-independent production of RS1Φ in strain 55V71 was supported by additional genes or a phage genome other than CTXΦ. We therefore analyzed total phage nucleic acid preparations from strain 55V71 and discovered the presence of a 7.5-kb DNA that was resistant to restriction endonuclease digestion but sensitive to both DNaseI and the single-strand-specific nuclease, mung bean nuclease (Fig. 1). The 7.5-kb single-stranded DNA (ssDNA) was apparently present in particles, because it was protected from nuclease digestion during the phage preparation. These findings suggested that the 7.5-kb ssDNA was the genome of a filamentous phage that we designated KSF-1Φ. The presence of a RF of the ssDNA was also revealed by using probes derived from the 7.5-kb ssDNA for Southern hybridization analysis of plasmid preparations from 55V71. A preliminary restriction analysis of pKSF-1 showed that the KSF-1 genome is distinct from that of the CTXΦ genome and those of other previously reported filamentous phages of V. cholerae (22–26). Neither the ssDNA nor the corresponding RF DNA (pKSF-1) hybridized with probes representing the entire CTXΦ genome, indicating that KSF-1Φ is very different from CTXΦ.
Figure 1.
Plasmid and phage DNA preparations of strain 55V71 were digested with the restriction endonuclease HindIII or single-strand-specific endonuclease mung bean nuclease, electrophoresed in agarose gel (A), and analyzed by Southern blot hybridization (B) using a gene probe derived from KSF-1Φ genome. Lane 2, undigested plasmid DNA; lane 3, plasmid DNA digested with HindIII; lane 4, plasmid DNA digested with mung bean nuclease; lane 5, undigested phage DNA; lane 6, phage DNA digested with HindIII; lane 7, phage DNA digested with mung bean nuclease. Lanes 1 and 8 show the 1-kb DNA ladder used as molecular size markers (Invitrogen).
CTXΦ-Independent Transfer of RS1 Is Mediated by KSF-1Φ and Does Not Require TCP.
When recipient strains were infected with RS1-KmΦ, the cells maintained the RS1Φ genome either as a plasmid or as an integrated RS1 element, but no CTX-negative strain except 55V71 produced RS1-KmΦ in the culture supernatant (Table 3). The discovery of KSF-1Φ in strain 55V71 suggested that it might support CTXΦ-independent morphogenesis of RS1Φ.
To test this hypothesis, we examined whether the KSF-1Φ genome from 55V71 could complement CTX-independent production of RS1Φ in other nontoxigenic strains. Because the wild-type KSF-1Φ did not have a selectable marker, we cloned the entire RF DNA of KSF-1Φ into the XbaI site of pUC18 to construct pKSF-1.1. Nontoxigenic V. cholerae strains initially infected with RS1-KmΦ were electroporated with pKSF-1.1, and recipient strains containing both pKSF-1.1 and pRS1-Km were selected by their resistance to both ampicillin and kanamycin. Analysis of RS1-KmΦ production showed that strains that simultaneously carried pRS1-Km (or its integrated form) and pKSF-1.1, produced pRS1-KmΦ in the culture supernatant. However, strains carrying either pRS1-Km alone, or pRS1-Km and pUC18 simultaneously, did not produce detectable pRS1-KmΦ in the culture supernatant (Table 3). These results confirmed that the KSF-1Φ genome can mediate production of RS1Φ in the absence of CTXΦ.
By using strains RV508 and native 55V71 as recipients, 55V71(pRS1-Km) supernatants were titered for KmR-transducing activity. These assays detected at least 3.2 × 103 transducing particles per ml of the supernatant fluid (Table 3). The susceptibility of the strains to 55V71-derived transducing particles varied between 1.1 × 10−5 and 2.4 × 10−3 in in vitro assays, and between 2.8 × 10−6 and 1.9 × 10−3 in rabbit ileal loops (Table 2), indicating that there was no major difference between the frequency of infection in vivo and under laboratory conditions.
