Abstract
CD4 binds to MHC class II molecules and enhances T-cell activation. The CD4-related transmembrane protein LAG-3 (lymphocyte activation gene-3, CD223) binds to the same ligand but inhibits T-cell proliferation. We have previously shown that LAG-3 cell surface expression is tightly regulated by extracellular cleavage in order to regulate its potent inhibitory activity. Given this observation and the contrasting functions of CD4 and LAG-3, we investigated the cell distribution, location and transport of these related cell surface molecules. As expected, the vast majority of CD4 is expressed at the cell surface with minimal intracellular localization, as determined by flow cytometry, immunoblotting and confocal microscopy. In contrast, nearly half the cellular content of LAG-3 is retained in intracellular compartments. This significant intracellular storage of LAG-3 appears to facilitate its rapid translocation to the cell surface following T-cell activation, which was much faster for LAG-3 than CD4. Increased vesicular pH inhibited translocation of both CD4 and LAG-3 to the plasma membrane. While some colocalization of the microtubule organizing center, early/recycling endosomes and secretory lysosomes was observed with CD4, significantly greater colocalization was observed with LAG-3. Analysis of CD4:LAG-3 chimeras suggested that multiple domains may contribute to intracellular retention of LAG-3. Thus, in contrast with CD4, the substantial intracellular storage of LAG-3 and its close association with the microtubule organizing center and recycling endosomes may facilitate its rapid translocation to the cell surface during T-cell activation and help to mitigate T-cell activation.
Keywords: CD4, Cellular activation, Lymphocyte activation gene-3, Protein trafficking, T lymphocytes
Introduction
Lymphocyte activation gene-3 (LAG-3; CD223) is a type I transmembrane protein that is expressed on the cell surface of activated T cells and a subpopulation of NK cells [1]. It has been reported that LAG-3 plays an important role in negatively regulating T-cell activation and proliferation [2]. Adoptively transferred Lag3−/− T cells or T cells co-transferred with anti-LAG-3 mAb exhibited enhanced homeostatic proliferation in lymphopenic hosts [3]. Both natural and induced Treg express increased LAG-3, which is required for their maximal suppressive function [3, 4]. Furthermore, ectopic expression of LAG-3 on CD4+ effector T cells reduced their proliferative capacity and conferred on them regulatory potential against third party T cells [4]. Finally, recent studies have shown that high LAG-3 expression on exhausted LCMV (lymphocytic choriomeningitis virus)-specific CD8+ T cells contributes to their unresponsive state and limits CD8+ T-cell anti-tumor responses [5, 6]. Thus, LAG-3 is an important global regulatory molecule that controls many aspects of T-cell proliferation and homeostasis.
LAG-3 is closely related to CD4, which is a coreceptor for T helper cell activation. Both molecules have four extra-cellular Ig-like domains and require binding to their ligand, MHC class II, for their functional activity [1, 7, 8]. Although the mechanism of LAG-3 function remains unclear, a conserved KIEELE motif in the cytoplasmic domain of LAG-3 is essential [2]. In contrast to CD4, LAG-3 is only expressed on the cell surface of activated T cells [1, 7–10]. LAG-3 surface expression is further regulated by two metalloproteases, ADAM10 and ADAM17, which cleave surface LAG-3, a proportion of which is both constitutive and TCR-ligation induced [11]. Importantly, prevention of LAG-3 cleavage blocks T-cell proliferation and cytokine secretion [11] suggesting that LAG-3 surface expression is under tight regulatory control. This observation raised the question of whether other mechanisms are used to control the expression and distribution of LAG-3. Cytotoxic T-Lymphocyte Antigen 4 (CTLA-4), which is another inhibitory molecule for T-cell activation, is mainly stored in intracellular compartments such as the trans-Golgi network, endosomes and lysosomes [12–17]. Surface expression is tightly regulated by controlled internalization and trafficking to the plasma membrane. This raised the possibility that LAG-3 surface expression might also be regulated by modulating its intracellular storage and trafficking.
