Abstract
Human immunodeficiency virus (HIV-1) develops resistance to 3′-azido-2′,3′-deoxythymidine (AZT, zidovudine) by acquiring mutations in reverse transcriptase that enhance the ATP-mediated excision of AZT monophosphate from the 3′ end of the primer. The excision reaction occurs at the dNTP-binding site, uses ATP as a pyrophosphate donor, unblocks the primer terminus and allows reverse transcriptase to continue viral DNA synthesis. The excision product is AZT adenosine dinucleoside tetraphosphate (AZTppppA). We determined five crystal structures: wild-type reverse transcriptase–double-stranded DNA (RT–dsDNA)–AZTppppA; AZT-resistant (AZTr; M41L D67N K70R T215Y K219Q) RT–dsDNA–AZTppppA; AZTr RT–dsDNA terminated with AZT at dNTP- and primer-binding sites; and AZTr apo reverse transcriptase. The AMP part of AZTppppA bound differently to wild-type and AZTr reverse transcriptases, whereas the AZT triphosphate part bound the two enzymes similarly. Thus, the resistance mutations create a high-affinity ATP-binding site. The structure of the site provides an opportunity to design inhibitors of AZT-monophosphate excision.
The development of effective anti-HIV drugs and their use in combination therapies has been estimated to have saved more than 3 million years of life1. However, drug therapies do not cure HIV infections, and drug therapy is life-long. Prolonged therapies lead to problems with drug toxicity and drug resistance. Understanding the molecular mechanisms of resistance is important for developing more effective drugs and treatment strategies. Many anti-AIDS drugs target the viral enzyme reverse transcriptase (EC 2.7.7.49; gag-pol gene). HIV-1 reverse transcriptase is a heterodimer of p66 and p51 subunits. The p66 subunit comprises the N-terminal polymerase domain and the C-terminal RNase H domain2,3. The polymerase domain resembles a right hand and has four subdomains: fingers, palm, thumb and connection. The p51 subunit, which contains the N-terminal 440 amino acids of p66, has a structural role.
There are two broad classes of reverse transcriptase inhibitors, nucleoside (or nucleotide) reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs). NNRTIs are allosteric inhibitors of DNA polymerization. All of the approved NRTIs lack a 3′ OH and, when incorporated into the nascent DNA primer strand by reverse transcriptase, act as chain terminators. Resistant reverse transcriptases can discriminate against NRTIs by two mechanisms. The exclusion mechanism is exemplified by (i) M184V or M184I, which replaces a flexible side chain near the polymerase active sites with a β-branched amino acid that selectively discriminates against NRTIs with an oxathiolane ring but allows incorporation of dNTPs with normal deoxyribose rings, thus reducing the incorporation of 2′,3′-dideoxy-3′-thiacytidine (3TC, lamivudine)4,5 and (ii) K65R, which forms a molecular platform with the conserved Arg72; this platform allows reverse transcriptase to discriminate tenofovir diphosphate from dATP6. In contrast, AZT-resistant7 HIV-1 reverse transcriptase excises AZT monophosphate (AZTMP) from the blocked 3′ end of the DNA primer strand (Fig. 1a), thus freeing the end of the DNA and allowing reverse transcriptase to continue DNA synthesis8-10. The primary excision substrate in vivo is ATP10,11.
Figure 1.
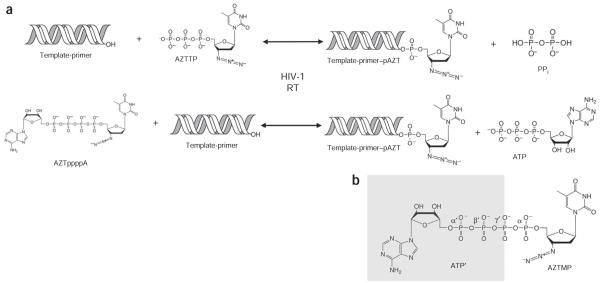
Chemical reactions of DNA polymerization and ATP-mediated pyrophosphorolytic excision catalyzed by HIV-1 reverse transcriptase (RT). (a) DNA polymerization incorporates nucleotides and NRTIs into the DNA primer strand (top row). Incorporated NRTIs can be removed by pyrophosphorolysis, a reverse reaction of polymerization, in which the ATP-mediated excision of AZTMP (pAZT) produces AZTppppA and unblocks the DNA primer strand (bottom row, right to left). (b) Chemical structure of AZTppppA. The two-headed nucleotide AZTppppA was chemically synthesized19 and used for structural studies of excision-product complexes of wild-type and AZTr reverse transcriptases; ATP portion, ATP’; β’ and γ’ phosphates of ATP’ are γ and β phosphates of AZTTP, respectively.
The mutations commonly associated with excision-mediated AZT resistance are M41L, D67N, K70R, L210W, T215Y, T215F, K219Q and K219E. We refer to the protein containing the five mutations (M41L, D67N, K70R, T215Y and K219Q) used in this study as AZTr; these substitutions are also widely referred to as thymidine analog mutations (TAMs). The primary mutations (K70R and T215Y) appear at early stages and can confer resistance to AZT12. The secondary mutations (M41L, D67N, L210W and K219Q) improve AZT resistance in combination with one or two primary mutations. Additional mutations, for example the fingers insertions, can enhance the ability of the mutant reverse transcriptase to excise other NRTIs13-15. Mutations such as K65R and K70E reduce ATP-mediated excision16,17. Pyrophosphorolysis, the pyrophosphate-mediated excision reaction that is equivalent to the polymerization reaction run backwards, generates a nucleoside (or NRTI) triphosphate from the 3′ end of the DNA primer strand (Fig. 1a). In ATP-mediated excision, the β and γ phosphates of ATP take the place of pyrophosphate; if the DNA primer is terminated with AZTMP, the excision reaction produces an AZTppppA, freeing the end of the DNA primer strand for continued polymerization. Reverse transcriptase must exhibit higher preference for polymerization than for pyrophosphorolysis to incorporate nucleotides efficiently; therefore, mutations emerge that enhance the use of ATP as an excision substrate. The AZTr (or TAM) mutations enhance the ability of the AZT-resistant reverse transcriptase to bind ATP and carry out the excision reaction11 without affecting the incorporation of AZT triphosphate (AZTTP) or dNTPs. The mutant reverse transcriptase has a higher binding affinity for AZTppppA than for AZTTP as a substrate18. The primary resistance mutation T215F or T215Y may have π-π interactions with the adenine ring of ATP11. Although the other AZT-resistance mutations may participate in ATP binding, the exact roles of individual mutations and how they, in various combinations, enhance AZT resistance are not well understood.
To elucidate the molecular basis of ATP-mediated excision, we determined five crystal structures: wild-type and AZTr HIV-1 reverse transcriptases in complexes with a dideoxy-terminated dsDNA template-primer and a chemically synthesized excision product, AZTppppA (Fig. 1b)19; AZTr RT–dsDNA terminated with AZTMP at the dNTP-binding or pretranslocation (N) and primer-binding or post-translocation (P) sites; and AZTr apo reverse transcriptase. These structures reveal (i) why an ATP binds wild-type reverse transcriptase with low specificity; (ii) how AZTr (or TAM) mutations create an ATP-binding site distinct from the ATP-binding site in wild-type reverse transcriptase and (iii) the specific interactions of the mutated residues with ATP. Based on their specific roles in enhancing the excision, we have categorized the fingers and palm mutations known to directly or indirectly support excision as excision-enhancing mutations (EEMs; Table 1).