In previous studies, when RS1Φ was produced by using the CTXΦ morphogenesis genes, the RS1Φ particles could infect only recipient cells that expressed TCP, the receptor for CTXΦ (4, 12). Because RS1Φ can evidently be produced by using KSF-1Φ genes without the participation of the CTXΦ genome, we examined whether RS1Φ derived from a strain bearing KSF-1Φ required TCP for infection. RS1-KmΦ isolated from strain 55V71 (pRS1-Km) was used to test the susceptibility of different TCP-positive and TCP-negative strains, both in vitro and in rabbit ileal loops. Twelve V. cholerae strains belonging to the O1, O139 or non-O1, non-O139 serogroups were tested, and all were susceptible to infection by RS1-KmΦ (Table 2). Non-O1 non-O139 strains that were negative for the TCP pathogenicity island were infected by RS1-KmΦ with similar efficiency as TCP-positive recipients. There was also no significant difference in susceptibility when comparing infections performed in vitro and in rabbit ileal loops. In studies of CTXΦ, the efficiency of transduction was considerably higher inside the intestines of infant mice or in rabbit ileal loops compared with in vitro conditions, and this was attributed to more efficient expression of the phage receptor, TCP, in vivo (4, 15, 21, 27). Therefore, the absence of a difference in susceptibility between in vitro and in vivo conditions supports the hypothesis that TCP is not the receptor for RS1Φ produced by a CTXΦ-negative strain.
Distribution of KSF-1Φ Sequences Among Environmental Strains of V. cholerae.
Examination of the distribution of KSF-1Φ among naturally occurring V. cholerae strains showed that besides strain 55V71, DNA homologous to KSF-1Φ genomic DNA was present in another 3 of 26 (11.5%) environmental non-O1 non-O139 strains. When three strains carrying KSF-1 homologous sequences were electroporated with pRS1-Km, all produced RS1-KmΦ particles.
Discussion
In this study, we describe a previously uncharacterized filamentous vibriophage (designated KSF-1Φ) that is capable of packaging the RS1 element of V. cholerae and transmitting it to diverse V. cholerae strains. KSF-1Φ-mediated transfer of RS1 requires neither CTXΦ nor TCP. The genetic structure of KSF-1Φ differs from that of previously identified filamentous vibriophages, and the KSF-1 genome is present in a significant fraction of environmental V. cholerae strains. Our results show that the RS1 element can be packaged into at least two kinds of bacteriophage particles, which have different receptors.
At present, little is known about the structure and biology of KSF-1Φ, and further studies are necessary to characterize this phage further and identify its receptor. However, because RS1Φ elaborated by KSF-1Φ-positive strains is presumably produced by using KSF-1Φ virion proteins, it seems possible that the receptor for this RS1Φ is the same as KSF-1Φ receptor. This receptor is not TCP, but is apparently ubiquitous among V. cholerae strains because all strains examined in this study were susceptible to RS1Φ produced from 55V71. Filamentous phages are known to use pili as their receptors, and V. cholerae produces several types of pili and also carries genes for many pilin-like proteins (9). Hence, it is likely that the KSF-1 phage receptor is a pilus other than TCP. It may be mentioned that at least one other filamentous vibriophage has been shown to use the mannose-sensitive hemagglutinin pilus as its receptor instead of TCP (28).
If we are correct that the receptor for the KSF-1 phage is widely distributed among V. cholerae clinical and environmental strains, then it is apparent that KSF-1Φ (and RS1Φ, if present in the same cells) could spread rapidly among these strains. We found that besides strain 55V71, KSF-1Φ related sequences were detected in another 3 of 26 environmental non-O1, non-O139 strains tested in this study. This level of distribution of KSF-1Φ among non-O1, non-O139 V. cholerae population may be significant, particularly in view of the ability of the phage to package alternative DNA elements such as RS1. It remains to be shown that KSF-1Φ particles can mediate transfer of its own genome to recipient cells, but given that KSF-1 DNA can be detected as single-stranded DNA in cell-free particles, and that the RF of KSF-1 also packages RS1 DNA into infectious particles, we tentatively conclude that KSF-1 is a viable phage capable of self-transmission in nature.