In this study, we addressed the following questions. First, what is the extent of intracellular storage and localization of LAG-3 versus its relative CD4? Second, what is the sub-cellular localization of LAG-3 and CD4 in activated T cells? Third, what is the fate of intracellular LAG-3?
Results
Substantial intracellular storage of LAG-3 in activated murine CD4+ T cells
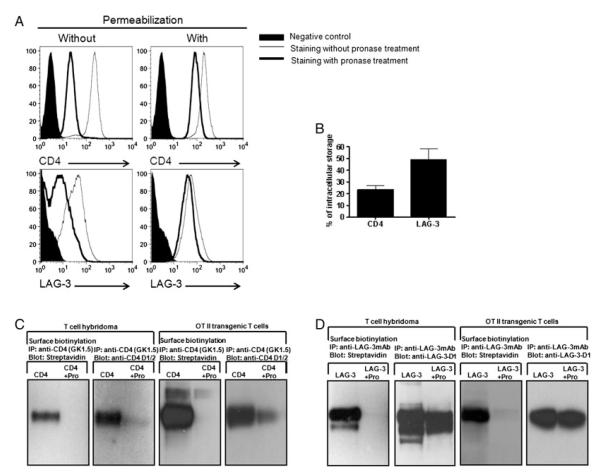
In order to determine cellular distribution of CD4 and LAG-3, we performed intracellular staining for CD4 or LAG-3 using flow cytometry. Freshly isolated naïve CD4+ T cells do not express LAG-3 [10]; so naïve T cells were first stimulated with plate-bound anti-CD3 and anti-CD28 for 72 h and then treated with pronase to remove cell surface CD4 and LAG-3 from activated CD4+ T cells. Pronase treatment removed most of the surface CD4 and LAG-3 on activated T cells (Fig. 1A). While intracellular staining revealed that a relatively small amount (23%) of CD4 is present inside cells, in contrast a greater amount (49%) of LAG-3 appears to be retained intracellularly (Fig. 1A and B). One might speculate that the slightly lower LAG-3 surface expression compared with CD4 following T-cell activation and the increased percentage of intracellular LAG-3 versus CD4 is due to its continuous cleavage by the metalloproteases ADAM10 and ADAM17 that limits surface LAG-3 expression [11, 18]. However, when T cells were treated with the metalloproteinase inhibitor TAPI (Calbiochem), cell surface LAG-3 expression was only slightly increased (data not shown). While prevention of LAG-3 cleavage by TAPI slightly changed the ratio of surface and intracellular LAG-3, the effect was small and not sufficient to account for the differences observed between LAG-3 and CD4. The extent of intracellular LAG-3 storage was also examined by Western blot analysis. Hybridomas ectopically expressing CD4 or LAG-3 were treated with pronase to remove cell surface CD4 and LAG-3, and the intracellularly stored molecules pulled down with either anti-CD4 or anti-LAG-3 Ab. Consistent with the flow cytometry data, there was a small amount of CD4 stored inside cells while a substantial amount of intracellular LAG-3 was detected (Fig. 1C and D). To exclude the possibility that this is an overexpression artifact of T-cell hybridomas, splenocytes from OTII TCR transgenic mice were stimulated with OVA326–339 peptide to induce LAG-3 expression and subjected to the same analysis. These data clearly show that a substantially greater proportion of LAG-3 is stored intracellularly, compared with CD4, in normal T cells (Fig. 1C and D).
Figure 1.