Table 1.
Biochemical analysis and structural interpretation of individual resistance mutations in binding of ATP to AZTr reverse transcriptase
| Mutations | Biochemical analysis | Structural interpretation |
|---|---|---|
| M41L | Usually associated with T215Y, conferring
higher phenotypic resistance to AZT than T215Y alone50 |
Leu41 is next to Phe116 and may help
stabilize AZTMP binding at the N site. |
| E44A or E44D | Accessory AZT-resistance mutation; present
in background of TAMs; enhances excision of AZT39 |
Glu44 interacts with adenine N6 of ATP;
this would interfere with the π-π stacking of Tyr215 and ATP. E44A and E44D may prevent Glu44 from interfering with stacking of Tyr215 and ATP. |
| K65R | Discriminates against NRTIs by causing
slower incorporation of NRTIs than natural substrates; antagonistic to TAMs40–43,51,52 |
Restricts structural adaptability of
enzyme by forming a molecular platform through guanidinium stacking between K65R and Arg72 (ref. 6). |
| D67N | Preferentially associated with K70R; enhances excision of AZT53 | Residue 67 faces the phosphate groups of
ATP; D67N may help binding of ATP by neutralizing negative charge of Asp67. |
| Δ67 | Present in background of TAMs; increases excision of NRTIs34 | No direct structural evidence. Mutation
seems to destabilize the dead-end complex formed with an incoming dNTP, allowing mutant reverse transcriptases to excise an array of NRTIs15,34. |
| Thr69 insertions | Present in background of TAMS; enhance
excision of a range of NRTIs13–16,33 |
No direct structural evidence. Mutations
have effects analogous to the Δ67 complex. |
| K70R | Primary AZT-resistance mutation | Interacts with the α phosphate and
3′-OH of ATP, directly enhancing binding and positioning of ATP. |
| K70E | Substantially impairs the ability of
AZT-resistant reverse transcriptase to excise AZT17 |
Interferes with proper positioning of phosphates of ATP. |
| L210W | Strongly associated with T215F;
contributes substantially to AZT resistance35,36 |
Indole ring of Trp210 stacks with T215Y,
enhancing the π-π stacking between T215Y and adenine of ATP11. |
| T215Y or T215F | Primary AZT-resistance mutation | Aromatic side chain π-π stacks with adenine of ATP11. |
| K219Q | Secondary AZT-resistance mutation;
preferentially associated with K70R54,55 |
Interacts with ribose-ring O4′ of
ATP; may help stabilize binding of ribose ring. |
EEMs are bold.
RESULTS
Structures of RT–dsDNA–AZTppppA ternary complexes
We determined the structure of the wild-type excision product complex at a resolution of 3.15 Å and refined to Rwork of 0.264 and Rfree of 0.288 (Table 2). The structure of the AZTr HIV-1 reverse transcriptase excision product complex was determined at a resolution of 3.2 Å and refined to Rwork of 0.280 and Rfree of 0.308 (Table 2). The two excision product complexes had HIV-1 reverse transcriptase cross-linked4,20 to a 27:21-mer dsDNA template-primer and a bound excision product, AZTppppA (Fig. 1b). Although the crystal unit cell parameters of both excision product complexes were similar to that of a ternary complex crystal of HIV-1 RT–dsDNA–dTTP4 that crystallized with the symmetry of space group P212121, our data analysis and structure solutions revealed the breakdown of higher-order symmetry. Both reverse transcriptase excision ternary complex crystals have the symmetry of the monoclinic space group P21 with four independent RT–dsDNA–AZTppppA complexes per asymmetric unit.
Table 2.
Crystallographic data and refinement statistics
| AZTr excision product complex |
Wild-type excision product complex |
AZTr N-site complex | AZTr P-site complex | Unliganded AZTr RT | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P21 | P21 | P212121 | P3212 | C2 |
| Cell dimensions | |||||
| a, b, c(Å) | 78.7, 283.4, 155.2 | 78.5, 274.8, 152.4 | 79.1, 154.4, 277.9 | 166.5, 166.5, 220.8 | 237.1, 71.2, 94.6 |
| α, β, γ(°) | 90.0, 89.7, 90.0 | 90.0, 90.1, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 106.0, 90.0 |
| Resolution (Å) | 30–3.20 (3.31–3.20) | 30–3.10 (3.21–3.10) | 40–3.62 (3.75–3.62) | 40–2.90 (3.00–2.90) | 30–2.60 (2.69–2.60) |
| R merge | 0.122 (0.517) | 0.129 (0.539) | 0.137 (0.786) | 0.127 (0.558) | 0.051 (0.378) |
| I / σI | 8.7 (1.5) | 11.1 (2.0) | 9.2 (1.0) | 7.3 (1.5) | 20.7 (1.9) |
| Completeness (%) | 98.1 (95.1) | 90.3 (83.5) | 83.9 (49.3) | 94.2 (87.1) | 93.1 (73.4) |
| Redundancy | 3.3 (2.6) | 5.3 (4.6) | 6.7 (2.6) | 4.5 (2.5) | 3.4 (1.8) |
| Refinement | |||||
| Resolution (Å) | 25–3.20 | 25–3.15 | 30–3.70 | 25.0–2.90 | 20.0–2.65 |
| No. reflections | 111,149 | 98,589 | 31,625 | 73,103 | 41,862 |
| Rwork/Rfree | 0.280/0.308 | 0.264/0.288 | 0.312/0.350 | 0.260/0.294 | 0.260/0.294 |
| No. atoms | |||||
| Protein | 32,140 | 31,652 | 15,868 | 11,420 | 8,016 |
| DNA/ligand/ion | 3,839 | 3,816 | 1,804 | 918 | ND |
| Water | 93 | 76 | ND | 138 | 38 |
| B-factors | |||||
| Protein | 89.7 | 72.4 | 118.4 | 72.8 | 95.6 |
| DNA/ligand/ion | 93.2 | 75.5 | 159.8 | 100.2 | ND |
| Water | 46.5 | 34.8 | ND | 54.7 | 67.9 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.011 | 0.010 | 0.012 | 0.011 | 0.009 |
| Bond angles (°) | 1.41 | 1.42 | 1.45 | 1.50 | 1.33 |
Values in parentheses are for highest-resolution shell. ND, not determined. RT, reverse transcriptase.
In both structures, all four noncrystallographic symmetry–related copies in an asymmetric unit are nearly identical in the conformation of the enzyme and DNA, but they differ in their overall structural order. In the crystals of the wild-type and AZTr excision product complexes, the four noncrystallographic symmetry–related copies in the asymmetric unit vary in average temperature factor (Bav) from 49 to 103 Å2 and from 59 to 128 Å2, for the best-to the least-ordered copies, respectively. This wide range of structural order may explain the anisotropic diffraction observed for the two excision product crystals (see Online Methods). Differences in average isotropic temperature factors or amino acid conformations between noncrystallographic symmetryrelated copies have been observed in the structure of an HIV-1 reverse transcriptase precatalytic ternary complex4 and many other protein crystal structures21,22. Unless otherwise noted, all of the analyses were done with the best-ordered copy from each of the crystals.
