Abstract
Estimation of the covariance structure of longitudinal processes is a fundamental prerequisite for the practical deployment of functional mapping designed to study the genetic regulation and network of quantitative variation in dynamic complex traits. We present a nonparametric approach for estimating the covariance structure of a quantitative trait measured repeatedly at a series of time points. Specifically, we adopt Huang et al.’s (2006a) approach of invoking the modified Cholesky decomposition and converting the problem into modeling a sequence of regressions of responses. A regularized covariance estimator is obtained using a normal penalized likelihood with an L2 penalty. This approach, embedded within a mixture likelihood framework, leads to enhanced accuracy, precision and flexibility of functional mapping while preserving its biological relevance. Simulation studies are performed to reveal the statistical properties and advantages of the proposed method. A real example from a mouse genome project is analyzed to illustrate the utilization of the methodology. The new method will provide a useful tool for genome-wide scanning for the existence and distribution of quantitative trait loci underlying a dynamic trait important to agriculture, biology and health sciences.
Keywords: Functional Mapping, Quantitative Trait Loci, Covariance Estimation, Longitudinal Data, Multivariate Normal Mixture
1 INTRODUCTION
The past two decades have witnessed extensive growth in an effort to map quantitative trait loci (QTLs) in a variety of organisms using statistical methodologies. A QTL refers to a gene or a region of chromosome that is associated with a quantitative trait, such as height, weight, or body mass (Lander & Botstein, 1989; Zeng, 1994; Kao et al., 1999; Lynch & Walsh, 1998; Wu et al., 2007). However, most mapping existing strategies, such as simple, composite, and multiple interval mapping, can only make use of phenotypic measurements at a single time point to estimate the genetic effects of QTLs. While many traits undergo developmental or dynamic changes in time course, these strategies fall short in capturing the temporal pattern of QTL expression. Many attempts to model this type of phenomenon are hindered by complexity in structure and intensive computation. Fortunately, a novel approach, called functional mapping by Ma et al. (2002), provides a useful framework for genetic mapping through mean and covariance modeling of multi- or longitudinal traits. The mean is typically modeled using a biologically relevant parametric function, such as a logistic curve for growth data (Bertalanffy, 1957; West et al., 2001), and the covariance is assumed to follow an AR(1) structure – a common choice in longitudinal data modeling (Diggle et al., 2002). The EM algorithm is used to estimate the model parameters. Functional mapping has the advantage of fully capturing the temporal change of effects of a QTL on an organism’s trait. Because it requires a small number of model parameters to estimate, it is computationally efficient and can be used on data that have limited sample sizes. Functional mapping has shown potential as a powerful statistical method in QTL mapping. It has been used as a modeling tool in a number of areas such as allometric scaling (Wu et al., 2002; Long et al., 2006), thermal reaction norm (Yap et al., 2007), HIV-1 dynamics (Wang & Wu, 2004), tumor progression (Li et al., 2006), biological clock (Liu et al., 2007), and drug response (Lin et al., 2007).
In this paper, we investigate the covariance structure of functional mapping. The covariance is assumed to be identical among different genotypes or segregating groups of a QTL. The assumption of an AR(1) structure in functional mapping, like many longitudinal models, is more of a convenience issue rather than a meaningful approximation. The AR(1) has a simple structure, with only two parameters, and its inverse and determinant have closed forms. This makes computation easier and faster. Furthermore, the EM algorithm formulas for all model parameters at the M-step are easily derived. However, an AR(1) model assumes the data has variance and covariance stationarity. Approximate variance stationarity can usually be achieved by making use of the so-called transform-both-sides (Carroll & Ruppert, 1984) method which does an optimal power transformation of the data (Wu et al., 2004b). The AR(1) model can then be used on the transformed data. But covariance stationarity will still be a problem. Another parametric approach is by using structured antedependence models (SADs) (Zimmerman & Núñez-Antón, 2001; Zhao et al., 2005) which can model both nonstationary variance and correlation functions. Zhao et al. (2005) incorporated SAD in functional mapping and recommended using it in conjunction with an AR(1)-structured model.
The problem with assuming a parametric structure for the covariance matrix in likelihood-based models is that the underlying structure can be significantly different which can lead to considerable bias in parameter estimates. An alternative is to model the covariance matrix nonparametrically. We adopt the method proposed by Huang et al. (2006a) in estimating longitudinal covariance matrices. Their approach is based on the modified Cholesky decomposition (Newton, 1988) wherein the positive-definite covariance matrix Σ of a zero-mean random longitudinal vector y = (y1,…,ym)′, can be uniquely diagonalized as
| (1) |
where T is a lower triangular matrix with ones in the diagonal, D is a diagonal matrix, and ′ denotes matrix transpose. This diagonalization allows modeling of T and D instead of Σ directly. That is, if we can find estimates T̂ and D̂ of T and D, respectively, then an estimator of Σ is Σ̂ = T̂−1 D̂(T̂−1)′ which is positive-definite. It is possible to model T and D because their nonredundant entries have statistical interpretation (Pourahmadi, 1999): the subdiagonal entries of T are the regressions coefficients when each yt (t = 2,…,m) is regressed on its predecessors yt−1,…,y1 and the entries of D are the corresponding prediction error variances. More precisely, y1 = ε1 and for t = 2,…,m,
| (2) |
where −ϕtj is the (t, j)th entry (for j < t) of T, and is the tth diagonal element of D. {ϕtj, j = 1,…,t − 1; t = 2,…,m} and are referred to as generalized autoregressive parameters (GARPs) and innovation variances (IVs), respectively. This implies that modeling the covariance matrix, through T and D, is equivalent to modeling a sequence of regressions. Therefore, variable selection and ridge regression types of procedures can be employed to shrink the regression coefficients to produce a regularized covariance estimator. These techniques are built within a normal penalized likelihood framework by using L1, SCAD, and L2 penalties, respectively (Fan and Li, 2001). In this paper, we adopt the L2 penalty approach and propose an extension of this method to covariance estimation in the mixture likelihood framework of functional mapping. Such an extension is possible by capitalizing on the posterior probability representation of the mixture log-likelihood used in the implementation of the EM algorithm, as will be seen in Section 3. Estimation is then carried out by using the ECM algorithm (Meng & Rubin, 1993) with two CM-steps.
This paper will be organized in the following way. In Section 2, we briefly describe functional mapping. In Section 3, we discuss the nonparametric procedure by Huang et al. (2006a) and describe how it can be integrated into functional mapping. Sections 4 and 5 are devoted to simulation results and analysis of a real data, respectively. Section 6 concludes with a discussion.
2 FUNCTIONAL MAPPING
2.1 Model Formulation
Suppose there is a mapping population of n individuals. Each individual is typed for a panel of molecular markers used to construct a genetic linkage map for the genome. The genetic and statistical principles for linkage analysis and map construction with molecular markers were given in Wu et al. (2007). The mapping population is measured for a phenotypic trait at m time points, with a phenotypic observation vector for individual i expressed as yi = (yi1,…,yim)′. Assume that the trait is controlled by a set of QTLs that form a total of J genotypes. Under the assumption of a multivariate normal density, the phenotype of individual i that carries a QTL genotype k (k = 1,…,J) is expressed as
| (3) |
where the mean genotype value gk is modeled by a logistic curve
| (4) |
and Σ is modeled accordingly, such as by an AR(1), SAD, etc.