A greater portion of LAG-3, compared with CD4, is stored intracellularly. (A) Freshly isolated CD4+ T cells from C57BL/6 mice were activated with plate-bound anti-CD3 and anti-CD28 for 72 h. Activated T cells were harvested and treated, or not, with pronase to remove surface CD4 and LAG-3. Cells were fixed and permeabilized before staining for intracellular expression analysis following pronase treatment. Treated cells were stained with anti-CD4 or anti-LAG-3 and analyzed by flow cytometry. (B) Normal naïve CD4+ T cells isolated from spleens and lymph nodes of C57BL/6 mice were activated with plate-bound anti-CD3 and anti-CD28 for 72 h. T cells were collected, stained for CD4 and LAG-3 and analyzed by flow cytometry. For surface plus intracellular staining, cells were fixed and permeabilized before staining. Percentage of intracellular CD4 or LAG-3 was calculated by dividing total (surface plus intracellular) CD4 or LAG-3 from the amount of intracellular CD4 or LAG-3. Results are expressed as mean +SEM of three independent experiments. (C and D) LAG-3+ CD4+ 3A9 T-cell hybridomas and OTII TCR transgenic T cells (previously stimulated with APC plus OVA326–339 peptide) were surface-biotinylated and then treated with pronase, or not. Whole cell lysates were immunoprecipitated with (C) anti-CD4 mAb or (D) anti-LAG-3 mAb and probed either with streptavidin, anti-CD4.D1/2 antisera (upper panels) or anti-LAG-3.D1 antisera (lower panels).
To further investigate the localization of CD4 and LAG-3 in activated CD4+ T cells, we used confocal microscopy to visualize intracellularly stored CD4 and LAG-3. CD4 were mainly expressed on the cell surface with only a small portion observed in intracellular locations. While LAG-3 was also expressed on the cell surface, there appeared to be substantially more LAG-3 in the small amount of T-cell cytoplasm that can be observed by confocal microscopy (Fig. 2A and B). After pronase treatment of activated CD4 T cells, most of membrane CD4 and LAG-3 was removed and intracellular storage of CD4 and LAG-3 was observed by confocal microscopy (Fig. 2A). Importantly, Lag3−/− T cells were used to ensure Ab specificity.
Figure 2.
Cell surface and intracellular expression of CD4 and LAG-3. (A) Freshly isolated CD4+ T cells from WT C57BL/6 mice were incubated in 6-well plates coated with anti-CD3 and anti-CD28 mAb. After 72 h, activated T cells were harvested, fixed, stained for CD4 or LAG-3 with or without permeabilization and analyzed by confocal microscopy. Some activated T cells were treated with pronase and stained for CD4 or LAG-3 as described above. (B) Activated T cells were fixed, permeabilized and stained for LAG-3 (anti-LAG-3 mAb and anti-mouse IgG Alexa 555), followed by staining for CD4 (anti-CD4.Alexa 647 mAb).
Rapid translocation of LAG-3 to the T-cell surface
We next investigated the role of intracellular LAG-3 in T cells. We hypothesized that intracellular LAG-3 might facilitate its rapid translocation to the T-cell surface. We first examined the kinetics of surface LAG-3 restoration after pronase treatment. Activated T cells were treated with pronase and surface recovery assessed by flow cytometry following incubation at different time points at 37°C. Surprisingly, restoration of LAG-3 cell surface expression was more rapid than CD4 (Fig. 3). One hour after pronase treatment, 30% of the starting cell surface expression of LAG-3 had been restored in contrast with 10% for CD4. For both molecules, this re-expression was partially blocked within the first hour by the protein synthesis inhibitor cycloheximide and to a slightly greater extent by the protein transport inhibitor Brefeldin A (Fig. 3). Re-expression essentially plateaus after 1 h in the presence of both inhibitors suggesting that the continued increase in LAG-3 and CD4 expression beyond the first hour is due to new protein synthesis. It is noteworthy that this plateau was higher for LAG-3 compared with CD4. In the presence of Brefeldin A for 3 h only 4% of the total surface CD4 compared with 14% of LAG-3 was restored suggesting that a greater proportion of LAG-3 was stored intracellularly, consistent with our previous observations (Fig. 3B and C). Overall, these results suggest that intracellular storage of LAG-3 facilitates its rapid translocation to the cell surface.
Figure 3.