Superposition of the structures of both excision product ternary complexes showed considerable overall similarity with an r.m.s. deviation of 0.8 Å for all Cα atoms; the polymerase active sites and the individual sites of the resistance mutations in the two structures superimpose well. The ternary complex of wild-type RT–dsDNA–dTTP4 also superimposes well on both structures of AZT excision complexes (Supplementary Fig. 1); the AZTTP portion of the AZTppppA superimposes well with dTTP, the natural nucleotide counterpart, as do the surrounding residues. The facts that (i) the AZTr mutations have no noticeable effect on the binding of AZTTP versus dTTP and (ii) the mutated residues have no direct interaction with AZTTP explain why the AZTr mutations have no appreciable effect on either the binding of AZTTP or its incorporation23-25.
Binary complexes of AZTr RT with AZTMP-terminated dsDNA
To understand the preexcision states of reverse transcriptase, we determined crystal structures of AZTr HIV-1 reverse transcriptase in complexes with an AZTMP-terminated dsDNA; the primer-terminating AZTMP was positioned either in the pretranslocated or the post-translocated sites in the N-site and P-site structures, respectively. The structure of AZTr RT–dsDNA P-site complex was determined at a resolution of 2.9 Å and refined to Rwork of 0.260 and Rfree of 0.294 (Table 2). This structure was determined in the presence of the monoclonal antibody fragment Fab3 and resembles the structure of wild-type RT–dsDNA–AZTMP-terminated P-site complex20. An AZTMP at the P site is not accessible for excision and the AZTr mutations have no direct or substantial indirect effect on AZTMP at the P site. The AZTMP must reside in the N site for excision to occur, thus an ATP must bind to the N-site complex.
We determined the crystal structure of the binary complex of AZTr reverse transcriptase with AZT-terminated (N-site) dsDNA at a resolution of 3.7 Å and refined it to an Rwork of 0.312 and Rfree of 0.350 (Table 2). Even at this moderate resolution, the electron density maps clearly define the position of the mobile β3-β4 fingers loop of p66 (Supplementary Fig. 2). In contrast to all known structures of reverse transcriptase–DNA binary complexes, including our reported wild-type RT–dsDNA–AZTMP-terminated N-site complex20 and the AZTMP P-site complex discussed above, the structure of the N-site complex has the p66 fingers subdomain in a ‘closed’ configuration, which is analogous to that observed in ternary polymerase complexes and the excision product complexes of reverse transcriptase. The structure suggests that (i) the closed-fingers subdomain helps the fingers to interact with the AZTMP at the primer terminus, and (ii) the ATP excision substrate could bind to the closed-fingers N-site complex. Structural similarities among the N-site complex and the reverse transcriptase ternary complexes (both with closed fingers) permit a direct comparison between the pre- and post-incorporation and pre- and post-excision states of reverse transcriptase (Fig. 1a). The N-site complex represents the state after incorporation and before excision for AZTMP, whereas the AZTr RT–dsDNA–AZTppppA ternary complex represents the post-excision state.
Structure of apo AZTr reverse transcriptase
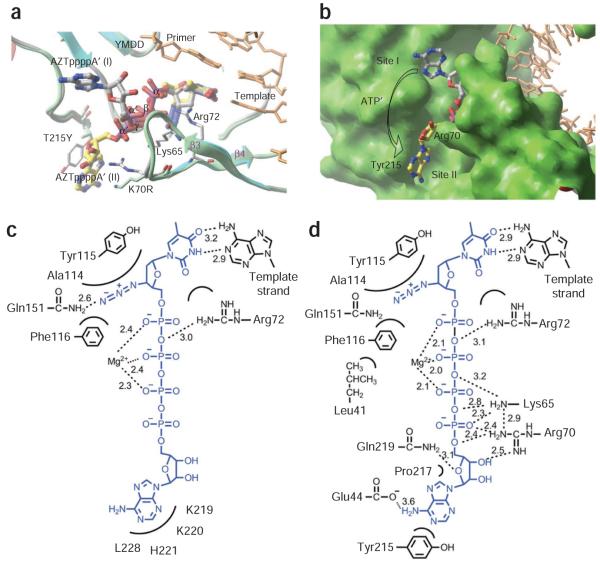
We also determined the crystal structure of apo AZTr HIV-1 reverse transcriptase at a resolution of 2.65 Å and refined to Rwork of 0.260 and Rfree of 0.294 (Table 2). The overall structure closely resembles the structure of wild-type apo reverse transcriptase26. A comparison of AZTr apo reverse transcriptase, P-site, and excision product complexes shows that the side chains of primary mutant residues Arg70 and Tyr215 assume different conformations before and after ATP binding (Supplementary Fig. 3). The side chains of Tyr215 and Pro217 are stacked in the apo AZTr reverse transcriptase structure. This stacking opens up during ATP binding, creating the ATP-binding cleft in the structure of the AZTppppA-bound AZTr reverse transcriptase ternary complex (Fig. 2).
Figure 2.
Binding of AZTppppA’ to AZTr HIV-1 reverse transcriptase. (a) Interactions of AZTppppA’ (carbons, green; phosphates, orange) with the mutated residues (cyan) and with the active site residues; hydrogen bonds, dotted lines; nucleic acid, thin lines. (b) Molecular surface representing the azido-binding cleft at the N site. (c) π-π stacking between Tyr215 and the adenine ring, and the hydrophobic interaction between Pro217 and the ribose ring stabilize the binding of ATP’ to AZTr reverse transcriptase. (d) Primary mutation K70R enhances ATP’ binding by interacting with the ribose ring and α phosphate. Structure images were made using PyMOL (http://www.pymol.org/).
DISCUSSION
AZTr mutations have no effect on binding or incorporation of AZTTP
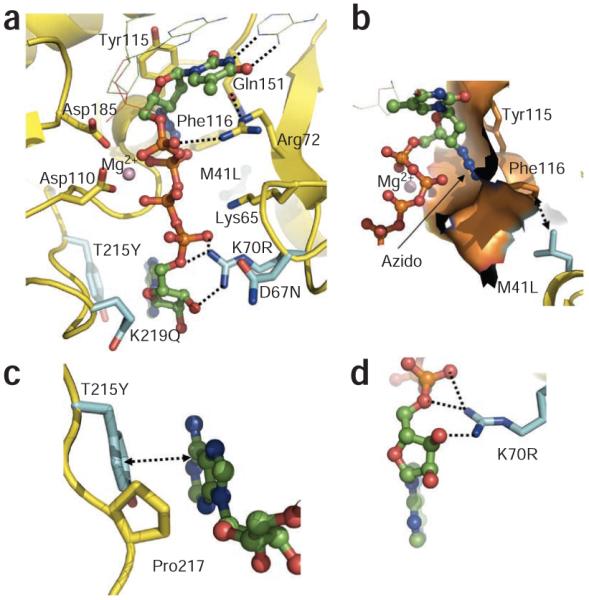
The binding of the AZTTP part of AZTppppA is markedly similar to the binding of dTTP to reverse transcriptase (Supplementary Fig. 1), and both are incorporated in the same catalytic DNA polymerization reaction. AZTTP has a characteristic 3′-azido group that is substituted for the 3′ OH of dTTP. Comparison of the eight conformations of AZTTP (four independent reverse transcriptase complexes in each of the wild-type and AZTr structures) shows that the azido group can adopt different positions; the azido group is disordered in three of the four noncrystallographic symmetry–related copies. The ordered azido group is positioned over an α-helix and interacts primarily with the helix residues Ala114, Tyr115 and Phe116 (Fig. 2b). The ‘Q151M complex’ (Q151M, A62V, V75I, F77L and F116Y)27,28 causes resistance to AZT by an exclusion mechanism; Q151M and F116Y may interfere with the binding of the azido group and thereby discriminate AZTTP from dTTP.
