Abstract
HIV-1 replication in simian cells is restricted at an early postentry step because of the presence of an inhibitory cellular factor. This block reduces the usefulness of HIV-1-based lentiviral vectors in primate animal models. Here, we demonstrate that substitution of the cyclophilin A (CyPA) binding region in the capsid of an HIV-1-based lentiviral vector (LV) with that of the macrophage tropic HIV-1 Ba-L resulted in a vector that was resistant to the inhibitory effect and efficiently transduced simian cells. Notably, the chimeric gag LV efficiently transduced primary simian hematopoietic progenitor cells, a critical cellular target in gene therapy. The alterations in the CyPA binding region did not affect CyPA incorporation; however, transduction by the gag chimeric LV seemed to be relatively insensitive to cyclosporin A, indicating that it does not require CyPA for early postentry steps. In dual infection experiments, the gag chimeric LV failed to remove the block to transduction of the WT LV, suggesting that the gag chimeric LV did not saturate the inhibitory simian cellular factor. These data suggest that the CyPA binding region of capsid contains a viral determinant involved in the postentry restriction of HIV-1-based lentiviral vectors. Overall, the findings demonstrate that the host range of HIV-1-based LV can be altered by modifications in the packaging construct.
Keywords: gag‖cyclophilin A‖gene therapy
Replication of HIV-1 depends on host cell factors, and differential expression and species variation of these cellular factors play an important role in determining the tropism of the virus. The first barrier to HIV-1 tropism is at virus entry, which is mediated by binding of the viral gp120 to CD4 together and the chemokine receptor, typically either CCR5 and CXCR4 (1, 2). Cell type-specific expression of CD4 and the chemokine receptors is a primary determinant of HIV-1 cellular tropism. Postentry, cell type-specific restrictions in the HIV-1 replication cycle have also been reported. Reverse transcription is blocked in quiescent T lymphocytes and nondividing macrophages (3–6) by a mechanism that is not yet known. Another rate-limiting step in HIV-1 replication is nuclear translocation of the preintegration complex, which is governed through the host cell karyopherin pathway in an ATP-dependent manner (7). Restrictions at this level have been observed in particular in quiescent cells (8, 9).
Host cell factors that are required for the production of progeny virus are also potential points of virus restriction. Cyclophilin A (CyPA) is encapsidated in HIV-1 virions [but not simian immunodeficiency virus (SIV) or HIV-2] as a result of its interaction with the viral gag protein at a binding site on capsid (CA) at positions 221 and 222 (10, 11). HIV-1 virions produced in the absence of CyPA fail to establish infection of target cells because of a block at an early postentry step. Other cellular cofactors for HIV-1 have been described. CEM15/Apobec 3G interferes with the production of infectious virions in the absence of a functional vif allele (12, 13); TSG101 interacts with the PTAPP motif in gag p6, where it facilitates release of virus from cells (14–16).
HIV-1 exhibits a restricted species tropism and is unable to replicate in several nonhuman primate species (17–23). Virus replication is restricted at reverse transcription and seems to result from an inhibitory activity in the nonhuman primate cells, as it was overcome by high multiplicity of infection (moi) (21–23).
In this study, we analyzed whether the block to transduction in simian cells by an HIV-1-based lentiviral vector (LV) could be overcome by altering the viral gag protein. We found that a gag chimeric LV that contained the CyPA binding region of HIV-1 Ba-L, a macrophage tropic primary isolate, efficiently transduced simian cells. In coinfection experiments, the gag chimeric LV was unable to override the block of transduction by the WT vector, implying that the simian inhibitory factor cannot be saturated by an LV containing the CyPA binding site of HIV-1 Ba-L. The chimeric gag LV also transduced primary baboon CD34+ hematopoietic progenitor cells efficiently, resulting in transduced cell numbers comparable to that of human CD34+ hematopoietic progenitor cells. These results have significant implications for use of LVs in gene therapy.
Materials and Methods
Construction and Production of HIV-1-Based LVs.
HIV-1-based LVs expressing the GFP, yellow fluorescence protein (YFP), and cyan fluorescence protein (CFP) (CLONTECH) were constructed by cloning of the respective cDNAs into the LV construct (24), in which expression is under control of the human cytomegalovirus promoter. In addition, the LV construct contains the following cis-acting sequences: the packaging signal (Ψ) comprising the 5′ UTR and the 5′ sequence of the gag ORF; the rev responsive element (RRE) essential for nuclear export of unspliced viral RNA in the presence of rev; a polypurine tract (cPPT) from the HIV-1 pol gene, which enhances nuclear translocation of the viral DNA in the target cell (24, 25); and the woodchuck hepatitis virus posttranscriptional regulatory element sequence, which improves translation of the transgene (26) (Fig. 1A).