The likelihood function can be represented by a multivariate mixture model
| (5) |
where Ω is the parameter vector which we will specify shortly, and pij is the conditional probability of a QTL genotype given the genotypes of flanking markers with . The conditional probability is expressed in terms of the recombination fraction between a putative QTL and the flanking markers that bracket the QTL. Its value is known if the position of a QTL between the two flanking markers is given. In practical computations, a QTL is searched at every 1 or 2 centi-Morgans (cM) on each marker interval throughout a linkage map so that pij is known beforehand. Thus, Ω consists of the mean parameters plus the parameters for Σ, ΩΣ. That is, Ω = (Ωμ, ΩΣ). The reader is referred to Wu et al. (2007) for more about QTL interval mapping.
2.2 Parameter Estimation
The log-likelihood function can be written as
| (6) |
Taking derivatives on equation (6) yields
| (7) |
where
is interpreted as the posterior probability that individual i has QTL genotype k and θ ∈ Ω.
Let P = {Pik, k = 1,…,J; i = 1,…,n}. The maximum likelihood estimates (MLEs) are computed using the EM algorithm (Dempster et al., 1977; Lander & Botstein, 1989; Zeng, 1994; Ma et al., 2002) on the expanded parameter set {Ω, P} as follows:
Initialize Ω.
E-Step: Update P.
- M-Step: Conditional on P, solve for Ω in
Repeat steps (2)–(3) until some convergence criterion is met.
The values at convergence are the MLEs of Ω. Ma et al. (2002) and Yap et al. (2007) provide formulas for updating Ω in the case when the mean is modeled by logistic and rational curves, respectively, and Σ has an AR(1) structure in a backcross population.
After obtaining the MLEs, we can formulate a hypothesis about the existence of a QTL affecting genotype mean patterns as
H0 : a1 = … = aJ, b1 = … = bJ, r1 = … = rJ
H1 : at least one of the inequalities above does not hold,
where H0 is the reduced (or null) model so that only a single logistic curve can fit the phenotype data and H1 is the full (or alternative) model in which case there exist more than one logistic curves that fit the phenotype data due to the existence of a QTL. A number of other important hypotheses can be tested, as outlined in Wu et al. (2004a).
The evidence for the the existence of a QTL can be displayed graphically using the log-likelihood ratio (LR) test statistic
plotted over the entire linkage map, where Ω̃ and Ω̂ denote the MLEs under H0 and H1, respectively. The peak of the LR plot, which we shall from hereon refer to as maxLR, would suggest a putative QTL because this corresponds to when H1 is the mostly likely over H0. The distribution of LR is difficult to determine. However, a nonparametric method called permutation tests by Doerge and Churchill (1996) can be used to find an approximate distribution and a significance threshold for the existence of a QTL. In permutation tests, the functional mapping model is applied to several random permutations of the phenotype data on the markers and a threshold is determined from the set of maxLR values obtained from each permutation test run. The idea here is to disassociate the markers and phenotypes so that repeated application of the model on permuted data will produce an approximate empirical null distribution.
3 COVARIANCE ESTIMATION
3.1 Modified Cholesky Decomposition and Penalized Likelihood
If
where , then equation (7) becomes
by equation (1) where for t = 2,…,m. It is implicitly assumed, therefore, that for k = 1,…,J. Note that if and then εk = Tyk so that var(εk) = TΣT′=D.
For a given tuning parameter λ > 0, define the penalized negative log-likelihood as
| (8) |
where is the L2 penalty function. Conditional on P and Ωμ, minimization of (8) gives the penalized likelihood estimators of T and D and consequently, Σ. The case when λ = 0 gives the maximum likelihood estimator. Other penalty functions can also be used (lam and fan, 2007), but we use the L2-penalty to facilitate the computation.
3.2 ECM Algorithm
If no structure is imposed on the covariance matrix, it is difficult to find closed form M-step solutions in the EM algorithm for the mean parameters in functional mapping. Hence, estimation of the mean parameters is carried out by using an optimization procedure such as the simplex method (Nelder & Mead, 1965) which can be implemented by a built-in function in Matlab. We partition the parameter space according to mean and covariance parameters (Ωμ and ΩΣ) and then use the ECM algorithm (Meng & Rubin, 1993) with two CM-steps. Our general algorithm is outlined as follows:
Initialize Ω = (Ωμ, ΩΣ).
E-Step: Update P.
- CM-Steps:
- Conditional on P and Ωμ, solve for ΩΣ using equations (11) – (13) (Section 3.3) to get .
- Conditional on P and , estimate Ωμ using an optimization procedure.
Repeat steps (2) – (3) until some convergence criterion is met.
3.3 Computing the Penalized Likelihood Estimates
The penalty likelihood, where P and Ωμ are given as in the first CM step of the ECM algorithm, can be written as
Thus, we need to minimize
| (9) |
and
| (10) |
for t = 2,…,m.
The minimizer of (9) is simply
| (11) |
For t = 2,…,m, (10) can be minimized by an alternating minimization over and ϕtj, j = 1,…,t − 1:
- For fixed ϕtj, j = 1,…,t − 1, (10) is minimized with respect to by
(12) - Letting ϕt(t) = (ϕt1, ϕt2,…,ϕt,t−1)′ and , minimization of (10) for fixed , leads to the closed form solution
(13)
where
and It is a (t − 1) × (t − 1) identity matrix.
Note that in formulas (11)–(13), the posterior probabilities, Pik’s, are the weights for the genotype groups, k = 1,…,J.
The preceding calculations were based on the L2 penalty, . If the L1 penalty, , is used instead, a closed form solution like (13) cannot be obtained and an iterative algorithm is needed. This is carried out by using an iterative local quadratic approximation of (Fan and Li, 2001; Öjelund et al., 2001). The reader is referred to Huang et al.(2006a) for additional details.
3.4 Selection of the Tuning Parameter
The tuning parameter λ is selected using a K-fold cross-validation procedure, where K = 5 or 10, but generalized cross-validation can alternatively be used. The criterion is the log-likelihood function (6). The full data set Z is randomly split into K subsets of about the same size. Each subset, say Zs (s = 1,…,K), is used to validate the log-likelihood based on the parameters estimated using the data Z \ Zs. The value of λ that maximizes the average of all cross-validated log-likelihoods is used to select an estimate for Σ.
Note that there really are two sets of tuning parameters in our setting - one under the null model and another under the alternative. However, because the log-likelihood under the null model is constant throughout a marker interval, we shall assume that the corresponding tuning parameter has been estimated accordingly and in the succeeding sections simply refer to the tuning parameters as the ones for the alternative model. This is important for constructing a meaningful and valid test as demonstrated in the generalized likelihood tests by Fan et al. (2001).
4 SIMULATIONS
In this section, the performance of the nonparametric covariance estimator is assessed and compared to an AR(1)-structured estimator. We investigate data generated from both multivariate normal and t-distributions. We begin with the former.
Consider an F2 population in which there are three different genotypes at a single marker or QTL. Since the purpose of the simulation is to investigate the statistical properties of nonparametric modeling for the covariance structure in functional mapping, we will simulate only one linkage group in which a single QTL for a longitudinal trait is located. The simulated linkage group of length 100 cM contains six equally-spaced markers. A QTL is located between the second and third markers, 12 cM from the second marker. Each phenotype associated with the simulated QTL had m = 10 measurements and was sampled from a multivariate normal distribution, using logistic curves as expected mean vectors for three different QTL genotypes and three different types of covariance structure as given below. The curve parameters for three genotypes were a1 = 30, a2 = 28.5, a3 = 27.5 for QQ, b1 = b2 = b3 = 5 for Qq, and r1 = r2 = r3 = 0.5 for qq and the covariance structures were assumed as
Σ1 = AR(1) with σ2 = 3, ρ = 0.6;
Σ2 = σ2{(1−ρ)I + ρ1)}, with σ2 = 3, ρ = 0.5, where 1 is a matrix of 1’s, and I is the identity matrix (Compound Symmetry);
- An unstructured covariance matrix
Σ1 and Σ2 were considered previously by Huang et al. (2006a) and Σ1 by Levina et al. (2008). Σ3 has increasing diagonal elements and decreasing long term dependence which is typical of longitudinal growth data. It is based on the sample covariance matrix of a real data set.