The AZTTP moiety of AZTppppA does not interact with any of the EEMs (Table 1) or AZTr mutations, suggesting that AZTr mutations do not affect the binding or incorporation of AZTTP. The AZTr mutation M41L is proximal to Glu44 and Phe116, which interacts with the azido group of AZT; M41L is 3.9 Å and 3.4 Å away from Phe116 and Glu44, respectively. The side chain of Phe116 is about 3.1 Å and 3.0 Å away from the side chain of Gln151 and the 3′-azido group of the AZT, respectively. A leucine at position 41 may stabilize the interactions of Phe116 and surrounding residues with the 3′-azido group (Fig. 2b); the interaction with the 3′-azido group would make AZTMP more likely to occupy the N site after it is incorporated. In contrast to AZTMP excision, an incorporated 3′-azidopurine analog is not excised efficiently29. Notably, the interactions of the 3′ azido (Fig. 2b) seem to be accessible for an incorporated AZTMP but not for a 3′-azidopurine analog. Apart from M41L, the other AZTr mutations do not have any noticeable structural impact on binding of the AZTTP portion to reverse transcriptase.
ATP binds differently to wild-type and AZTr reverse transcriptase
The excision product AZTppppA is a two-headed nucleotide with AZTTP on one end and ATP on the other; both nucleotides share the two middle phosphate groups (Fig. 1b). To avoid confusion, hereafter we refer to the ATP part of AZTppppA as ATP’. The AMP’ moiety is positioned quite differently in the two excision-product complexes (Fig. 3a,b); the conformational switching primarily occurs by torsional rotations about the bonds linking the β’ and α’ phosphates, which position the two AMPs ~10 Å apart.
Figure 3.
AZTppppA’ binds differently to AZTr reverse transcriptase than to wild-type reverse transcriptase. (a) Superposition of the wild-type HIV-1 RT–dsDNA–AZTppppA’ structure (gray) on the AZTr HIV-1 RT–dsDNA–AZTppppA’ structure (cyan). The AMP’ portion of AZTppppA’ binds differently to AZTr reverse transcriptase (yellow) than to wild-type reverse transcriptase (gray). (b) Molecular surface of AZTr reverse transcriptase showing ATP’-binding sites. Site I, where ATP’ binds to wild-type reverse transcriptase, remains almost unchanged in AZTr reverse transcriptase; however, a new ATP’-binding site (site II) is created by AZTr mutations that substantially enhance the binding of ATP’ to AZTr reverse transcriptase. (c) Summary of interactions of AZTppppA’ (blue) with wild-type HIV-1 reverse transcriptase. (d) Summary of interactions of AZTppppA’ with AZTr HIV-1 reverse transcriptase. Hydrogen bonds and metal coordinations, dotted lines. Ribbon and molecular representations were drawn using Schrodinger (http://www.schrodinger.com/).
In the wild-type excision product complex, ATP’ is located away from the sites of the AZTr mutations and there are no distinctive interactions between the AMP’ and reverse transcriptase (Fig. 3c). In fact, the conformation of the AMP’ shows large positional variations when the four independent wild-type RT–dsDNA–AZTppppA’ complexes in the crystallographic asymmetric unit are compared (Supplementary Fig. 4). The binding of ATP’ seems to rely on the interactions of the β’ and γ’ pyrophosphates with metal ion B and with reverse transcriptase (Fig. 3c). This explains why wild-type reverse transcriptase shows no appreciable selectivity among nucleoside triphosphates, nucleoside diphosphates and pyrophosphate in vitro excision reactions9.
All the AZTr mutations, except M41L, surround the AMP’ of AZTppppA’ in the AZTr RT–dsDNA–AZTppppA’ structure (Fig. 2a). The primary resistance mutations T215Y and K70R interact extensively with the AMP’, and thereby substantially enhance the binding of AZTppppA’ to AZTr reverse transcriptase18,30. Thus, the AZT-resistance mutations do not alter a preexisting ATP’-binding site (site I), but rather create a new binding site that has additional interactions with the ATP’ moiety (site II). The secondary AZTr mutations contribute to the binding of ATP’ to the new site created by the primary mutations. This explains why the secondary mutations do not, by themselves, confer appreciable resistance to AZT. ATP’ can bind to both sites in AZTr reverse transcriptase, but because of the additional interactions with the mutated side chains it preferentially binds to site II. However, the positions of the β’ and γ’ phosphates, which chelate the catalytic metal ions, remain unchanged even though the AMP’ part binds differently to AZTr reverse transcriptase than to wild-type reverse transcriptase. This common mode of metal ion chelation and the spatial positioning of the pyrophosphate moiety of the ATP’ (Fig. 3a) suggest that the excision reaction follows a common catalytic reaction path for both AZTr reverse transcriptase and wild-type reverse transcriptase using either ATP or pyrophosphate as the excision substrate. In this common catalytic platform, the efficiency of excision depends on the ability of reverse transcriptase to bind and appropriately position the excision substrates to carry out the pyrophosphorolytic reaction.
AZTr reverse transcriptase can use ATP as an excision substrate more efficiently than wild-type reverse transcriptase; this is the basis of AZT resistance. Two kinetic studies31,32 have reported increased catalytic efficiency in the AZTr (or TAM) background, however no appreciable enhancement in ATP-binding affinity was detected. The efficiency of catalysis must depend on proper positioning of the pyrophosphate moiety, which is favored in AZTr reverse transcriptase. AZTr reverse transcriptase interacts better with the AMP part (Fig. 3c,d). The binding of ATP has several structural requirements, including having the primer-terminated AZTMP at the N site, closing of the β3-β4 loop, opening of Tyr215 and Pro217 stacking (Supplementary Fig. 3b), and precise metal-ion chelation. These events may contribute to the measured lower and inconsistent binding affinity31,32 of ATP to AZTr reverse transcriptase; for example, a higher probability that the primer-terminating NRTI occupies the P site would result in lower ATP-binding affinity. However, once a primer-terminating AZTMP is at the N site and the AZTr mutations stabilize ATP binding, the excision is favored.
AZTr mutations create a new binding site for ATP
In the AZTr reverse transcriptase complex, AMP’ interacts extensively with the side chains of primary mutant residues Arg70 and Tyr215 (Figs. 2 and 3d). The guanidinium group of Arg70 forms hydrogen bonds with the 3′ OH and 5′ O atoms of the ribose ring and the α’ phosphate. The π-π stacking of Tyr215 with a purine base would be more extensive than with a pyrimidine base, which may explain why ATP and GTP are favored substrates for AZTMP excision10,11. Pro217 makes hydrophobic contacts with the ribose ring. Tyr215 and Pro217 help position the adenosine moiety of ATP’ at site II.
The secondary AZT-resistance mutations also contribute to binding of ATP’. The side chain of the mutated Gln219 is ~3.1 Å away from the O4′ of the ribose ring (Fig. 3d) and the interaction may help stabilize the position and conformation of the ribose ring. The exact role of D67N is not clear from the structure. This residue is at the tip of the flexible β3-β4 loop with its side chain facing the ATP’ (Fig. 3a). D67N could provide a better environment for the binding of ATP’ by eliminating the negatively charged environment of the original Asp67. In the β3-β4 fingers loop, 69-insertion and Δ67-deletion mutations emerge13-16,33,34 that enhance the ability of the enzyme to excise other NRTIs. These mutations destabilize the dead-end complex formed with an incoming dNTP, allowing the mutant reverse transcriptases to excise a broad array of NRTIs15,34. Also, rearrangement of the fingers may facilitate stronger interactions between ATP’ and primary AZTr mutations.