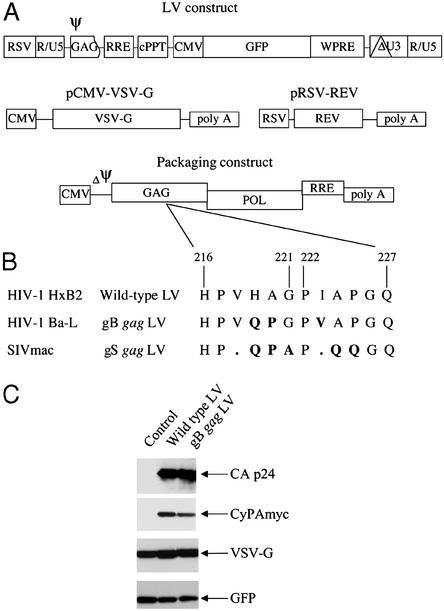
Figure 1.
(A) Schematic presentation of the HIV-1-based LV packaging construct (27). LVs are produced by cotransfections of four different plasmid constructs into 293T cells. The viral RNA genome is produced from the LV construct and contains the promoter and transgene sequences. In addition, the LV construct contains the following cis-acting sequences: packaging signal (Ψ) comprising the 5′ UTR and the 5′ sequence of the gag ORF, the RRE, the cPPT, and the woodchuck hepatitis virus posttranscriptional regulatory element. The 3′ LTR contains a large deletion in the U3 region (depicted as ΔU3). The LV packaging system consists of three constructs: the packaging construct, pRSV-rev, and pCMV-VSV-G. The packaging construct contains in addition the cis-acting RRE and lacks a packaging signal (ΔΨ). (B) Amino acid sequence alignment of the gag CyPA binding region of the WT, gB, and gS gag LV packaging constructs. The WT LV was generated from HIV-1 HxB2. The gB gag LV contains the CyPA binding region of the macrophage tropic HIV-1 Ba-L, and the gS gag LV contains the corresponding region of SIVmac. (C) Western blot analysis for CyPA incorporation into LV virions. LVs were produced in 293T cells by transient transfection of the LV construct expressing GFP, the packaging construct for WT or gB gag LV, pRSV-rev, and pCMV-VSV-G. Myc-tagged CyPA was expressed from an additional construct. In the control cells, the packaging construct was not cotransfected. Virions were isolated on a sucrose gradient, and virion-associated proteins were analyzed by using antibodies against p24, myc, VSV-G, and GFP.
The LV packaging system consists of three constructs encoding for gag/pol (packaging construct), vesicular stomatitis virus glycoprotein envelope (pCMV-VSV-G), and rev (pRSV-rev) (Fig. 1A) (27). The gag/pol packaging construct contains similar to the LV construct the cis-acting RRE, and it requires rev for efficient nuclear export.
LVs were produced by transient transfection in 293T cells by using the calcium phosphate method, as described (27). Infectious LV was harvested at 48 and 72 h posttransfection and filtered through 0.22-μm-pore cellulose acetate filters. The infectious LVs were concentrated by ultracentrifugation (2 h at 50,000 × g) and subsequently purified by ultracentrifugation on a sucrose 20% gradient (2 h at 46,000 × g).
Mutations in the packaging construct were constructed by using a site-directed mutagenesis kit (Stratagene). In the gB gag LV, the CypA binding region of the WT construct was replaced by the CypA of the macrophage tropic HIV-1 variant Ba-L (Fig. 1B), and in gS gag LV, the CyPA binding region of the WT construct was replaced by the corresponding region of SIVmac (Fig. 1B).
Vector concentrations were analyzed by using an immunocapture p24-gag ELISA (Alliance; DuPont/NEN). To determine the infectious titer, 293T cells were plated in 24-well plates at a density of 1 × 105 cells per well and were transduced with serial dilutions of the vector. Four days after inoculation, transduction efficiency was analyzed by fluorescence-activated cell sorter (FACS).
Cell Cultures and Infection Assays.