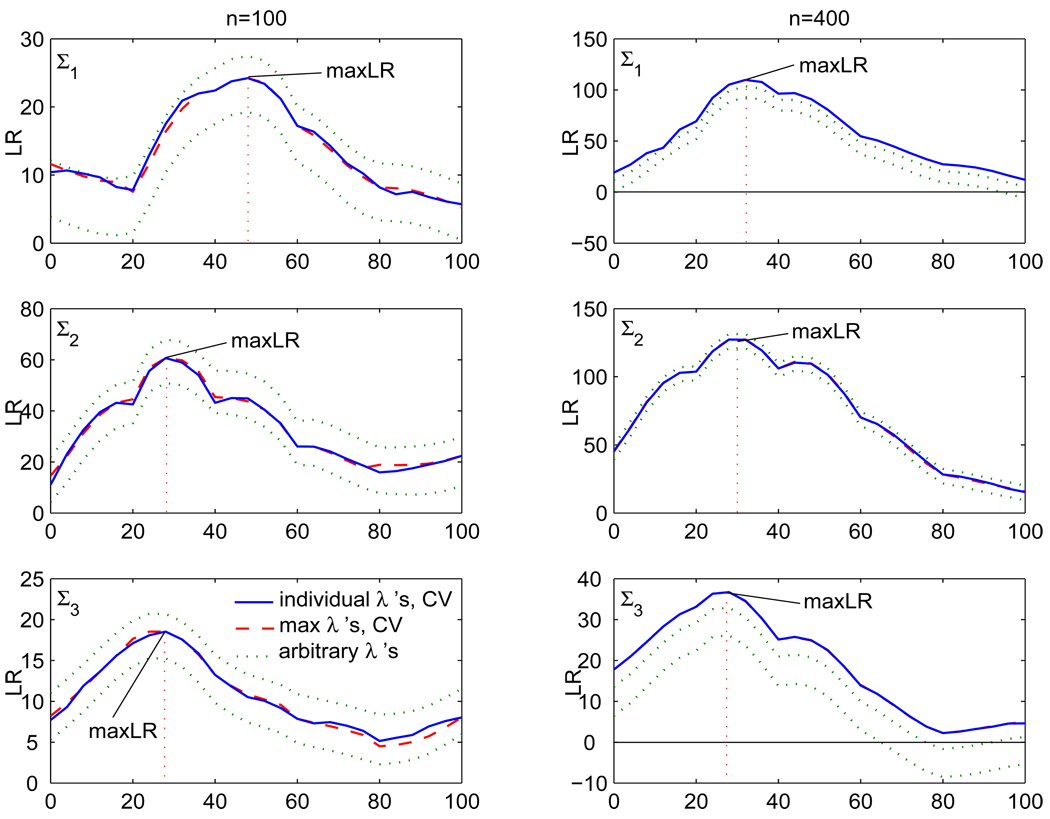
Functional mapping was applied to the simulated data, with n = 100 and 400 samples, using a logistic model for the mean, and the proposed nonparametric estimator and an AR(1) structured estimator for the covariance matrix. The simulated linkage group was searched at every 4 cM (i.e. 0, 4, 8,…,100) for a total of 26 search points across 5 marker intervals. The estimated model parameters at each point were used to construct an LR plot for the QTL linkage map. For the nonparametric covariance estimator, the LR plot is constructed from parameter estimates obtained out of individual tuning parameters λc (c = 1,…,26), that are separately cross-validated. However, we focused our attention only on those λ’s corresponding to the maximum LR at each marker interval. An initial LR plot was constructed using an arbitrary λ0 (λc = λ0 for all c = 1,…,26), and the maximum on each marker interval was located. At the point corresponding to each maximum, λ = λ̂d (d = 1,…,5) was selected using 5-fold cross-validation. The final model parameter estimates were based on the λ̂d that produced the maximum LR or maxLR. In Figure 1, the broken line LR plot is the result of our procedure while the solid one is based on individual λc’s that have each been separately cross-validated. For n = 400, these two plots are indistinguishable. The reason for this is that, the cross-validated λ’s at each search point within a marker interval are not that different from one another. Thus, using one λ for each marker interval (the one that produces the maximum LR) will not significantly alter the general shape of the LR plot. The two dotted line plots were based on λc, for all c = 1, 2,…,26, set to two different arbitrary values of λ. They all have the same location of the maximizer.
Figure 1.
Log-likelihood ratio (LR) plots based on simulated data under three different covariance structures. The solid line plot is based on cross-validated (CV) tuning parameters at each search point (individual λ’s). The broken line plot is based on cross-validated tuning parameters (max λ’s) corresponding to the maximum LR in each marker interval. The dotted line plot is based on two different arbitrary tuning parameter values, each assumed at all search points.
To measure the fit of the estimate Σ̂l (l = 1, 2, 3) of the true covariance structure Σl, we used the nonnegative functions
which correspond, respectively, to entropy and quadratic losses. Each of these is 0 when Σ̂l = Σl and large values suggest significant bias. These functions were also used by Wu & Pourahmadi (2003), Huang et al. (2006a & b), and Levina et al. (2008) to assess the performance of covariance estimators.
A hundred of simulation runs were carried out and the averages on all runs of the estimated QTL location, logistic mean parameter estimates, maxLR, entropy and quadratic losses, including the respective Monte carlo standard errors (SE), were recorded. The results are shown in Tables 1 and 2. For Σ1, the AR(1) estimator performs well as expected, but the nonparametric estimator also does a good job. Both provide better precision with increased sample size. The maxLR values are comparable, i.e., 38.52 and 112.03 from Table 1 versus 37.78 and 128.21 from Table 2, respectively, are not too different from each other.
Table 1.
The averaged QTL position, mean curve parameters, maximum log-likelihood ratios (maxLR), entropy and quadratic losses and their standard errors (given in parentheses) for three QTL genotypes in an F2 population under different sample sizes (n) based on 100 simulation replicates (Nonparametric Estimator, Normal Data).