L210W occurs in combination with T215Y, and the addition of L210W to reverse transcriptases carrying T215Y substantially augments AZT resistance35,36. Modeling based on the AZTr excision product complex shows that the aromatic side chains of Trp210 and Tyr215 stack; this would enhance the π-π stacking between Tyr215 and ATP’ on the opposite face of Tyr215 (refs. 11,35). E44A and E44D are strongly associated with the primary mutation T215Y but not with K70R37-39. In the AZTr excision product complex, the Glu44 side chain is ~3.6 Å from the N6 atom of adenine (Fig. 3d). Tyr215 and Glu44 lie on opposite sides of the adenine ring of ATP’, and E44A and E44D could disrupt the interaction of Glu44 with the adenine, permitting more favorable π-π stacking between the adenine and Tyr215.
K65R and K70E interfere with excision
The four independent copies of the AZTr excision complex in the asymmetric unit provide different snapshots of the interactions of Lys65 and Arg70 side chains with AZTppppA’ (Supplementary Fig. 5). The different sets of interactions of Lys65 with the phosphates of AZTppppA’ in the different copies provide direct structural evidence for the antagonistic relationship between K65R and K70R6,40,41. In the first and fourth copies, Arg70 interacts with the α’ phosphate of ATP’, whereas Lys65 is primarily disordered. The second copy has relatively weaker electron density and a weaker interaction of Arg70 with the α’ phosphate, whereas Lys65, with stronger electron density, interacts with β’ phosphate of ATP’. In the third copy, both Arg70 and Lys65 interact with AZTppppA’, however with a different hydrogen-bonding pattern than in the second copy (Supplementary Fig. 5). These snapshots show that (i) Arg70 is not required for ATP’ binding if T215Y is present and (ii) the interaction of Arg70 with the α’ phosphate interferes with the interaction between Lys65 and the β’ phosphate, and vice versa. K65R was originally isolated from patients treated with dideoxycytidine (ddC) or dideoxyinosine (ddI), and confers resistance to tenofovir and foscarnet16,42,43. K65R forms a molecular platform by guanidinium stacking with Arg72 (ref. 6) and is antagonistic to ATP-mediated pyrophosphorolysis40,41. The interplay between the positioning of Arg70 and Lys65 in the current structures confirms that the K65R-induced molecular platform (Arg65 and Arg72)7 would interfere with the positioning of Arg70 and vice versa. K70E impairs ATP-mediated excision in association with the AZTr mutations (M41L, L210W and T215Y)17. It seems that a K70E substitution (an oppositely charged, shorter substitution than K70R) would reposition the phosphates away from their catalytically relevant positions even if the base of ATP’ were well accommodated in site II (Figs. 2a and 3).
Excision versus DNA polymerization by reverse transcriptase
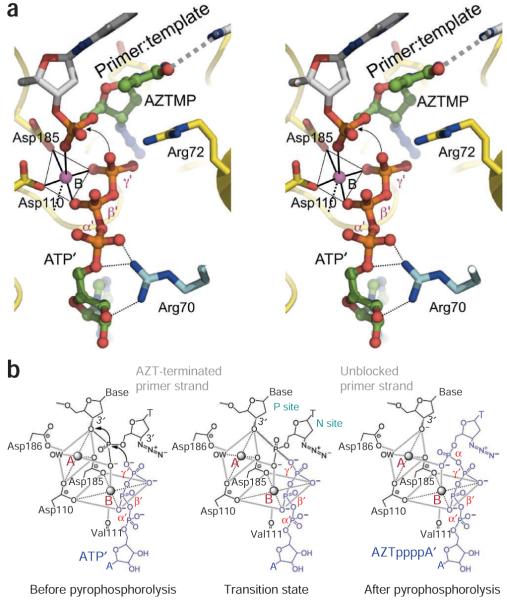
The catalytic reactions of both polymerization and excision (Fig. 1a) are carried out by the same catalytic triad: Asp110, Asp185 and Asp186. We compare the crystal structures of the AZT excision complex and the N-site complex (Fig. 4) to elucidate excision and its relationship to DNA polymerization by HIV-1 reverse transcriptase (Fig. 5). A superposition of the N-site structure on that of the excision complex shows that the α-phosphorus atom of AZTTP shifts ~1.4 Å after incorporation (Fig. 4), while maintaining the metal ion coordination of the α-phosphate oxygen. On the basis of the relative positions of N-site AZTMP and the ATP’ part of AZTppppA’ obtained based on the above superposition (Fig. 5a), we created a structural model of the reverse transcriptase ternary complex preceding the excision of AZTMP by ATP’ (Fig. 5b). The coordination environment for excision is similar to that for polymerization44; there is almost perfect octahedral coordination geometry for metal ion B involving a γ’-phosphate oxygen of the ATP’ and a phosphate oxygen of terminal AZTMP; the other four coordinating atoms are from Asp110, Asp185, carboxyl of Val111 and β’ phosphate of ATP’. The plane of coordination for metal ion B is defined by the oxygen atoms from Asp110, Asp185 and β’ and γ’ phosphates of ATP’. The phosphate oxygen from AZTMP, which occupies an axial position in the metal ion B coordination environment, should also coordinate with metal ion A. Our analysis suggests that the overall catalytic mechanism (Fig. 5b) in the ATP-mediated excision is the reverse of the catalytic mechanism of DNA polymerization by reverse transcriptase. The model shows that attacking γ’-phosphate oxygen of ATP’ is ~3 Å from the phosphorus of AZTMP at the DNA-primer terminus; the attacking γ’-phosphate oxygen is almost collinear with the scissile phosphate bond of AZTMP (Figs. 4 and 5a). The catalytic reactions of polymerization (forward) and excision (backward) by reverse transcriptase should follow the same transition states, but in reverse order. Excision is accessible only when an incorporated NRTI (like AZT) is at the N site and when a phosphate oxygen of an excision substrate (ATP or pyrophosphate) acts as a nucleophile (Fig. 5b).
Figure 4.
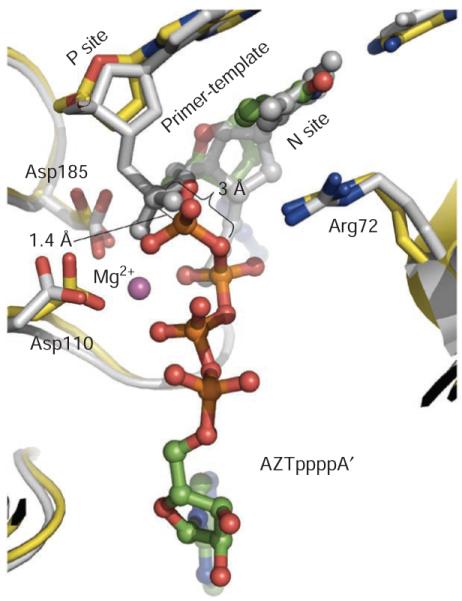
Superposition of AZTr reverse transcriptase excision product complex (yellow) and AZTr reverse transcriptase–template-primer–AZTMP (N site) complex (gray) structures. AZTppppA’ has green carbon and orange phosphorus atoms. The AZT portions superimpose well; the α-phosphorus atoms are ~1.4 Å apart in the two superimposed structures, indicating the position of the α phosphate before and after AZTTP incorporation; the metal coordination of an α-phosphate oxygen is maintained before and after incorporation of AZTMP. Figure generated using PyMOL.