293T, CV-1, and FrhL2 cells were cultured in DMEM supplemented with 10% FBS (HyClone), antibiotic/antimycotic (Life Technologies, Rockville MD). Twenty-four hours before inoculation, cells were plated in 24-well plates at a density of 1 × 104 per well. Cells were inoculated with different amounts of LV as indicated in the results. The number of infected cells was quantified by FACS analysis 4–6 days after inoculation. Where indicated, cells were treated with cyclosporin A (CsA) (1 μg/ml, Sigma) starting 1 h before inoculation.
Primary human bone marrow CD34+ hematopoietic progenitor cells were purchased from CAMBREX (Baltimore). Purified primary CD34+ G-CSF mobilized peripheral blood hematopoietic progenitor cells from baboon were obtained from the National Heart, Lung, and Blood Institute program of excellence in gene therapy nonhuman primate hematopoietic cell transplantation core. Hematopoietic progenitor cells were inoculated with the indicated LV in serum-free medium (StemSpan; StemCell Technologies, Vancouver) for 2 h. Subsequently, cells were washed and cultured in H5100 medium (StemCell Technologies) supplemented with recombinant human IL-3 (50 ng/ml; StemCell Technologies), recombinant human IL-6 (25 ng/ml; StemCell Technologies), and recombinant human stem cell factor (100 ng/ml; StemCell Technologies). Transduction efficiency was analyzed at day 6 after inoculation by FACS analysis.
CyPA Incorporation by LVs.
Human CyPA was cloned by PCR from a 293T cDNA library and was cloned into the pcDNA3.1/myc-his expression vector (Invitrogen). LVs were produced by transient transfection in 293T cells by using the calcium phosphate method, as described (27), using the following plasmid DNA concentration: 22.5 μg of LV construct, 15.5 μg of packaging construct, 5.5 μg of pRSV-REV, 8 μg of pCMV-VSV-G, and 12.5 μg of pcDNA-CyPA. Twenty-four hours after transfection, the culture medium was replaced, and infectious LV was harvested at 48 and 72 h posttransfection. The culture supernatant was filtered through 0.22-μm-pore cellulose acetate filters and layered on a sucrose gradient consisting of 7.5 ml of 25% sucrose over 7.5 ml of 45% sucrose in Hanks' buffered saline. Gradients were subjected to ultracentrifugation for 2 h at 60,000 × g in a Beckman SW28 rotor. The interphase was harvested and diluted with Hanks' buffered saline to lower the sucrose concentration. Virions were pelleted at 55,000 × g in a Beckman SW41 rotor. The virion pellets were dissolved in Hepes lysis buffer (20 mM Hepes/0.5 M NaCl/1 mM EDTA/0.25% Triton X-100/1 mM EGTA) supplemented with a protease inhibitor mixture (Roche Biochemicals). Samples were run on 12% Tris-glycine gels (NuPAGE, Invitrogen) and blotted onto poly(vinylidene difluoride) membranes according to the manufacturer's instructions. CA proteins were identified by using a murine monoclonal anti-HIV-1 p24 antibody, a gift from H. Schuitemaker (CLB/Sanquin, Amsterdam); myc-tagged CyPA was detected by using the murine anti-myc mAb 9E10; the murine monoclonal anti-VSV-G antibody was obtained from Sigma; and the rabbit anti-GFP antibody was obtained from Novus Biologicals (Littleton, CO). To visualize the specific antibody binding, horseradish peroxidase-conjugated antibodies against rabbit or murine Igs (Amersham Pharmacia) and ECL detection reagent (Amersham Pharmacia) were used.
Results
Generation of gag Chimeric LVs.
Because cell type-specific postentry restrictions have been observed within the wide variety of biological HIV-1 variants (28), we tested whether the CyPA binding region from HIV-1 variants with distinct biological properties could influence the host range of the virus. Comparison of the amino acid sequences of the CA region of the gag proteins of the macrophage tropic HIV-1 variant Ba-L and the LV packaging construct, which is generated from the T-tropic HIV-1 HxB2, showed that amino acids critical for CyPA binding (glycine at position 221 and proline at position 222; refs. 10 and 11) were conserved but that three adjacent amino acids of HIV-1 Ba-L (glutamine 219, proline 220, and valine 223; Fig. 1B) differed. Based on this sequence, we constructed a chimeric gag packaging construct containing the CyPA binding region of HIV-1 Ba-L (gB gag LV). To determine whether the gB gag LV was still able to encapsidate CyPA, LVs were produced in 293T cells expressing myc-tagged CyPA. Virions were isolated on sucrose gradient, and CyPA incorporation was analyzed by Western blotting. Both the WT and the gB gag LV were able to package CyPA (Fig. 1C). CyPA incorporation occurred only in the presence of the packaging construct that was cotransfected, whereas the membrane-bound VSV-G and the cytosolic GFP were also observed in the control lacking the packaging construct.