| Covariance | n | QTL Location |
QTL genotype 1 |
QTL genotype 2 |
QTL genotype 3 |
maxLR | LE | LQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| â1 | b̂1 | r̂1 | â2 | b̂2 | r̂2 | â3 | b̂3 | r̂3 | ||||||
| Σ1 | 100 | 32.84 (0.99) | 30.11 (0.07) | 5.04 (0.04) | 0.50 (0.00) | 28.52 (0.05) | 4.97 (0.03) | 0.50 (0.00) | 27.47 (0.07) | 5.06 (0.04) | 0.50 (0.00) | 38.52 (1.27) | 0.53 (0.01) | 1.00 (0.02) |
| 400 | 31.52 (0.28) | 30.00 (0.03) | 4.99 (0.01) | 0.50 (0.00) | 28.49 (0.02) | 5.01 (0.01) | 0.50 (0.00) | 27.52 (0.03) | 4.97 (0.02) | 0.50 (0.00) | 112.03 (1.80) | 0.14 (0.00) | 0.28 (0.01) | |
| Σ2 | 100 | 32.56 (0.76) | 30.07 (0.06) | 4.98 (0.03) | 0.50 (0.00) | 28.55 (0.04) | 4.99 (0.02) | 0.50 (0.00) | 27.38 (0.06) | 5.07 (0.04) | 0.51 (0.00) | 47.05 (1.32) | 0.44 (0.01) | 0.83 (0.02) |
| 400 | 31.68 (0.26) | 30.04 (0.02) | 4.97 (0.01) | 0.50 (0.00) | 28.48 (0.02) | 5.01 (0.01) | 0.50 (0.00) | 27.54 (0.02) | 4.98 (0.01) | 0.50 (0.00) | 145.83 (2.00) | 0.13 (0.00) | 0.26 (0.01) | |
| Σ3 | 100 | 33.24 (2.22) | 30.07 (0.10) | 5.04 (0.03) | 0.50 (0.00) | 28.59 (0.06) | 5.01 (0.02) | 0.50 (0.00) | 27.66 (0.09) | 5.01 (0.02) | 0.50 (0.00) | 19.57 (0.59) | 0.56 (0.01) | 1.09 (0.02) |
| 400 | 32.32 (1.19) | 29.99 (0.04) | 5.00 (0.01) | 0.50 (0.00) | 28.50 (0.03) | 5.00 (0.01) | 0.50 (0.00) | 27.62 (0.05) | 5.01 (0.01) | 0.50 (0.00) | 38.90 (1.06) | 0.14 (0.00) | 0.29 (0.01) | |
| True values: | 32 | 30 | 5 | 0.5 | 28.5 | 5 | 0.5 | 27.5 | 5 | 0.5 | ||||
Table 2.
The averaged QTL position, mean curve parameters, maximum log-likelihood ratios (maxLR), entropy and quadratic losses and their standard errors (given in parentheses) for three QTL genotypes in an F2 population under different sample sizes (n) based on 100 simulation replicates (AR(1) Estimator, Normal Data).
| Covariance | n | QTL Location |
QTL genotype 1 |
QTL genotype 2 |
QTL genotype 3 |
maxLR | LE | LQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| â1 | b̂1 | r̂1 | â2 | b̂2 | r̂2 | â3 | b̂3 | r̂3 | ||||||
| Σ1 | 100 | 33.24 (0.77) | 29.99 (0.06) | 5.03 (0.04) | 0.50 (0.00) | 28.48 (0.05) | 4.99 (0.03) | 0.50 (0.00) | 27.57 (0.07) | 5.04 (0.05) | 0.50 (0.00) | 37.78 (1.09) | 0.02 (0.00) | 0.04 (0.00) |
| 400 | 31.80 (0.32) | 30.01 (0.03) | 4.97 (0.02) | 0.50 (0.00) | 28.50 (0.02) | 5.02 (0.01) | 0.50 (0.00) | 27.51 (0.03) | 4.98 (0.02) | 0.50 (0.00) | 128.21 (1.98) | 0.01 (0.00) | 0.01 (0.00) | |
| Σ2 | 100 | 35.28 (1.57) | 30.36 (0.09) | 4.63 (0.05) | 0.48 (0.00) | 28.54 (0.07) | 5.04 (0.04) | 0.50 (0.00) | 27.12 (0.09) | 5.51 (0.07) | 0.52 (0.00) | 64.68 (2.53) | 2.15 (0.06) | 6.57 (0.38) |
| 400 | 31.96 (0.54) | 30.51 (0.04) | 4.62 (0.02) | 0.48 (0.00) | 28.42 (0.03) | 5.08 (0.02) | 0.50 (0.00) | 27.14 (0.04) | 5.35 (0.03) | 0.51 (0.00) | 193.84 (4.65) | 2.66 (0.04) | 9.94 (0.25) | |
| Σ3 | 100 | 46.48 (2.74) | 30.39 (0.38) | 5.33 (0.09) | 0.51 (0.00) | 28.01 (0.35) | 4.99 (0.07) | 0.52 (0.00) | 27.85 (0.39) | 5.20 (0.09) | 0.51 (0.00) | 112.66 (2.83) | 9.64 (0.13) | 73.15 (2.06) |
| 400 | 43.64 (2.64) | 30.60 (0.30) | 5.28 (0.06) | 0.51 (0.00) | 27.64 (0.34) | 4.93 (0.07) | 0.52 (0.00) | 28.38 (0.33) | 5.34 (0.08) | 0.50 (0.00) | 288.87 (6.09) | 10.14 (0.07) | 80.36 (1.12) | |
| True values: | 32 | 30 | 5 | 0.5 | 28.5 | 5 | 0.5 | 27.5 | 5 | 0.5 | ||||
For Σ2 and Σ3, the nonparametric estimator performs better than the AR(1) estimator. The AR(1) estimator shows high values for both averaged losses which translates to significantly biased estimates in QTL location and poor mean parameter estimates, particularly for Σ3 at the second and third genotype group. Increased sample size does not help and even makes mean parameter estimates worse in the case of Σ3. Values of maxLR for nonparametric and AR(1) estimators are very different in these cases. But because the averaged losses for the nonparametric estimator are much smaller, we would expect that the corresponding maxLR values must be close to the true ones.
To assess the robustness of our proposed nonparametric estimator, we modeled simulated data from a t-distribution with five degrees of freedom. The results are presented in Tables 3 and 4. The results show that despite inflated average losses, the nonparametric estimator still outperforms the AR(1) estimator. Notice that the quadratic loss is severely inflated because of the fat tails of the t-distribution. It may not be a reliable measure of performance but we present the results here for illustration.
Table 3.
The averaged QTL position, mean curve parameters, maximum log-likelihood ratios (maxLR), entropy and quadratic losses and their standard errors (given in parentheses) for three QTL genotypes in an F2 population under different sample sizes (n) based on 100 simulation replicates (Nonparametric Estimator, Data from t-distribution).
| Covariance | n | QTL Location |
QTL genotype 1 |
QTL genotype 2 |
QTL genotype 3 |
LE | LQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| â1 | b̂1 | r̂1 | â2 | b̂2 | r̂2 | â3 | b̂3 | r̂3 | |||||
| Σ1 | 100 | 32.52 (1.34) | 30.07 (0.08) | 5.02 (0.04) | 0.50 (0.00) | 28.58 (0.06) | 5.02 (0.04) | 0.50 (0.00) | 27.53 (0.09) | 5.07 (0.06) | 0.50 (0.00) | 2.56 (0.12) | 10.51 (0.75) |
| 400 | 32.88 (0.49) | 30.03 (0.04) | 5.01 (0.02) | 0.50 (0.00) | 28.46 (0.03) | 5.00 (0.02) | 0.50 (0.00) | 27.59 (0.03) | 4.99 (0.02) | 0.50 (0.00) | 1.84 (0.06) | 6.24 (0.25) |
|
| Σ2 | 100 | 32.56 (1.08) | 30.15 (0.07) | 4.94 (0.03) | 0.50 (0.00) | 28.54 (0.05) | 5.02 (0.03) | 0.50 (0.00) | 27.47 (0.08) | 5.09 (0.04) | 0.50 (0.00) | 2.27 (0.11) | 8.81 (0.66) |
| 400 | 32.84 (0.03) | 30.06 (0.03) | 4.97 (0.02) | 0.50 (0.00) | 28.48 (0.03) | 5.01 (0.01) | 0.50 (0.00) | 27.53 (0.03) | 5.02 (0.02) | 0.50 (0.00) | 1.78 (0.05) | 5.86 (0.22) |
|
| Σ3 | 100 | 40.92 (2.76) | 29.95 (0.13) | 5.03 (0.03) | 0.50 (0.00) | 28.63 (0.09) | 5.00 (0.02) | 0.50 (0.00) | 27.78 (0.14) | 5.05 (0.04) | 0.50 (0.00) | 2.68 (0.14) | 11.82 (1.26) |
| 400 | 33.08 (1.37) | 29.95 (0.06) | 5.00 (0.01) | 0.50 (0.00) | 28.56 (0.04) | 5.02 (0.01) | 0.50 (0.00) | 27.51 (0.06) | 4.99 (0.02) | 0.50 (0.00) | 1.90 (0.06) | 6.55 (0.26) |
|
| True values: | 32 | 30 | 5 | 0.5 | 28.5 | 5 | 0.5 | 27.5 | 5 | 0.5 | |||
Table 4.