Figure 5.
Mechanism of ATP-mediated excision by HIV-1 reverse transcriptase. (a) Stereo view showing relative locations of DNA-primerterminated AZTMP and the ATP excision substrate; locations based on superposition (Fig. 4) of structures of AZTr RT–dsDNA–AZTppppA’ and AZTr RT–dsDNA (with AZTMP at the primer terminus occupying the N site). The metal A position was not observed in the current structures, therefore only metal B is shown. (b) Scheme for ATP-mediated excision. Catalysis involves two Mg2+ ions, on the basis of our structural results and the reported coordination geometry at the catalytic active site of HIV-1 reverse transcriptase4,44. The catalytic reactions of polymerization and excision should use the same cations (A and B) in the identical coordination environment. Planes and axes of coordination for both metal ions are gray boxes and lines, respectively. The excision substrate (ATP’) and the product (AZTppppA’) are blue; the prepyrophosphorolysis state was derived by positioning ATP’ in N-site complex structure, and the excision-complex structures represent the post-pyrophosphorolysis state.
In general, viral polymerases lack a proofreading mechanism, and the AZTr reverse transcriptases do not have higher fidelity than wild-type reverse transcriptase45, suggesting that excision or pyrophosphorolysis is not a standard mechanism to correct nucleotide misincorporations. However, pyrophosphorolysis has been observed for hepatitis B virus (HBV) reverse transcriptase, which has polymerase active site structure46 similar to that of HIV-1 reverse transcriptase; HBV reverse transcriptase is inhibited by selected HIV-1 NRTI drugs. 3TC-monophosphate (MP) is not excised by either HIV-1 or HBV reverse transcriptase, but HBV reverse transcriptase can excise 2′-3′dideoxycytidine (ddC)-MP at physiological concentrations of pyrophosphate47. The RNA-dependent RNA polymerase NS5B of hepatitis C virus (HCV) also exhibits pyrophosphorolysis of chain-terminating nucleotides at physiological concentrations of pyrophosphate48. Studies on EEMs and relevant structures of these polymerases will help elucidate excision-based resistance by other viral polymerases.
Conclusions
The structures presented here help to elucidate the molecular basis of ATP-mediated AZTMP excision by HIV-1 reverse transcriptase, a story that has been developing through nearly two decades of biochemical, biophysical and virological investigations49 since AZT-resistance mutations were first identified in patients7. The structures of excision product complexes show that the AMP’ part binds differently to AZTr reverse transcriptase than to wild-type reverse transcriptase; the AMP’ part binds to a new site created by the AZT-resistance mutations. The AZTTP part of AZTppppA’ and dTTP share a common mode of binding and divalent metal cation chelation, suggesting that wild-type and AZTr reverse transcriptase share a common catalytic mechanism for the excision reaction that is the reverse of the incorporation of AZTTP or dTTP by reverse transcriptase.
The structure of the N-site AZTr reverse transcriptase binary complex demonstrates that the fingers subdomain can assume a closed conformation if the primer-terminated AZTMP is at the N site, which also facilitates the binding of an ATP. AZTppppA is a better substrate for AZTr reverse transcriptase than AZTTP18,30, suggesting that appropriately designed excision-product (AZTppppA) mimics would bind AZTr reverse transcriptase with high affinity and also inhibit dNTP binding. Small molecules targeting only the AMP’-binding site would inhibit the excision reaction; such inhibitors could be used as co-drugs with excision-susceptible NRTIs like AZT.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge personnel at the Cornell High Energy Synchrotron Source and the Advanced Photon Source for support of data collection, the members of our laboratories, including R. Bandwar and S. Martinez, for valuable conversations and assistance, and P. Clark for assistance with protein preparation. We are grateful to the US National Institutes of Health (NIH; grants R37 MERIT Award AI 27690 to E.A. and P01 GM 066671 to E.A. and R.A.J.) for support of reverse transcriptase structural studies. S.H.H. was supported by the Intramural Research Program of NIH, US National Cancer Institute (NCI), Center for Cancer Research and US National Institute of General Medical Sciences. This research was supported, in part, by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Appendix
ONLINE METHODS
Protein expression and purification
Recombinant HIV-1 reverse transcriptases were produced by coexpressing the p66 and p51 subunits in Escherichia coli strain BL21(DE3)(pLysE). Expressed reverse transcriptases were purified by nickel-affinity and anion exchange chromatograph. In AZTr reverse transcriptase, the p66 subunit contained five AZT-resistance mutations (M41L, D67N, K70R T215Y and K219Q), along with C280S and Q258C. The p51 subunit contained C280S and was truncated at Gln428; a GGHHHHHHH tag was added after Gln428 for nickel-affinity purification. The wild-type reverse transcriptase had C280S and Q258C in p66 and C280S in p51. Six histidines and an HRV14 C3 protease cleavage site were added at the N terminus of p51 in the wild-type reverse transcriptase construct; this tag was proteolytically removed after the DNA cross-linking and nickel-affinity purification.
Synthesis of cross-linkable DNA primer and AZTppppA
The cross-linkable DNA primer was synthesized as described56. The primer sequence is 5′-ACAGTCCCTGTTCGGG*CGCC-3′ (G* has a thioalkyl tether covalently attached to the N2 group of the guanosine). AZTppppA was synthesized by reaction of AMP with a trimetaphosphate derivative of AZT19.
Preparation and purification of RT–dsDNA complex
The reverse transcriptase complexes were trapped using a strategy of covalent cross-linking of DNA to reverse transcriptase56. The cross-linkable primer was first annealed with a 27-mer template at an equimolar ratio. The sequence of the template for making excision product complexes and the N-site complex is 5′-ATGCATGGCGCCCGAACAGGGACTGTG-3′. The sequence of the template for making the P-site complex is 5′-ATGCTAGGCGCCCGAACAGGGACTGTG-3′.
For the excision product complexes, the annealed nucleic acid substrates were reacted with reverse transcriptase in a 3:1 ratio in the presence of 150 mM NaCl, 50 μM β-mercaptoethanol, 100 μM dideoxy-ATP and 50 mM Tris-HCl, pH 8.0. Polymerization was initiated by addition of 5 mM MgCl2, and the reaction was allowed to proceed for 2 h at room temperature. After dideoxy-AMP incorporation, 2 mM AZTppppA was added to the reaction mixture, and then incubated overnight at 4 °C; this produced higher cross-linking efficiencies, typically 70–90%. For the pretranslocation and post-translocation complexes, the cross-linking reaction and purification were carried out as reported20.
Crystallization and data collection
Crystallization of wild-type and AZTr HIV-1 reverse transcriptase excision product complexes was done as follows. The initial crystallization samples contained 8–10 mg ml−1 RT–dsDNA, 75 mM NaCl, 5 mM AZTppppA and 10 mM Tris-HCl, pH 8.0. Crystallization sample (2 μl) was mixed with 2 μl of reservoir solution containing 50 mM Bis-Tris propane, pH 6.8, 10–11% (w/v) PEG8000, 0.3 M (NH4)2SO4, 5% (v/v) glycerol, 5% (w/v) sucrose, 20 mM MgCl2 and 5 mM spermidine. Crystals were grown at 4 °C using the hanging drop vapor diffusion method. Crystals of both the excision product complexes appeared as fragile thin-plate clusters. Crystals were carefully separated and stabilized in a holding buffer (the reservoir solution with PEG8000 concentration elevated to 12% (w/v)) for 5 min. Crystals were then soaked with cryoprotective solution (holding buffer containing 20% (v/v) glycerol) for ~30 s and flash-cooled in liquid nitrogen.