A second chimera was constructed that contained the analogous region of SIVmac gag (gS gag LV, Fig. 1B). This sequence does not bind CyPA and results in virions that are cyclophilin independent (29). Recombinant LV virions containing the WT and chimeric gag proteins were generated by transfection of 293T cells (27). The WT and chimeric gag virions had comparable infectious vector titer and p24 concentrations (≈1–2 × 105 infectious particles per ng of p24).
Sensitivity to CsA.
Because CyPA is incorporated into the virion by binding to the gag CA protein and is required for an early postentry step in replication, we tested whether CsA, which has high affinity for CyPA, had any effect on the transduction efficiency of the WT and chimeric gag LVs. GFP expressing WT and chimeric gag LVs were produced in the presence of CsA (5 μg/ml) to prevent CyPA incorporation, and their transduction efficiency was analyzed in 293T cells at an inoculum of 5 ng of p24. Five days after inoculation, GFP expression was analyzed. CsA treatment during LV production had no effect on the transduction efficiency of the WT and chimeric gag LVs in 293T cells (Fig. 2A), which may indicate that high concentrations of CyPA present in the target cells supported efficient transduction of CyPA-deficient LVs as described (30, 31).
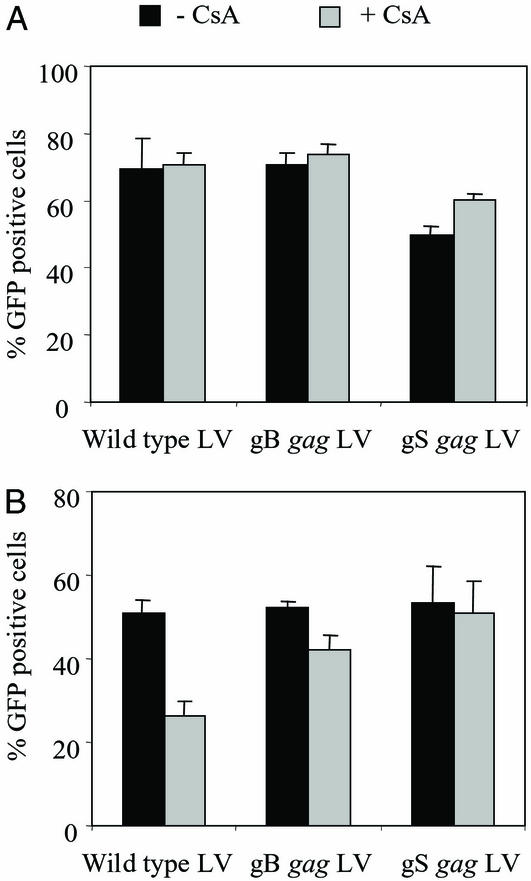
Figure 2.
The effect of CsA treatment on the transduction efficiency of the WT and gag chimeric LVs. (A) Producer cells. WT and chimeric gag LVs expressing GFP were produced in the presence of CsA (5 μg/ml), and their transduction efficiency was analyzed in 293T cells at an inoculum of 5 ng of p24. The average transduction efficiency of two independent experiments is given. Black bar, LVs produced in the absence of CsA; gray bar, LVs produced in the presence of CsA. (B) Target cells. 293T cells were inoculated with 5 ng of p24 of GFP expressing LV in the absence or presence of CsA (1 μg/ml). Five days after inoculation, transduction efficiencies were analyzed by FACS. The average inhibition observed in six independent experiments is given. Black bar, no CsA treatment; gray bar, CsA treatment.
Next, we tested whether CsA treatment of the target cells had any effect on the transduction efficiency of WT and chimeric gag LVs. 293T cells were inoculated with 5 ng of p24 of the WT and chimeric gag LVs expressing GFP in the presence or absence of CsA (1 μg/ml). Five days after inoculation, GFP expression was analyzed. CsA reduced the transduction efficiency of the WT LV by 24.5% but reduced the gB gag LV by only 10.2% (Fig. 2B). Transduction efficiency of gS gag LV was reduced by 2.6% in the presence of CsA (Fig. 2B). The gB and gS chimeric gag LVs were less sensitive to CsA, suggesting that it does not depend on CyPA for early postentry events.
The gB gag LV Efficiently Transduced Simian Cells.