The averaged QTL position, mean curve parameters, maximum log-likelihood ratios (maxLR), entropy and quadratic losses and their standard errors (given in parentheses) for three QTL genotypes in an F2 population under different sample sizes (n) based on 100 simulation replicates (AR(1) Estimator, Data from t-distribution).
| Covariance | n | QTL Location |
QTL genotype 1 |
QTL genotype 2 |
QTL genotype 3 |
LE | LQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| â1 | b̂1 | r̂1 | â2 | b̂2 | r̂2 | â3 | b̂3 | r̂3 | |||||
| Σ1 | 100 | 34.00 (1.12) | 30.04 (0.08) | 5.01 (0.04) | 0.50 (0.00) | 28.61 (0.06) | 5.00 (0.03) | 0.50 (0.00) | 27.51 (0.08) | 5.06 (0.06) | 0.51 (0.00) | 1.65 (0.10) |
5.03 (0.39) |
| 400 | 33.04 (0.40) | 29.98 (0.03) | 4.99 (0.02) | 0.50 (0.00) | 28.48 (0.03) | 5.01 (0.02) | 0.50 (0.00) | 27.61 (0.03) | 4.98 (0.02) | 0.50 (0.00) | 1.61 (0.07) |
4.75 (0.28) |
|
| Σ2 | 100 | 38.92 (1.91) | 30.57 (0.13) | 4.62 (0.06) | 0.48 (0.00) | 28.48 (0.09) | 5.09 (0.05) | 0.50 (0.00) | 27.13 (0.14) | 5.58 (0.09) | 0.52 (0.00) | 6.24 (0.25) |
35.25 (2.50) |
| 400 | 32.16 (0.48) | 30.61 (0.05) | 4.55 (0.02) | 0.48 (0.00) | 28.35 (0.04) | 5.13 (0.02) | 0.51 (0.00) | 27.22 (0.05) | 5.30 (0.03) | 0.51 (0.00) | 7.35 (0.17) |
45.86 (1.75) |
|
| Σ3 | 100 | 49.12 (2.96) | 29.71 (0.50) | 5.23 (0.11) | 0.58 (0.06) | 28.80 (0.38) | 5.21 (0.08) | 0.51 (0.00) | 27.04 (0.49) | 5.37 (0.19) | 0.53 (0.01) | 22.04 (0.56) | 301.53 (14.94) |
| 400 | 42.64 (2.39) | 30.78 (0.38) | 5.38 (0.09) | 0.51 (0.00) | 28.21 (0.35) | 5.08 (0.08) | 0.52 (0.00) | 27.12 (0.36) | 5.05 (0.08) | 0.52 (0.00) | 24.45 (0.49) | 366.54 (15.65) | |
| True values: | 32 | 30 | 5 | 0.5 | 28.5 | 5 | 0.5 | 27.5 | 5 | 0.5 | |||
5 DATA ANALYSIS
We study a real mouse data set from an experiment by Vaughn et al. (1999). Briefly, the data consists of an F2 population of 259 male and 243 female progeny with 96 markers located on a total of 19 chromosomes. The mice were measured for their body mass at 10 weekly intervals starting at age 7 days. Corrections were made for the effects due to dam, litter size at birth, parity, and sex (Cheverud et al., 1996; Kramer et al., 1998).
Functional mapping was first used to analyze this data in Zhao et al. (2004), who investigated QTL × sex interaction. They used a logistic curve to model the genotype means and employed the transform-both-sides (TBS) technique for variance stabilization in order to utilize an AR(1) structure. Their method identified 4 of 19 chromosomes that each had significant QTLs and they concluded that there were sex differences of body mass growth in mice. However, Zhao et al. (2005) applied an SAD covariance structure in functional mapping and found three QTLs. Liu and Wu (2007) likewise analyzed the same data using a Bayesian approach in functional mapping and detected only three significant QTLs.
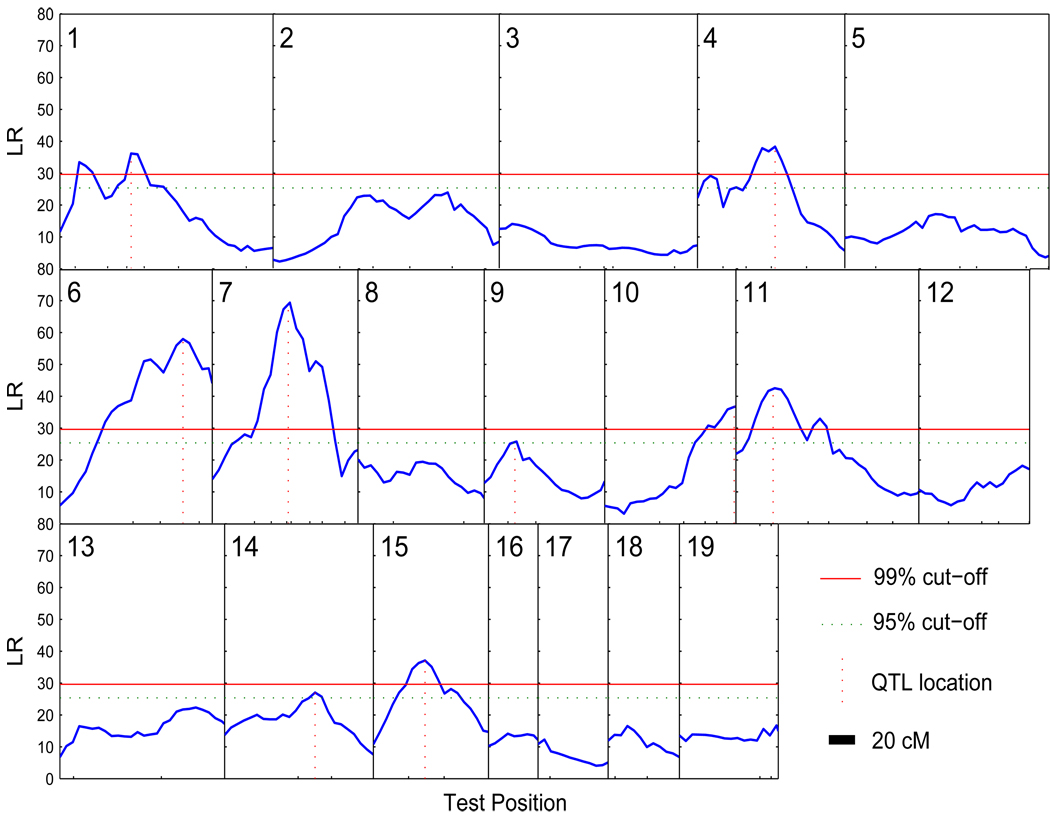
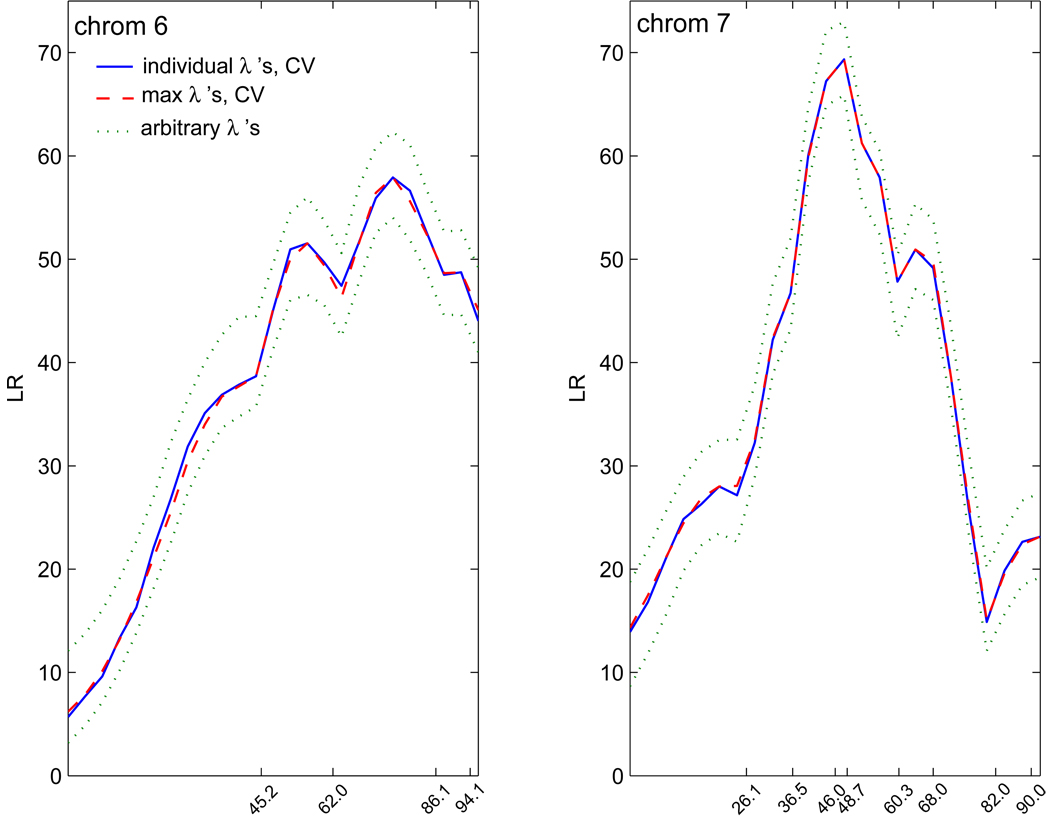
Here, we applied our proposed nonparametric model in a genome-wide scan for growth QTLs without regard to sex. We scanned the linkage map at intervals of 4 cM. Figure 2 shows the LR plots for all 19 chromosomes. They were obtained using λ’s that were cross-validated at each search point. We conducted a permutation test (Doerge and Churchill, 1996; also briefly described in Section 2.2) to identify significant QTLs. For every permutation run, we calculated maxLRe for chromosome e = 1,…,19 using the same general procedure as in the simulations (section 4). In this mouse data set, however, some markers were either missing or not genotyped and we used only the available markers (Table 5). Thus, every marker interval had different sets of available phenotype data. But we believe this did not affect the results because of the large sample size of the available