Rapid translocation of LAG-3 to the cell surface. (A) CD4+ T cells 72 h post-activation were treated with pronase and incubated in complete media in the presence or absence of cycloheximide (50 μM) or Brefeldin A (35 μM) at 37°C. At the indicated times, cells were collected, stained with anti-CD4 and anti-LAG-3 and surface expression determined by flow cytometry. Histograms are representative of three independent experiments, which are averaged in the graphs. (B and C) T cells were treated as above and at the time points indicated, surface re-expression of CD4 and LAG-3 was calculated by percentage of surface expression of CD4 and LAG-3 compared with those of untreated cells. Results are mean±SEM of three independent experiments.
Vesicular pH affects movement of CD4 and LAG-3 to the plasma membrane
We next assessed which mechanisms were involved in CD4 or LAG-3 delivery to the plasma membrane in T cells. After removal of cell surface CD4 and LAG-3 with pronase treatment, cells were incubated with colchicine (tubulin polymerization inhibitor) or cytochalasine D (actin polymerization inhibitor) for 3 h and the restoration of cell surface CD4 and LAG-3 was measured. Brefeldin A, which was shown above to block the restoration of LAG-3/CD4 expression, was included as a positive control. Surprisingly, actin and tubulin depolymerization did not affect restoration of cell surface CD4 and LAG-3 (Fig. 4). To confirm disruption of actin and tubulin after inhibitor treatment, we stained actin and tubulin in inhibitor-treated cells and verified disruption of actin and tubulin by confocal microscopy (data not shown). We then incubated pronase-treated T cells with different vesicular acidification/function inhibitors (NH4Cl, chloroquine, concanamycin A). Interestingly, all three inhibitors decreased CD4 and LAG-3 cell surface restoration in T cells (Fig. 4), suggesting that vesicular acidification/function was required for the restoration of both molecules.
Figure 4.
Vesicular pH affects movement of CD4 and LAG-3 to plasma membrane. CD4+ T cells 72 h post-activation were treated with pronase and incubated in complete media in the presence or absence of (A) colchicine (100 μM), cytochalasine D (5 μM), brefeldin A (35μM) or (B) NH4Cl (10 mM), chloroquine (20 μM), concanamycin A (500 nM) or brefeldin A (35 μM) at 37°C. At the indicated times, cells were collected and stained with anti-CD4 and anti-LAG-3 and surface expression determined by flow cytometry. Surface recovery of CD4 and LAG-3 was calculated as described in Fig. 3. Results are mean±SEM of three independent experiments.
Intracellular location and retention of LAG-3 and CD4
To assess the subcellular location of LAG-3 and CD4, we treated activated T cells with pronase, and then permeabilized and stained with anti-CD4 or anti-LAG-3 in conjunction with Ab against different subcellular markers for analysis by confocal microscopy. A significant proportion of LAG-3 appeared to colocalize with the microtubule organizing center (MTOC), using γ-tubulin as a marker (Fig. 5A). While the colocalization of γ-tubulin with LAG-3 was statistically greater than with CD4, as determined using Pearson coefficient analysis (Fig. 5C), some CD4/γ-tubulin colocalization was still evident. A significant proportion of both intracellular CD4 and LAG-3 appeared to colocalize with the early and recycling endosome marker, early endosomal antigen 1 (EEA1), which likely represents newly synthesized protein that is on route to the cell surface and/or in the process of recycling (Fig. 5B and D). To further investigate subcellular location and possible intracellular trafficking pathway of CD4 and LAG-3, we used Rab11b, which is a marker of the endosomal recycling compartment, and Rab27a, which plays a critical role in secretory lysosome-dependent exocytosis. In the staining of both markers, a significantly higher proportion of LAG-3 appeared to colocalize with Rab11b and Rab27a than CD4, although this was most evident with Rab11b:LAG-3 colocalization (Fig. 6). These observations suggest that CD4 and LAG-3 have partially overlapping but distinct patterns of intracellular location and trafficking mechanisms that might play an important role in regulating LAG-3 membrane expression in activated T cells.
Figure 5.