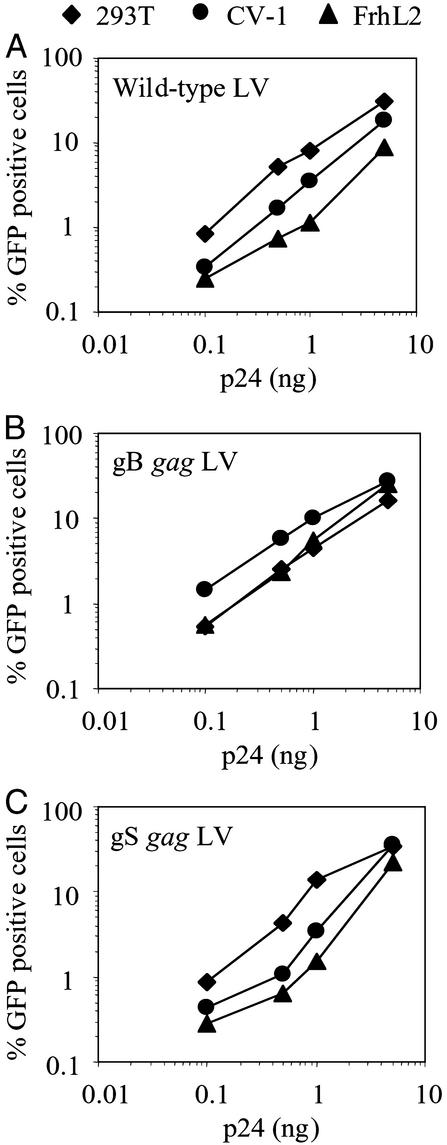
Next, we tested whether the chimeric gag LVs could transduce simian cells. African green monkey kidney cell line CV-1 and the rhesus Macaque fibroblast line FrhL2 were inoculated with increasing amounts of WT and chimeric gag LVs expressing GFP. For comparison, the human 293T cell line was inoculated in parallel. The WT LV efficiently transduced human 293T cells, whereas transduction was reduced in the simian cell lines (Fig. 3A). In contrast, the gB gag LV transduced the simian cell lines as efficiently as it transduced the 293T cells (Fig. 3B). Thus, the chimeric vector escaped the effects of the simian inhibitory factor. Transduction by the gS gag chimeric LV was more efficient in human 293T cells as compared with simian cells, indicating that the SIVmac gag region in the HIV-1 CA is unable to support efficient transduction of simian cells (Fig. 3C). Because the gS gag LV did not alter the tropism of the WT LV, it was not included in subsequent experiments.
Figure 3.
Efficient transduction of simian cells by gB gag LV. 293T (⧫), CV-1 (●), and FrhL2 (▴) cells were transduced with increasing amounts of WT (A), gB (B), and gS (C) gag LVs expressing GFP. Six days after inoculation, transduction efficiencies were analyzed by FACS.
The Inhibitory Activity in Simian Cells Does Not Act on the gB gag Chimera.
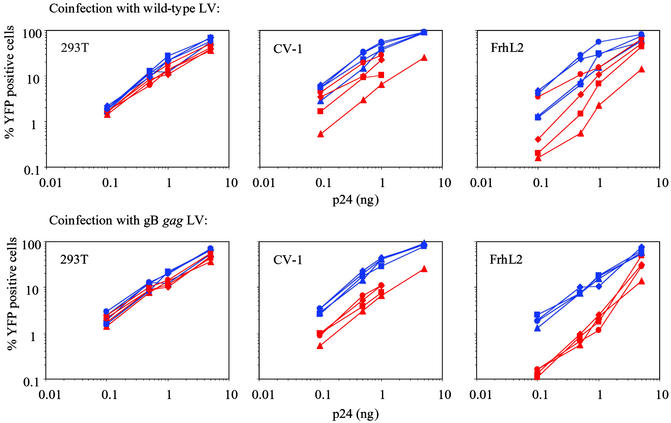
HIV-1 replication is blocked at reverse transcription in simian cells because of the presence of a dominant inhibitor (21–23). The block in HIV-1 infection can be overcome by coinfection and preincubation of the simian target cells with virions at high moi, suggesting that the block is mediated by a saturable inhibitory activity. Because transduction of simian cells by the gB gag LV was unaffected by the inhibitory factor, we surmised that the factor fails to act on chimeric gag. To test this, 293T, CV-1, and FrhL2 cells were coinfected with increasing concentrations of WT and gB gag LVs expressing YFP (0.1–5 ng p24) and WT and gB gag LVs expressing CFP (1, 5, and 10 ng of p24). As in the previous experiment, we found that transduction of the WT LV was impaired in CV-1 and FrhL2 as compared with the human 293T cells, whereas the gB gag LV transduced these cell lines at similar levels. As demonstrated in Fig. 4 Upper, coinfection with a CFP expressing WT vector resulted in a dose-dependent increase of YFP expression from the WT vector in CV-1 and FrhL2. At a dose of 10 ng of p24, transduction efficiency of the WT vector was comparable to that of gB gag LV. YFP expression in CV-1 and FrhL2 from the gB gag LV was also enhanced by coinfection with CFP expressing WT vector, albeit to a lesser extent (Fig. 4 Upper).