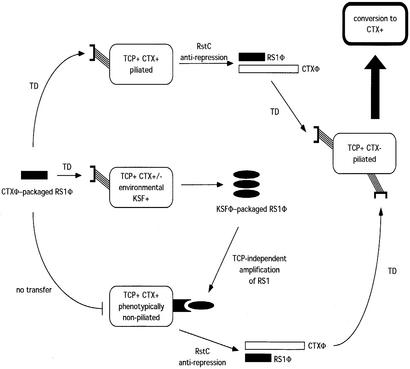
The findings presented here define a mechanism by which the RS1 element can be easily transmitted to both TCP-negative and TCP-positive recipients, thus maximizing the horizontal transfer of RS1 element in the natural habitat. Perhaps more critically, it indicates that RS1 can be acquired by toxigenic TCP+ strains that are not expressing TCP. Little is known about the expression of TCP in the aquatic habitats of V. cholerae, but it is reasonable to assume that its expression would be low compared with the intraintestinal environment. If so, there may be relatively few opportunities for toxigenic, TCP+ strains to acquire RS1 because they might be exposed to CTXΦ-assembled RS1Φ particles only within the intestine. However, because KSF-1Φ-assembled RS1Φ particles use a different receptor, they can infect such strains long after they have stopped expressing TCP (for example, after being shed from the host). The vast majority of V. cholerae clinical isolates, which include O1 El Tor and O139 V. cholerae, carry one or more copies of RS1 (29–32), suggesting that acquisition of the satellite phage RS1Φ is probably quite efficient in nature and occurs by multiple mechanisms. A hypothetical model of KSF-1Φ-enhanced dissemination of RS1 satellite phage and its probable impact on the dissemination of CTXΦ is presented in Fig. 2.
Figure 2.
Hypothetical model of KSF-1Φ-enhanced dissemination of CTXΦ. Infectious particles carrying the RS1 genome are indicated by black shading. RS1Φ particles packaged by CTXΦ use TCP as the receptor for infecting recipient cells, whereas RS1Φ particles packaged by KSF-Φ1 use an unidentified but ubiquitous receptor for infecting recipient cells. TD, transduction.
The consequences of acquisition of RS1Φ by toxigenic TCP-positive strains is only just beginning to be understood in molecular detail. For example, it is known that the RstC protein encoded by the RS1 element modulates the expression of several CTXΦ genes (including ctxAB) by activating promoters controlled by the CTXΦ and RS1Φ lysogenic repressor RstR (13). RstC-mediated RstR antirepression appears to be a mechanism by which RS1Φ assures some production of RS1Φ phage particles even in lysogenized cells. However, RstC enhancement of RS1Φ particle production also enhances CTXΦ particle production, thus promoting the dissemination of both the satellite and the helper phage (13).
Furthermore, because RstC promotes expression of CT via activation of RstR-repressed phage promoters, RS1Φ potentially enhances the virulence of CTXΦ, RS1Φ double lysogens (13). If the double lysogens produce a larger or longer diarrheal purge, it seems likely that more bacterial as well as phage progeny will result from each clinical event. RS1Φ would thus contribute to the improved evolutionary fitness of V. cholerae as a pathogen, and this in turn may promote its more efficient seeding of the environment and establishment of endemicity. In this regard, it is worth noting that all V. cholerae strains associated with the 7th and 8th cholera pandemics carry RS1Φ, whereas the 6th pandemic strains they displaced do not (33). It is therefore tempting to speculate that RS1Φ-encoded properties such as RstC may be responsible for their phenomenal success as pathogens, though these strains do carry other unique chromosomal segments that may also contribute to their improved fitness as pandemic clones (34).