In contrast to the reported wild-type HIV-1 reverse transcriptase N-site complex20, the AZTr HIV-1 reverse transcriptase N-site complex was crystallized without the antibody fragment Fab. AZTTP was incorporated into the cross-linked primer by in vitro polymerization as described20. The initial crystallization samples contained 8–10 mg ml−1 RT–dsDNA, 75 mM NaCl and 10 mM Tris-HCl, pH 8.0. Crystallization sample (2 μl) was mixed with 2 μl of reservoir solution containing 50 mM Bis-Tris propane, pH 6.4, 10–11% (w/v) PEG8000, 0.3 M (NH4)2SO4, 5% (v/v) glycerol, 5% (w/v) sucrose and 20 mM MgCl2. Crystals were stabilized in a holding buffer (the reservoir solution with PEG8000 concentration elevated to 12% (w/v)) for 5 min, and then soaked with cryoprotective solution (holding buffer containing 20% (v/v) glycerol) for ~30 s and finally flash-cooled in liquid nitrogen.
The crystallization and cryocooling conditions for the AZTr reverse transcriptase P-site complex and apo AZTr HIV-1 reverse transcriptase have been described20,26. Diffraction data were collected using synchrotron radiation and processed, reduced and scaled using HKL2000 (ref. 57; Table 2). Indexing of the diffraction patterns was challenging and ellipsoidal truncation and anisotropic scaling58 was applied.
Structure determination and refinement
All crystal structures were solved by molecular replacement (Table 2). For the excision product complexes and the N-site complex, the reported wild-type ternary polymerization complex (PDB 1RTD4) was used as the starting model. For the P-site complex and apo AZTr reverse transcriptase, the reported wild-type HIV-1 reverse transcriptase P-site complex (PDB 1N5Y) and apo wild-type reverse transcriptase (PDB 1DLO) were used as starting models, respectively. Rotational and translational searches were carried out using Phaser as implemented in CCP4 (ref. 59). Structure refinement was conducted using CNS60. Rigid body refinement involved three stages. First, the whole asymmetric unit was treated as a rigid body; second, each molecule was treated this way; and third, each complex was subdivided into multiple rigid bodies according to subdomains, domains and DNA. For the excision product complexes and N-site complex, because the asymmetric unit contained multiple copies, noncrystallographic symmetry restraints were applied throughout refinement. Noncrystallographic symmetry–restraint groups were set up as follows. For low-resolution structures, main chain noncrystallographic symmetry restraints were separated from those of side chains because main chains are generally better defined than side chains. Main chains and side chains were divided into multiple noncrystallographic symmetry–restraint groups according to domains, subdomains and DNA. Each noncrystallographic symmetry–restraint group was assigned a weight according to the quality of the electron density map for the corresponding region. At the beginning, tight noncrystallographic symmetry restraints were used and as refinement progressed, these restraints were gradually relaxed. The ensuing refinement involved several rounds of torsion angle annealing, energy minimization, density modification, noncrystallographic symmetry averaging and iterative model building. The planarity of the bases and Watson-Crick base-pairing geometry were used to restrain DNA stereochemistry. The maximum likelihood refinement target was usually MLF (amplitudes); in the structure refinement process, the phases were sometimes constrained using Hendrickson-Lattman coefficients calculated from improved phases and figures of merit from density modifications by solvent flipping using CNS60.
- 56.Sarafianos SG, et al. Trapping HIV-1 reverse transcriptase before and after translocation on DNA. J. Biol. Chem. 2003;278:16280–16288. doi: 10.1074/jbc.M212911200. [DOI] [PubMed] [Google Scholar]
- 57.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 58.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 60.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Footnotes
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Accession codes. Protein Data Bank: Coordinates and structure factors for AZTr RT–dsDNA–AZTppppA’, wild-type RT–dsDNA–AZTppppA’, AZTr reverse transcriptase N-site and P-site complexes, and apo AZTr reverse transcriptase have been deposited with accession codes 3KLE, 3KLF, 3KLG, 3KLH and 3KLI, respectively.
Note: Supplementary information is available on the Nature Structural Molecular Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Fauci AS. 25 years of HIV/AIDS science: reaching the poor with research advances. Cell. 2007;131:429–432. doi: 10.1016/j.cell.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 3.Jacobo-Molina A, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 5.Sarafianos SG, et al. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc. Natl. Acad. Sci. USA. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das K, et al. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J. Biol. Chem. 2009;284:35092–35100. doi: 10.1074/jbc.M109.022525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 8.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 9.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant IV-1 reverse transcriptase. Mol. Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 11.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 2001;75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeeninga RE, Keulen W, Boucher C, Sanders RW, Berkhout B. Evolution of AZT resistance in HIV-1: the 41–70 intermediate that is not observed in vivo has a replication defect. Virology. 2001;283:294–305. doi: 10.1006/viro.2001.0888. [DOI] [PubMed] [Google Scholar]
- 13.Mas A, et al. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 2000;19:5752–5761. doi: 10.1093/emboj/19.21.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J. Virol. 2002;76:9143–9151. doi: 10.1128/JVI.76.18.9143-9151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer PR, Lennerstrand J, Matsuura SE, Larder BA, Scott WA. Effects of dipeptide insertions between codons 69 and 70 of human immunodeficiency virus type 1 reverse transcriptase on primer unblocking, deoxynucleoside triphosphate inhibition, and DNA chain elongation. J. Virol. 2003;77:3871–3877. doi: 10.1128/JVI.77.6.3871-3877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer PR, et al. Relationship between 3′-azido-3′-deoxythymidine resistance and primer unblocking activity in foscarnet-resistant mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 2003;77:6127–6137. doi: 10.1128/JVI.77.11.6127-6137.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sluis-Cremer N, et al. Molecular mechanism by which the K70E mutation in human immunodeficiency virus type 1 reverse transcriptase confers resistance to nucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 2007;51:48–53. doi: 10.1128/AAC.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dharmasena S, Pongracz Z, Arnold E, Sarafianos SG, Parniak MA. 3′-Azido-3′-deoxythymidine-(5′)-tetraphospho-(5′)-adenosine, the product of ATP-mediated excision of chain-terminating AZTMP, is a potent chain-terminating substrate for HIV-1 reverse transcriptase. Biochemistry. 2007;46:828–836. doi: 10.1021/bi061364s. [DOI] [PubMed] [Google Scholar]
- 19.Han Q, Gaffney BL, Jones RA. One-flask synthesis of dinucleoside tetra- and pentaphosphates. Org. Lett. 2006;8:2075–2077. doi: 10.1021/ol060491d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarafianos SG, et al. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carfí A, Smith CA, Smolak PJ, McGrew J, Wiley DC. Structure of a soluble secreted chemokine inhibitor vCCI (p35) from cowpox virus. Proc. Natl. Acad. Sci. USA. 1999;96:12379–12383. doi: 10.1073/pnas.96.22.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 23.Carroll SS, et al. Sensitivity of HIV-1 reverse transcriptase and its mutants to inhibition by azidothymidine triphosphate. Biochemistry. 1994;33:2113–2120. doi: 10.1021/bi00174a018. [DOI] [PubMed] [Google Scholar]