Figure 4.
The reduced transduction of WT LV in simian cells cannot be overcome by coinfection with the gB gag LV. 293T, CV-1, and FrhL2 cells were inoculated with increasing amounts of WT (red) and gB gag (blue) LV expressing YFP. Coinfection was performed with WT (Upper) and gB gag (Lower) LV expressing CFP at the following concentrations: 1 ng of p24 (■), 5 ng of p24 (⧫), 10 ng of p24 (●), and no coinfection (▴). Six days after inoculation, YFP expression was analyzed by FACS.
Coinfections with a CFP expressing gB gag LV showed no effect on YFP expression from either vector (Fig. 4 Lower). This finding indicated that the gB gag LV is unable to overcome the block in transduction of the WT vector. These findings are consistent with a model in which an inhibitor binds to the WT but not the gB gag CA. YFP expression from both vectors in 293T cells was not affected by coinfection (Fig. 4 Left).
Enhanced Transduction of Primary Hematopoietic Progenitor Cells from Old World Monkey.
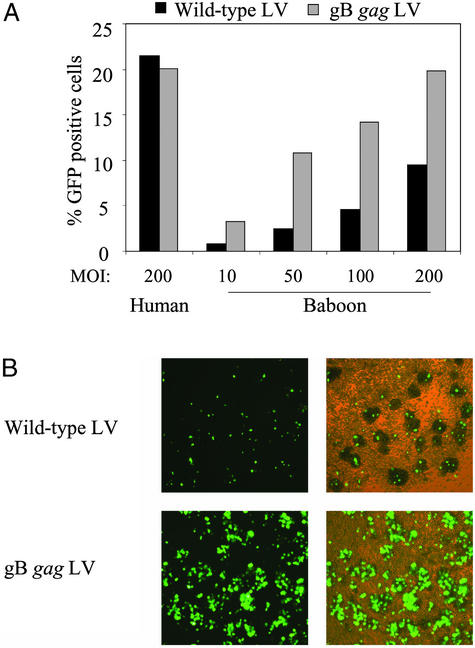
Next, we analyzed whether the gB chimeric gag LV was able to efficiently transduce baboon CD34+ hematopoietic progenitor cells. Baboon CD34+ cells were inoculated with WT and gB gag LV expressing GFP at an moi of 10, 50, 100, and 200. For comparison, human CD34+ hematopoietic progenitor cells were transduced in parallel at an moi of 200. After 2 h, the cells were washed and cultured in medium containing a cytokine mixture that allows expansion and differentiation of the progenitor cells. Human progenitor cells were transduced by the WT and the gB gag LV with similar efficiency. By day 6 postinoculation, ≈20% of the cells expressed GFP (Fig. 5A). Baboon hematopoietic progenitor cells were relatively resistant to transduction by the WT LV, and at an moi of 200, <10% of the cells expressed GFP. In contrast, the gB gag LV transduced baboon progenitor cells more efficiently, and >10% of the cells were transduced at an moi of 50 (Fig. 5). The number of transduced cells increased with the increasing moi, and at an moi of 200, GFP-expressing cell numbers were similar to that of the human progenitor cells.
Figure 5.
Enhanced transduction of primary baboon hematopoietic progenitor cells by gB gag LV. (A) Baboon CD34+ hematopoietic progenitor cells were inoculated with the WT (black bar) and the gB gag LV (gray bar) at an moi of 10, 50, 100, and 200. In parallel, human CD34+ hematopoietic progenitor cells were inoculated with the WT (black bar) and the gB gag LV (gray bar) at an moi of 200. GFP expression was analyzed at day 5 after inoculation by FACS. (B) GFP expression in baboon CD34+ hematopoietic progenitor cell cultures transduced with WT or gB gag LV at an moi of 200. (Left) Fluorescence microscope image. (Right) Merge of fluorescence and light microscopy image (×100).