data. We looked at chromosomes 6 and 7 and found this to be the case. Figure 3 shows LR plots based on tuning parameters cross-validated at each search point (solid line) and using the same tuning parameter for each search point as the one corresponding to the maximum LR in each marker interval (broken line; our procedure). The dotted line plots were again based on arbitrary tuning parameters and presented here to illustrate shape consistency. Each permutation run yielded the maximum maxLRe, for all e = 1,…,19, or the genome-wide maxLR. The two horizontal lines in Fig. 2 correspond to 95% (broken) and 99% (solid) thresholds based on 100 permutation test runs. There were nine chromosomes with significant QTLs (1, 4, 6, 7, 9, 10, 11, 14 and 15) based on the 95% threshold but only seven above the 99% threshold (1, 4, 6, 7, 10, 11 and 15). The two chromosomes that did not make the 99% threshold (9 and 14) barely made the 95%. For this mouse data set, we recommend using the 99% threshold because there were only 100 permutation test runs. Zhao et al. (2004) identified QTLs in chromosomes 6, 7, 11 and 15, and Zhao et al. (2005) and Liu and Wu (2007) found QTLs in chromosomes 6, 7 and 10. These were all at the 95% threshold. Our findings verified the results of these previous studies that made use of the functional mapping method and even detected more QTLs. Although there is a discrepancy in our results and others, it is inconclusive to say that these additional QTLs that our proposed model detected are nonexistent. In fact, Vaughn et al. (1999) identified 17 QTLs, although most of them are suggestive, using a simple interval mapping.
Figure 2.
The profile of the log-likelihood ratios (LR) between the full model (there is a QTL) and reduced (there is no QTL) model for body mass growth trajectories across the genome in a mouse F2 population. The genomic position corresponding to the peak of the curve is the optimal likelihood estimate of the QTL localization indicated by vertical broken lines. The ticks on the x-axis indicate the positions of markers on the chromosome. The map distances (in centi-Morgan) between two markers are calculated using the Haldane mapping function. The thresholds for claiming the genome-wide existence of a QTL are shown by horizontal lines.
Table 5.
Available markers and phenotype data of a linkage map in an F2 population of mice (data from Vaughn et al. (1999)).
| Marker Intervals |
||||||||
|---|---|---|---|---|---|---|---|---|
| Chromosome | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| 1 | 378 | 433 | 483 | 467 | 450 | 440 | 466 | |
| 2 | 414 | 404 | 453 | 465 | 430 | |||
| 3 | 477 | 491 | 489 | 476 | 475 | |||
| 4 | 461 | 475 | 481 | 481 | 491 | |||
| 5 | 441 | 439 | 449 | 381 | 385 | |||
| 6 | 467 | 483 | 485 | 481 | ||||
| 7 | 407 | 424 | 459 | 452 | 378 | 372 | 428 | 415 |
| 8 | 395 | 453 | 472 | |||||
| 9 | 498 | 496 | 498 | |||||
| 10 | 401 | 406 | 481 | 490 | 497 | |||
| 11 | 431 | 451 | 468 | 464 | 446 | |||
| 12 | 497 | 489 | 483 | 488 | ||||
| 13 | 450 | 443 | 466 | |||||
| 14 | 443 | 475 | 495 | |||||
| 15 | 491 | 494 | 468 | |||||
| 16 | 498 | |||||||
| 17 | 371 | 394 | ||||||
| 18 | 487 | 479 | 420 | |||||
| 19 | 445 | 468 | 468 | |||||
Figure 3.
Log-likelihood ratio (LR) plots for chromosomes 6 and 7 of the mice data. The solid line plot is based on cross-validated (CV) tuning parameters at each search point (individual λ’s). The broken line plot is based on cross-validated tuning parameters (max λ’s) corresponding to the maximum LR in each marker interval. The dotted line plot is based on two different arbitrary tuning parameter values, each assumed at all search points. Slight differences between the solid and broken line plots may be due to different sample sizes among marker intervals (see Table 5).