- 24.Kerr SG, Anderson KS. Pre-steady-state kinetic characterization of wild type and 3′-azido-3′-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry. 1997;36:14064–14070. doi: 10.1021/bi9713862. [DOI] [PubMed] [Google Scholar]
- 25.Krebs R, Immendorfer U, Thrall SH, Wohrl BM, Goody RS. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry. 1997;36:10292–10300. doi: 10.1021/bi970512z. [DOI] [PubMed] [Google Scholar]
- 26.Hsiou Y, et al. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 27.Ueno T, Mitsuya H. Comparative enzymatic study of HIV-1 reverse transcriptase resistant to 2′,3′-dideoxynucleotide analogs using the single-nucleotide incorporation assay. Biochemistry. 1997;36:1092–1099. doi: 10.1021/bi962393d. [DOI] [PubMed] [Google Scholar]
- 28.Deval J, et al. The molecular mechanism of multidrug resistance by the Q151M human immunodeficiency virus type 1 reverse transcriptase and its suppression using alpha-boranophosphate nucleotide analogues. J. Biol. Chem. 2002;277:42097–42104. doi: 10.1074/jbc.M206725200. [DOI] [PubMed] [Google Scholar]
- 29.Sluis-Cremer N, et al. The 3′-azido group is not the primary determinant of 3′-azido-3′-deoxythymidine (AZT) responsible for the excision phenotype of AZT-resistant HIV-1. J. Biol. Chem. 2005;280:29047–29052. doi: 10.1074/jbc.M503166200. [DOI] [PubMed] [Google Scholar]
- 30.Meyer PR, Smith AJ, Matsuura SE, Scott WA. Chain-terminating dinucleoside tetraphosphates are substrates for DNA polymerization by human immunodeficiency virus type 1 reverse transcriptase with increased activity against thymidine analogue-resistant mutants. Antimicrob. Agents Chemother. 2006;50:3607–3614. doi: 10.1128/AAC.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray AS, et al. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry. 2003;42:8831–8841. doi: 10.1021/bi034435l. [DOI] [PubMed] [Google Scholar]
- 32.Selmi B, et al. The Y181C substitution in 3′-azido-3′-deoxythymidine-resistant human immunodeficiency virus, type 1, reverse transcriptase suppresses the ATP-mediated repair of the 3′-azido-3′-deoxythymidine 5′-monophosphate-terminated primer. J. Biol. Chem. 2003;278:40464–40472. doi: 10.1074/jbc.M302928200. [DOI] [PubMed] [Google Scholar]
- 33.White KL, et al. Molecular mechanisms of tenofovir resistance conferred by human immunodeficiency virus type 1 reverse transcriptase containing a diserine insertion after residue 69 and multiple thymidine analog-associated mutations. Antimicrob. Agents Chemother. 2004;48:992–1003. doi: 10.1128/AAC.48.3.992-1003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer PL, Imamichi T, Sarafianos SG, Arnold E, Hughes SH. Effects of the Delta67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision. J. Virol. 2004;78:9987–9997. doi: 10.1128/JVI.78.18.9987-9997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hooker DJ, et al. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J. Virol. 1996;70:8010–8018. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrigan PR, et al. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J. Virol. 1996;70:5930–5934. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano L, et al. Broad nucleoside-analogue resistance implications for human immunodeficiency virus type 1 reverse-transcriptase mutations at codons 44 and 118. J. Infect. Dis. 2002;185:898–904. doi: 10.1086/339706. [DOI] [PubMed] [Google Scholar]
- 38.Marcelin AG, et al. Thymidine analogue reverse transcriptase inhibitors resistance mutations profiles and association to other nucleoside reverse transcriptase inhibitors resistance mutations observed in the context of virological failure. J. Med. Virol. 2004;72:162–165. doi: 10.1002/jmv.10550. [DOI] [PubMed] [Google Scholar]
- 39.Girouard M, Diallo K, Marchand B, McCormick S, Gotte M. Mutations E44D and V118I in the reverse transcriptase of HIV-1 play distinct mechanistic roles in dual resistance to AZT and 3TC. J. Biol. Chem. 2003;278:34403–34410. doi: 10.1074/jbc.M303528200. [DOI] [PubMed] [Google Scholar]
- 40.White KL, et al. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antivir. Ther. 2006;11:155–163. doi: 10.1177/135965350601100209. [DOI] [PubMed] [Google Scholar]
- 41.Parikh UM, Zelina S, Sluis-Cremer N, Mellors JW. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS. 2007;21:1405–1414. doi: 10.1097/QAD.0b013e3281ac229b. [DOI] [PubMed] [Google Scholar]
- 42.Gu Z, Fletcher RS, Arts EJ, Wainberg MA, Parniak MA. The K65R mutant reverse transcriptase of HIV-1 cross-resistant to 2′, 3′-dideoxycytidine, 2′,3′-dideoxy-3′-thiacytidine, and 2′,3′-dideoxyinosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro. J. Biol. Chem. 1994;269:28118–28122. [PubMed] [Google Scholar]
- 43.White KL, et al. A combination of decreased NRTI incorporation and decreased excision determines the resistance profile of HIV-1 K65R RT. AIDS. 2005;19:1751–1760. doi: 10.1097/01.aids.0000189851.21441.f1. [DOI] [PubMed] [Google Scholar]
- 44.Das K, et al. Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J. Mol. Biol. 2007;365:77–89. doi: 10.1016/j.jmb.2006.08.097. [DOI] [PubMed] [Google Scholar]
- 45.Mansky LM, Bernard LC. 3′-Azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J. Virol. 2000;74:9532–9539. doi: 10.1128/jvi.74.20.9532-9539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das K, et al. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC) J. Virol. 2001;75:4771–4779. doi: 10.1128/JVI.75.10.4771-4779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urban S, Fischer KP, Tyrrell DL. Efficient pyrophosphorolysis by a hepatitis B virus polymerase may be a primer-unblocking mechanism. Proc. Natl. Acad. Sci. USA. 2001;98:4984–4989. doi: 10.1073/pnas.091324398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deval J, Powdrill MH, D’Abramo CM, Cellai L, Gotte M. Pyrophosphorolytic excision of nonobligate chain terminators by hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 2007;51:2920–2928. doi: 10.1128/AAC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acosta-Hoyos AJ, Scott WA. The role of nucleotide excision by reverse transcriptase in HIV drug resistance. Viruses. 2010;2:372–394. doi: 10.3390/v2020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kellam P, Boucher CA, Larder BA. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc. Natl. Acad. Sci. USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deval J, et al. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 2004;279:509–516. doi: 10.1074/jbc.M308806200. [DOI] [PubMed] [Google Scholar]
- 52.Sluis-Cremer N, Arion D, Kaushik N, Lim H, Parniak MA. Mutational analysis of Lys65 of HIV-1 reverse transcriptase. Biochem. J. 2000;348:77–82. [PMC free article] [PubMed] [Google Scholar]
- 53.Arion D, Parniak MA. HIV resistance to zidovudine: the role of pyrophosphorolysis. Drug Resist. Updat. 1999;2:91–95. doi: 10.1054/drup.1999.0076. [DOI] [PubMed] [Google Scholar]
- 54.Yahi N, et al. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J. Clin. Microbiol. 1999;37:4099–4106. doi: 10.1128/jcm.37.12.4099-4106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cozzi-Lepri A, et al. Thymidine analogue mutation profiles: factors associated with acquiring specific profiles and their impact on the virological response to therapy. Antivir. Ther. 2005;10:791–802. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.