Subcellular location of LAG-3 and CD4 in activated T cells. Activated CD4+ T cells were harvested, treated with pronase, fixed, permeabilized, stained with anti-LAG-3 mAb or anti-CD4 mAb plus either (A) anti-murine γ-tubulin or (B) anti-EEA1 and analyzed by confocal microscopy. Images are representative of three independent experiments. (C and D) Pearson coefficient was calculated using SlideBook software and more than 25 randomly selected cells of each group was analyzed for quantification of colocalization. Student’s t test was used for statistical analysis (***p<0.0001).
Figure 6.
Endosomal localization of intercellular LAG-3 and CD4 in activated T cells. CD4+ T cells were treated as in Fig. 5 except that mAb against the endosomal markers Rab11b (A) and Rab27a (B) were used. Images are representative of three independent experiments. (C and D) Pearson coefficient was calculated as in Fig. 5. Student’s t test was used for statistical analysis (***p<0.0001).
Finally, we asked which domains of CD4 and LAG-3 are important for their differential intracellular retention. We generated a panel of LAG-3/CD4 chimeric constructs that were transduced into a LAG-3−/CD4− 3A9 T-cell hybridoma (Supporting Information Fig. 1A). No enhanced intracellular storage was observed with any of the transductants possessing the N-terminal CD4 D1/D2 domains, while significant intracellular storage was observed with most of the LAG-3 D1/D2 chimeras regardless of the composition of the remaining domains (Supporting Information Fig. 1B and C). This suggested that the LAG-3 D1/D2 domains may contribute to intracellular retention. However, some reduction in intracellular storage was seen with some of the LAG-3/CD4 constructs suggesting either other membrane proximal domains of LAG-3 may contribute or that some domains of CD4 may interfere with retention (Supporting Information Fig. 1C). Taken together, these data suggest that the control of retention is complex and may involve multiple motifs and domains.
Discussion
Like many cell surface molecules, the majority of CD4 is expressed on the cell surface and only a small portion is retained/resides in intracellular locations. Most of this appears to reside in early/recycling endosomes. In striking contrast, approximately half of the LAG-3 molecules are retained intracellularly and appear to reside close to the MTOC and recycling endosomes. Significant colocalization with Rab11b suggests that LAG-3 may be continuously recycled and/or may be poised for rapid plasma membrane translocation. Partial colocalization of LAG-3 with Rab27a, a marker for the secretory lysosomal pathway, may suggest that LAG-3 can reach the plasma membrane through the MTOC via the secretory lysosomal pathway as has been described for CTLA-4 [17]. While these data clearly indicate that the trafficking and cellular location of these two closely related molecules is distinct, further studies will be required to further elucidate this in more detail. It should also be noted that the studies detailed here were performed with murine T cells and it remains to be determined whether similar observations would be made with human T cells.
In resting cells, the rate of CD4 endocytosis is low [19]. T-cell activation by antigen or phorbol esters increases CD4 internalization, which is either recycled to the plasma membrane or degraded in lysosomes [20–22]. After T-cell activation, the MTOC and Golgi are reorientated to the immunological synapse [23]. While some intracellular CD4 molecules appear to reside in or near the MTOC, this is clearly less than observed for LAG-3 (although this may be less evident simply because there is less intracellular CD4). Thus we hypothesize that this concentration of LAG-3 at the MTOC facilitates its rapid translocation to the cell surface following T-cell activation. Indeed, expression of LAG-3 following cell surface pronase treatment appeared to be significantly greater for LAG-3 than CD4, consistent with this notion.