It is also possible that RS1Φ might alter properties of V. cholerae that are unconnected with CTXΦ per se. We are investigating this possibility through expression profiling of RS1Φ lysogens using a V. cholerae genomic microarray (34). However, we predict that the incremental improvement in the evolutionary fitness of V. cholerae afforded by acquisition of RS1Φ alone will be minor simply because very few environmental or clinical strains have been described that carry solely RS1Φ (35).
This and other studies (12, 13) indicate that some filamentous vibriophages have the ability to alternatively package different DNA elements, and mediate the horizontal transfer of genetic material. Because of their flexible capsid structure, filamentous phages may be uniquely suited to mobilizing genes and larger elements. In addition to the examples of CTXΦ and KSF-1Φ in V. cholerae, a gene (orf 8) associated with a filamentous vibriophage has been linked exclusively to strains associated with the “pandemic” O3:K6 clone of Vibrio parahemolyticus (36, 37). Finally, it should be noted that the TCP pathogenicity island is also believed to have been recently acquired in V. cholerae evolutionary history (5). A previous report concluded that the TCP pathogenicity island corresponds to the genome of another filamentous phage (designated VPIΦ), which uses TcpA as its coat protein (38). However, repeated attempts by several different laboratories, including our own, have failed to confirm this report (S.M.F., J. Zhu, M.K., Asadulghani, and J.J.M., unpublished data). In the light of the results presented here, it seems possible that another filamentous vibriophage might be capable of packaging the TCP island. Clearly, further studies are required to understand how vibriophages interact with V. cholerae to promote the latter's acquisition of the critical genes that alter its virulence or adaptation to its environmental niche.
Acknowledgments
We thank Su Chiang for assistance with the manuscript. This research was funded in part by the United States Agency for International Development under Grant HRN-5986-A-00-6005-00 with the International Centre for Diarrhoeal Disease Research, Bangladesh, and the Ellison Medical Foundation Grant ID-T-0007-01 under a subagreement between the Harvard Medical School and International Centre for Diarrhoeal Disease Research, Bangladesh. The International Centre for Diarrhoeal Disease Research, Bangladesh is supported by countries and agencies that share its concern for the health problems of developing countries.
Abbreviations
- CT
cholera toxin
- TCP
toxin coregulated pilus
- RF
replicative form
- KmR
kanamycin resistance
References
- 1.Faruque S M, Albert M J, Mekalanos J J. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper J B, Morris J G, Levine M M. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faruque S M, Nair G B. Microbiol Immunol. 2002;46:59–66. doi: 10.1111/j.1348-0421.2002.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 4.Waldor M K, Mekalanos J J. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 5.Taylor R, Shaw C, Peterson K, Spears P, Mekalanos J J. Vaccine. 1988;6:151–154. doi: 10.1016/s0264-410x(88)80019-7. [DOI] [PubMed] [Google Scholar]
- 6.Kovach M E, Shaffer M D, Peterson K M. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 7.Ogierman M A, Zabihi S, Mourtzios L, Manning P A. Gene. 1993;126:51–60. doi: 10.1016/0378-1119(93)90589-u. [DOI] [PubMed] [Google Scholar]
- 8.Karaolis D K, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldor M K, Rubin E J, Gregory D N, Kimsey H H, Makalanos J J. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 12.Faruque S M, Asadulghani, Kamruzzaman M, Nandi R K, Ghosh A N, Nair G B, Mekalanos J J, Sack D A. Infect Immun. 2002;70:163–170. doi: 10.1128/IAI.70.1.163-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis B M, Kimsey H H, Kane A V, Waldor M K. EMBO J. 2002;21:4240–4249. doi: 10.1093/emboj/cdf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaper J B, Morris J G, Jr, Nishibuchi M. In: DNA Probes for Infectious Disease. Tenover F C, editor. Boca Raton, FL: CRC Press; 1988. pp. 65–77. [Google Scholar]