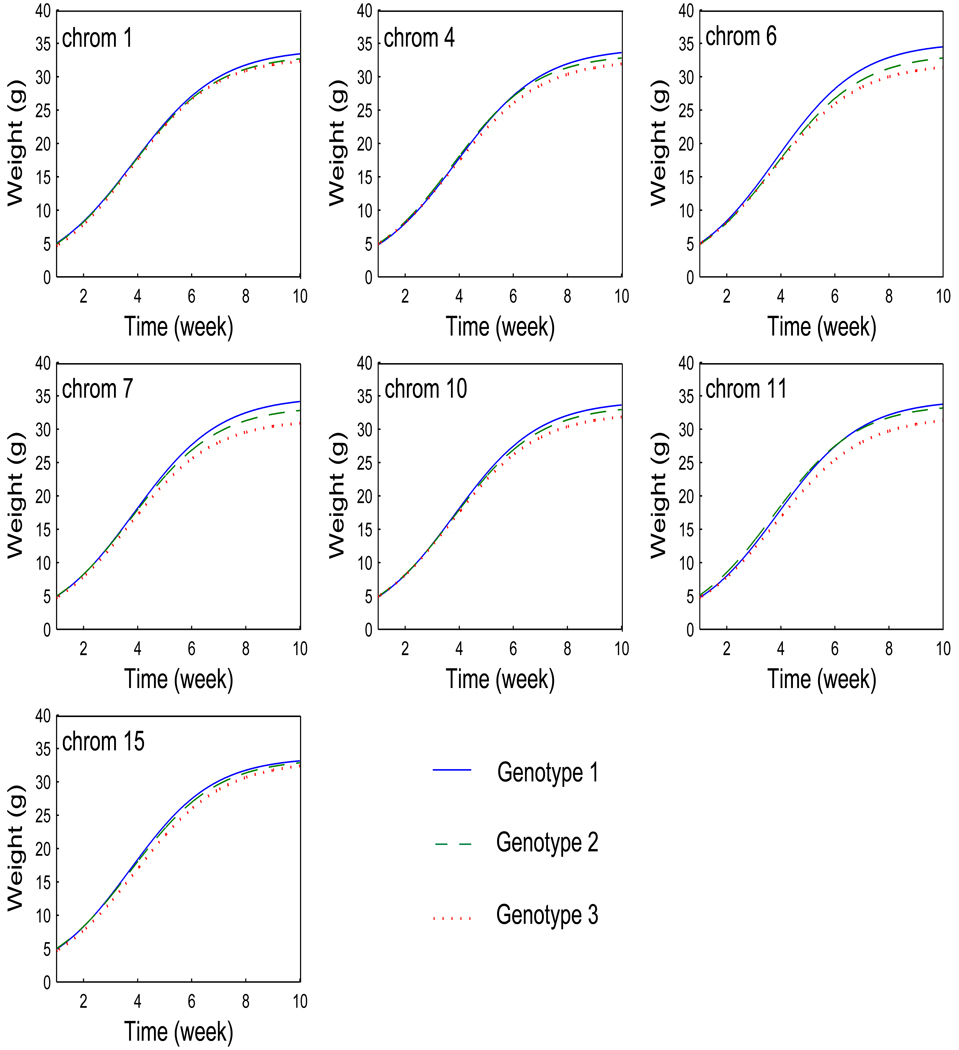
The estimated genotype mean curves for the detected QTLs are shown in Figure 4. Three genotypes at a QTL have different growth curves, indicating the temporal genetic effects of this QTL on growth processes for mouse body mass. Some QTLs, like those on chromosomes 6, 7 and 10, act in an additive manner because the heterozygote (Qq, broken curves) are intermediate between the two homozygotes (QQ, solid curves and qq, dot curves). Some QTL such as one on chromosome 11 are operational in a dominant way since the heterozygote is very close to one of the homozygotes.
Figure 4.
Three growth curves each presenting a genotype at each of seven QTLs detected on mouse chromosomes 1, 4, 6, 7, 10, 11, and 15 for growth trajectories of mice in an F2 population.
6 DISCUSSION
Covariance estimation is an important aspect in modeling longitudinal data. It is difficult, however, because of a large number of parameters to estimate and the positive-definite constraint. Many longitudinal data models resort to structured covariances which, although positive-definite and computationally favorable due to a reduced number of parameters, are possibly highly biased. However, Pourahmadi (1999, 2000) recognized that a positive-definite estimator can be found if modeling is done through the components of the modified Cholesky decomposition of the covariance matrix which converts the problem into modeling a set of regression equations. Wu & Pourahmadi (2003) and Huang et al. (2006b) proposed banding T, noting that terms in the regression farther away in time are negligible and can therefore be set to zero. Huang et al. (2006a) employed LASSO (Tibshirani, 1996) and ridge regression (Hoerl & Kennard, 1970) techniques through L1 and L2 penalties, respectively, in a normal penalized likelihood framework. Lam and Fan (2007) proposed a general penalized likelihood method on the covariance matrix, or precision matrix, or its generalized Cholesky decomposition and showed that the difficulty in estimating a large covariance matrix due to dimensionality increases merely by a logarithmic factor of the dimensionality. They also showed that the biases due to the use of L1-penalty can be significantly reduced by the SCAD penalty. Using these penalties allows shrinkage in the elements of T, even setting some of them to zero in the case of the L1 penalty. Levina et al. (2008) proposed using a nested lasso penalty instead. This type of penalty produces a sparse estimator for the inverse of the covariance matrix by adaptively banding each row of T. Their estimator provides better precision when the dimension is large. Smith & Kohn (2002) proposed a Bayesian approach by using hierarchical priors to allow zero elements in T.
In this paper, we adopted Huang’s L2 penalty approach to produce a regularized nonparametric covariance estimator in functional mapping. This penalty works best when the true T matrix has many small elements. Using the L1 or SCAD penalty gives a better estimator when some of the elements of T are actually zero. However, we believe that the differences in results between using either penalties will not be significant unless the dimension is very large. Nonetheless, the L1 or SCAD penalty can be easily incorporated into our scheme. We have shown how to integrate Huang’s procedure into the mixture likelihood framework of functional mapping. The key was to utilize the posterior probability representation of the derivative of the log-likelihood in (7) and apply an L2 penalty to the negative log-likelihood. Estimation was then carried out using the ECM algorithm with two CM-steps, based on a partition of the mean and covariance parameters. Our simulations have shown better accuracy and precision in estimates for genotype mean parameters, QTL location, and maxLR values, compared to using an AR(1) covariance structure. The maxLR values are important because the complete LR plot provides the amount of evidence for the existence of a QTL. LR values noticeably change when very different covariance structures are used. This is of course under the assumption of multivariate normal data. In our analysis of the mice data, although there were a few chromosomes that were found to have significant QTLs, chromosomes 6 and 7 seemed to have the largest evidence for QTL existence. The LR plots are also used in permutation tests to find a significance threshold. More precise estimates of the covariance structure means better estimates of the the peak of the LR plot and therefore more reliable permutation tests results.
With regards to the utilization of our proposed model, we suggest a preliminary analysis of the data by checking variance and covariance stationarity. If these latter conditions are satisfied then an AR(1) covariance structure may be appropriate. If covariance stationarity is not an issue then a TBS method coupled with an AR(1) model is applicable. If no stationarity is detected then an SAD or the nonparametric model may be more useful. Although we did not assess the comparative performance of these two models, we think that SAD becomes more computationally intensive if the data exhibits long-term dependence, in which case the nonparametric approach may be more appropriate. The nonparametric method should also be considered if other parametric structures are suspect. It can also be used to validate or suggest a family of parametric models.
A recent paper by Yang et al. (2007) proposed a model called composite functional mapping (Zeng 1994) which is an integration of composite interval mapping and functional mapping. Original functional mapping was based on simple interval mapping which searches for QTLs within one marker interval at a time and ignores potential marker effects beyond the marker interval. Composite functional mapping allows modeling of other markers by using partial regression analysis. This significantly improves the precision of functional mapping in QTL detection. However, composite functional mapping assumes an AR(1) covariance structure. It would be advantageous to incorporate our proposed method into this newly developed approach.