Discussion
HIV-1 fails to replicate in simian cells because of an early postentry block. Coinfection and heterokaryon experiments suggest that the block is caused by a yet identified dominant inhibitory activity (17–23). This effect reduces the usefulness of HIV-1-based LVs for gene therapy in simian animal models. Here, we demonstrate that alteration of the gag gene of the LV can remove the postentry block, significantly increasing the ability of HIV-1-based LV to transduce simian cell lines and primary baboon hematopoietic progenitor cells.
The mutations that enhanced LV transduction of simian cells were at the CyPA binding region of the gag CA protein. Three amino acids in the packaging construct of the LV, which was derived from the T-tropic HIV-1 HxB2, were substituted with the corresponding residues from the macrophage tropic HIV-1 Ba-L. These alterations did not affect the ability of the LV to encapsidate CyPA; however, transduction was relatively insensitive to CsA, indicating that it does not depend on CyPA for early postentry steps.
Münk et al. (23) recently reported that coinfection or preincubation of simian cells with a large amount of HIV-1 virions would saturate the block to infection of an HIV-1 reporter virus. This finding led to the suggestion that the simian cells contained a limiting quantity of an inhibitory factor that could be absorbed out by the presence of a sufficient quantity of virions. Here, we extend those findings by demonstrating that the WT LV could titrate out the inhibitory activity in simian cells but that gB gag LV could not. This finding is consistent with a model in which an inhibitor binds gag at the CyPA binding region of WT CA but not the gB gag CA. This finding suggests that the viral determinant involved in postentry restrictions of HIV-1 replication in simian cells is located at or near the CyPA binding region of the gag CA protein.
We observed that transduction by the gB gag LV is not affected by the presence of an inhibitor in simian cells and that this LV is unable to saturate the inhibitor. However, we did observe that the transduction efficiency of the gB gag LV in simian cells could be enhanced by coinfection with the WT LV, albeit only 2-fold. The inhibitor present in the simian cells has not been identified yet, and it is unknown whether saturation of the inhibitor by the WT LV has any effect at a cellular level. Therefore, it cannot be excluded that the stimulatory effect on transduction by gB gag LV may be the consequence of changes in intracellular conditions caused by saturation of the inhibitor.
In this study, we were able to relieve the postentry restriction for HIV-1 in simian cells by the generation of a chimeric gag HIV-1-based LV from two biologically different HIV-1 variants. A similar postentry-restricted tropism has been observed for murine leukemia virus (MLV) (32). The murine gene, Fv-1, involved in this restriction has been identified and encodes an endogenous retroviral gag-like protein (33). Fv1 interacts with the gag CA protein of the virus, restricting replication at a preintegration level, probably nuclear translocation. Two different forms of Fv1 have been identified in NIH 3T3 (Fv1N) and BALB/c (Fv1B) cells, thereby dividing MLV in two subclasses: N-tropic and B-tropic. The MLV tropism can be attributed to a single amino acid located in the gag CA of the virus. This single amino acid is also responsible for a similar restriction of N-tropic MLV replication in mammalian cells. Although the restriction of replication in mammalian cells is observed at a different level, before or at reverse transcription, it is possible that the inhibitor (Ref1) present in mammalian cells acts in a manner similar to Fv1 (34, 35). Whether the HIV-1 inhibitory factor present in simian cells is related to Fv1 and Ref1 is unclear.
Previously, it was demonstrated that SIV was restricted in African green monkey cells but not in rhesus macaque cells, whereas HIV-1 was restricted in both cell types (21–23). The restricted HIV-1 tropism could be transferred onto SIV by the HIV-1 CA-p2 domain (22, 36), indicating that this is the viral determinant involved. Here, we show that the CA region of SIV corresponding to the CyPA binding region of HIV-1 CA is unable to alter the cellular tropism of the HIV-1-based WT LV, which suggests that this region is not the primary viral determinant involved in the ability of SIV to replicate in simian cells.
The HIV-1 virion is able to encapsidate CyPA and depends on it for an early step in the virus replication (10, 11). So far, CyPA independent virus replication has not been observed for HIV-1 main group viruses (M-group) (29). We observed that the CyPA binding region of HIV-1 Ba-L made transduction of the WT LV insensitive to CsA, suggesting that replication of the primary HIV-1 Ba-L is independent of CyPA. Differences in the dependence on CyPA have been observed for HIV-1 outlier group (O-group) viruses. Like M-group viruses, O-group viruses incorporate CyPA; however, CyPA-dependent and independent replication has been observed (29, 37).