- 15.Faruque S M, Asadulghani, Saha M N, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 17.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg A, Vogelstein B. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 19.Keasler S P, Hall R H. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 20.Faruque S M, Rahman M M, Asadulghani, Islam K M N, Mekalanos J J. Infect Immun. 1999;67:5723–5729. doi: 10.1128/iai.67.11.5723-5729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faruque S M, Asadulghani, Rahman M M, Waldor M K, Sack D A. Infect Immun. 2000;68:4795–4801. doi: 10.1128/iai.68.8.4795-4801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikema M, Honma Y. Microbiology. 1998;144:1901–1906. doi: 10.1099/00221287-144-7-1901. [DOI] [PubMed] [Google Scholar]
- 23.Nakasone N, Honma Y, Toma C, Yamashiro T, Iganawa M. Microbiol Immunol. 1998;42:237–239. doi: 10.1111/j.1348-0421.1998.tb02277.x. [DOI] [PubMed] [Google Scholar]
- 24.Ehara M, Shimodori S, Kojima F, Ichinose Y, Hirayama T, Albert M, Supawat J K, Honma Y, Iwanaga M, Amako K. FEMS Microbiol Lett. 1997;154:293–301. doi: 10.1111/j.1574-6968.1997.tb12659.x. [DOI] [PubMed] [Google Scholar]
- 25.Kar S, Ghosh R K, Ghosh A N, Ghosh A. FEMS Microbiol Lett. 1996;145:17–22. doi: 10.1111/j.1574-6968.1996.tb08550.x. [DOI] [PubMed] [Google Scholar]
- 26.Jouravleva E A, McDonald G A, Garon C F, Boesman-Finkelstein M, Finkelstein R A. Microbiology. 1998;144:315–324. doi: 10.1099/00221287-144-2-315. [DOI] [PubMed] [Google Scholar]
- 27.Faruque S M, Asadulghani, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jouravleva E A, McDonald G A, Marsh J W, Taylor R K, Boesman-Finkelstein M, Finkelstein R A. Infect Immun. 1998;66:2535–2539. doi: 10.1128/iai.66.6.2535-2539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mekalanos J J. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 30.Basu A, Garg P, Datta S, Chakraborty S, Bhattacharya T, Khan A, Ramamurthy T, Bhattacharya S K, Yamasaki S, Takeda Y, Nair G B. Emerg Infect Dis. 2000;6:139–147. doi: 10.3201/eid0602.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu A, Mukhopadhyay A K, Sharma C, Jyot J, Gupta N, Ghosh A, Bhattacharya S K, Takeda Y, Faruque A S G, Albert M J, Nair G B. Microb Pathog. 1998;24:175–183. doi: 10.1006/mpat.1997.0186. [DOI] [PubMed] [Google Scholar]
- 32.Sharma C, Maiti S, Mukhopadhyay A K, Basu A, Basu I, Nair G B, Mukhopadhyaya R, Das B, Kar S, Ghosh R K, Ghosh A. J Clin Microbiol. 1997;35:3348–3350. doi: 10.1128/jcm.35.12.3348-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis B M, Moyer K E, Boyd E F, Waldor M K. J Bacteriol. 2000;182:6992–6998. doi: 10.1128/jb.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dziejman M, Balon E, Boyd D, Fraser C M, Heidelberg J F, Mekalanos J J. Proc Natl Acad Sci, USA. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukhopadhyay A K, Chakraborty S, Takeda Y, Nair G B, Berg D E. J Bacteriol. 2001;183:4737–4746. doi: 10.1128/JB.183.16.4737-4746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasu H, Iida T, Sugahara T, Yamaichi Y, Park K, Yokohama K, Makino K, Shinagawa H, Honda T. J Clin Microbiol. 2000;38:2156–2161. doi: 10.1128/jcm.38.6.2156-2161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iida T, Hattori A, Tagomori K, Nasu H, Naim R, Honda T. Emerg Infect Dis. 2001;7:477–478. doi: 10.3201/eid0703.010325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karaolis D K, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]