Acknowledgments
We thank Dr. James Cheverud for providing us with his mouse data to test our model. The preparation of this manuscript is partially supported by NSF grant (0540745) to R. Wu and NIH R01-GM072611 for NIGMS and NSF grant DMS-0714554 to J. Fan.
References
- Bertalanffy von L. Quantitative laws for metabolism and growth. Quart. Rev. Biol. 1957;32:217–231. doi: 10.1086/401873. [DOI] [PubMed] [Google Scholar]
- de Boor C. A Practical Guide to Splines. Revised ed. New York: Springer; 2001. [Google Scholar]
- Carrol RJ, Rupert D. Power transformations when fitting theoretical models to data. J. Am. Statist. Assoc. 1984;79:321–328. [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J. Roy. Statist. Soc. B. 1977;39:1–38. [Google Scholar]
- Diggle PJ, Heagerty P, Liang KY, Zeger SL. Analysis of Longitudinal Data. UK: Oxford University Press; 2002. [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Statist. Assoc. 2001;96:1348–1360. [Google Scholar]
- Fan J, Zhang C, Zhang J. Generalized likelihood ratio statistics and Wilks phenomenon. Ann. Statist. 2001;29:153–193. [Google Scholar]
- Green P. On use of the EM algorithm for penalized likelihood estimation. J. Roy. Statist. Soc. B. 1990;52:443–452. [Google Scholar]
- Hoerl A, Kennard R. Ridge regression: biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. [Google Scholar]
- Huang J, Liu N, Pourahmadi M, Liu L. Covariance selection and estimation via penalised normal likelihood. Biometrika. 2006a;93:85–98. [Google Scholar]
- Huang J, Liu L, Liu N. Estimation of large covariance matrices of longitudinal data with basis function approximations. J. Comput. Graph. Statist. 2006b;16:189–209. [Google Scholar]
- Kao C-H, Zeng Z-B, Teasdale RD. Multiple interval mapping for quantitative trait loci. Genetics. 1999;152:1203–1216. doi: 10.1093/genetics/152.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C, Fan J. Sparsistency and rates of convergence in large covariance matrices estimation. 2007 doi: 10.1214/09-AOS720. Manuscript (under review). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levina E, Rothman A, Zhu J. Sparse estimation of large covariance matrices via a nested lasso penalty. Ann. Appl. Statist. 2008;2:245–263. [Google Scholar]
- Li HY, Kim B-R, Wu RL. Identification of quantitative trait nucleotides that regulate cancer growth: A simulation approach. J. Theor. Biol. 2006;242:426–439. doi: 10.1016/j.jtbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Lin M, Hou W, Li HY, Johnson JA, Wu RL. Modeling sequence-sequence interactions for drug response. Bioinformatics. 2007;23:1251–1257. doi: 10.1093/bioinformatics/btm110. [DOI] [PubMed] [Google Scholar]
- Liu T, Liu XL, Chen YM, Wu RL. A unifying differential equation model for functional genetic mapping of circadian rhythms. Theor. Biol. Medical Model. 2007;4:5. doi: 10.1186/1742-4682-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Wu RL. A general Bayesian framework for functional mapping of dynamic complex traits. Genetics. 2007 (tentatively accepted). [Google Scholar]
- Long F, Chen YQ, Cheverud JM, Wu RL. Genetic mapping of allometric scaling laws. Genet. Res. 2006;87:207–216. doi: 10.1017/S0016672306008172. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- Ma C, Casella G, Wu RL. Functional mapping of quantitative trait loci underlying the character process: A theoretical framework. Genetics. 2002;161:1751–1762. doi: 10.1093/genetics/161.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X-L, Rubin D. Maximum likelihood estimation via the ECM algorithm: A general framework. Biometrika. 1993;80:267–278. [Google Scholar]
- Nelder JA, Mead R. A simplex method for function minimization. Comput. J. 1965;7:308–313. [Google Scholar]
- Newton HJ. TIMESLAB: A Time Series Analysis Laboratory. Pacific Grove, CA: Wadsworth & Brooks/Cole; 1988. [Google Scholar]
- Ö H Madsen, H, Thyregod P. Calibration with absolute shrinkage. J. Chemomet. 2001;15:497–509. [Google Scholar]
- Pourahmadi M. Joint mean-covariance models with applications to longitudinal data: Unconstrained parameterisation. Biometrika. 1999;86:677–690. [Google Scholar]
- Pourahmadi M. Maximum likelihood estimation of generalised linear models for multivariate normal covariance matrix. Biometrika. 2000;87:425–435. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J. Roy. Statist. Soc. B. 1996;58:267–288. [Google Scholar]
- Vaughn T, Pletscher S, Peripato A, King-Ellison K, Adams E, Erikson C, Cheverud J. Mapping of quantitative trait loci for murine growth: A closer look at genetic architecture. Genet. Res. 1999;74:313–322. doi: 10.1017/s0016672399004103. [DOI] [PubMed] [Google Scholar]
- Wang ZH, Wu RL. A statistical model for high-resolution mapping of quantitative trait loci determining human HIV-1 dynamics. Statist. Med. 2004;23:3033–3051. doi: 10.1002/sim.1870. [DOI] [PubMed] [Google Scholar]
- West GB, Brown JH, Enquist BJ. A general model for ontogenetic growth. Nature. 2001;413:628–631. doi: 10.1038/35098076. [DOI] [PubMed] [Google Scholar]
- Wu RL, Ma C-X, Casella G. Statistical Genetics of Quantitative Traits: Linkage, Maps, and QTL. New York: Springer-Verlag; 2007. [Google Scholar]
- Wu RL, Ma C, Lin M, Casella G. A general framework for analyzing the genetic architecture of developmental characteristics. Genetics. 2004a;166:1541–1551. doi: 10.1534/genetics.166.3.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RL, Ma C, Lin M, Wang Z, Casella G. Functional mapping of quantitative trait loci underlying growth trajectories using a transform-both-sides logistic model. Biometrics. 2004b;60:729–738. doi: 10.1111/j.0006-341X.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- Wu RL, Ma C, Littell R, Casella G. A statistical model for the genetic origin of allometric scaling laws in biology. J. Theor. Biol. 2002;217:275–287. [PubMed] [Google Scholar]
- Wu WB, Pourahmadi M. Nonparametric estimation of large covariance matrices of longitudinal data. Biometrika. 2003;90:831–844. [Google Scholar]
- Yang RQ, Gao HJ, Wang X, Zhang J, Zeng Z-B, Wu RL. A semiparametric model for composite functional mapping of dynamic quantitative traits. Genetics. 2007;177:1859–1870. doi: 10.1534/genetics.107.077321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap JS, Wang CG, Wu RL. A simulation approach for functional mapping of quantitative trait loci that regulate thermal performance curves. PLoS ONE. 2007;2(6):e554. doi: 10.1371/journal.pone.0000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Ma C, Cheverud JM, Wu RL. A unifying statistical model for QTL mapping of genotype × sex interaction for developmental trajectories. Physiol. Genomics. 2004;19:218–227. doi: 10.1152/physiolgenomics.00129.2004. [DOI] [PubMed] [Google Scholar]
- Zhao W, Chen Y, Casella G, Cheverud JM, Wu RL. A non-stationary model for functional mapping of complex traits. Bioinformatics. 2005;21:2469–2477. doi: 10.1093/bioinformatics/bti382. [DOI] [PubMed] [Google Scholar]
- Zimmerman D, Núñez-Antón V. Parametric modeling of growth curve data: An overview (with discussions) Test. 2001;10:1–73. [Google Scholar]