Postentry restrictions of HIV-1-based LVs have been observed in Old World monkeys and also in cells obtained from rodent, pig, rabbit, and cow (20). This finding has significant relevance for the use of HIV-1-based LVs in animal models for gene therapy. Species-specific cellular factors mediating resistance to the HIV-1-based LV may affect the outcome of these studies. We observed that the gB gag LV transduced primary baboon hematopoietic progenitor cells more efficiently as compared with the WT vector. It will be interesting to determine whether the gB gag LV is able to circumvent postentry restrictions in species other than Old World monkeys.
Acknowledgments
I.M.V. is an American Cancer Society Professor of Molecular Biology and is supported by the National Institutes of Health, the March of Dimes, the Wayne and Gladys Valley Foundation, the H. N. Frances C. Berger Foundation, and the Lebensfeld Foundation. N.R.L. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation and is funded by National Institutes of Health Grant R01 DA14494. C.M. is funded by Deutsche Forschungsgemeinschaft Grant MU 1608.
Abbreviations
- SIV
simian immunodeficiency virus
- LV
lentiviral vector
- CyPA
cyclophilin A
- CsA
cyclosporin A
- CA
capsid
- FACS
fluorescence-activated cell sorter
- RRE
rev responsive element
- moi
multiplicity of infection
- YFP
yellow fluorescence protein
- CFP
cyan fluorescence protein
References
- 1.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 2.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 3.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 4.Korin Y D, Zack J A. J Virol. 1998;72:3161–3168. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuitemaker H, Kootstra N A, Fouchier R A, Hooibrink B, Miedema F. EMBO J. 1994;13:5929–5936. doi: 10.1002/j.1460-2075.1994.tb06938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kootstra N A, Zwart B M, Schuitemaker H. J Virol. 2000;74:1712–1717. doi: 10.1128/jvi.74.4.1712-1717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Stanwick T L, Dempsey M P, Stevenson M. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke E K, Yuan H E, Luban J. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 11.Braaten D, Franke E K, Luban J. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon J H, Gaddis N C, Fouchier R A, Malim M H. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 13.Sheehy A M, Gaddis N C, Choi J D, Malim M H. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 14.Garrus J E, von Schwedler U K, Pornillos O W, Morham S G, Zavitz K H, Wang H E, Wettstein D A, Stray K M, Cote M, Rich R L, et al. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Serrano J, Zang T, Bieniasz P D. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 16.Demirov D G, Ono A, Orenstein J M, Freed E O. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. J Gen Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 18.Himathongkham S, Luciw P A. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 19.Kimball L E, Bosch M L. J Med Primatol. 1998;27:99–103. doi: 10.1111/j.1600-0684.1998.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besnier C, Takeuchi Y, Towers G. Proc Natl Acad Sci USA. 2002;99:11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowan S, Hatziioannou T, Cunningham T, Muesing M A, Gottlinger H G, Bieniasz P D. Proc Natl Acad Sci USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Münk C, Brandt S M, Lucero G, Landau N R. Proc Natl Acad Sci USA. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Follenzi A, Ailles L E, Bakovic S, Geuna M, Naldini L. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 25.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 26.Donello J E, Loeb J E, Hope T J. J Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dull T, Zufferey R, Kelly M, Mandel R J, Nguyen M, Trono D, Naldini L. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouchier R A, Brouwer M, Kootstra N A, Huisman H G, Schuitemaker H. J Clin Invest. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braaten D, Franke E K, Luban J. J Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ackerson B, Rey O, Canon J, Krogstad P. J Virol. 1998;72:303–308. doi: 10.1128/jvi.72.1.303-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin L, Braaten D, Luban J. J Virol. 1998;72:6430–6436. doi: 10.1128/jvi.72.8.6430-6436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goff S P. Cell. 1996;86:691–693. doi: 10.1016/s0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 33.Best S, Le Tissier P, Towers G, Stoye J P. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 34.Towers G, Bock M, Martin S, Takeuchi Y, Stoye J P, Danos O. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towers G, Collins M, Takeuchi Y. J Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorfman T, Gottlinger H G. J Virol. 1996;70:5751–5757. doi: 10.1128/jvi.70.9.5751-5757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiegers K, Kräusslich H-G. Virology. 2002;294:289–295. doi: 10.1006/viro.2001.1347. [DOI] [PubMed] [Google Scholar]