Interestingly, another T-cell inhibitory molecule, CTLA-4, also resides predominantly in intracellular regions [12–17]. Thus it may be important to tightly control the cell surface expression and location of potent inhibitory molecules such as LAG-3 and CTLA-4. Indeed, we have previously shown that LAG-3 surface expression is regulated by cleavage of the extracellular domains by two metalloproteases, ADAM10 and ADAM17, and that prevention of cleavage blocks T-cell activation and cytokine production [11, 18]. This rapid cleavage may suggest that only a small amount of LAG-3 is internalized, and thus a significant intracellular store of LAG-3 may compensate for the lack of a recycling pool of LAG-3. It has been suggested that CTLA-4 is delivered to the plasma membrane via the secretory lysosome pathway, which emanates from the MTOC [17]. It is possible that CTLA-4 and LAG-3 follow a similar pathway. Although we observed some colocalization of intracellular LAG-3 with Rab27a, such definitive analysis is obviously complex in cells with such a small amount of cytoplasm, and additional studies, such as electron microscopic analysis will be required to assess LAG-3 localization and transport further. Given the key role played by LAG-3 in regulating CD4+, CD8+ and Treg function [3–6], a greater understanding of LAG-3 expression, trafficking and function may lead to novel insight into this emerging therapeutic target.
Materials and methods
Mice
C57BL/6 mice were purchased from The Jackson Laboratory (BarHarbor, ME). Lag3−/− mice were provided by Y. H. Chien (Stanford University, PaloAlto, CA) with permission from C. Benoist and D. Mathis (Joslin Diabetes Center, Boston, MA) [24]. OT II TCR transgenic mice were kindly provided by S. Schoenberger (La Jolla Institute for Allergy and Immunology, La Jolla, CA with permission from W. Heath, Walter and Eliza Hall Institute, Parkville, Victoria, Australia) [25]. All animal experiments were performed in American Association for the Accreditation of Laboratory Animal Care-accredited, under specific pathogen-free facilities following national, state and institutional guidelines. Animal protocols were approved by the St. Jude institutional animal care and use committee.
Ab
A new mouse anti-LAG-3 mAb (4-10-C9) specific for the D3/D4 domains was generated. Briefly, 6-wk-old Lag-3−/− mice were given intraperitoneal injections on wk 0, 2 and 4 with a T-cell hybridoma (1 × 107) that ectopically expressed LAG-3. On week 6, the mice were injected intradermally with plasmid DNA that contained the LAG-3 cDNA in PBS. Following an initial screen, the mice with the highest anti-LAG-3 serum titers were hyperimmunized 3 days and 2 days prior to fusion with a murine LAG-3 Ig fusion protein in PBS (37.5 μg/mL). The spleens were fused and the clones screened by flow cytometry for anti-LAG-3 activity using a LAG-3+ T-cell hybridoma and donkey anti-mouse IgA PE (eBioscience, San Diego, CA). Positive clones were subcloned and re-screened. Supernatant from Clone 4-10-C9 was purified over protein G Sepharose (GE Healthcare, Piscataway, NJ).
The following Abs were used for immunoprecipitation and/or Western blotting: rat anti-LAG-3 mAb (C9B7W, specific for the D2 domain; BD-PharMingen, San Diego, CA), anti-CD4 mAb (GK1.5, specific for the D1 domain; BD-PharMingen), rabbit anti-LAG-3.D1 (generated against a D1 loop peptide (DSGQPTPIPALDLHQGMPSPRQPAPGRYTKLH) by Covance Immunology Service (Princeton, NJ) and rabbit anti-murine CD4.D1/D2 (kindly provided by K. Karjalainen, Instituto di Ricerca in Biomedicina, Bellinzona, Switzerland). For surface and intracellular LAG-3 staining by flow cytometry the following conjugates were used: rat anti-mouse LAG-3-AlexaFluor® 647 (AbD Serotec, Oxford, UK) and rat IgG1 isotype control-AlexaFluor® 647 (eBioscience). The following Ab were used for confocal microscopy: anti-CD4-AlexaFluor® 488 or 647 mAb (BD-PharMingen), anti-γ-tubulin Ab (clone Poly 6209) (BioLegend, San Diego, CA), anti-EEA1 (early endosome antigen 1) polyclonal Ab, anti-Rab11b and anti-Rab27a polyclonal Ab (Santa Cruz Biotech, Santa Cruz, CA). Secondary Ab: goat anti-rabbit IgG-AlexaFluor® 555, donkey anti-goat-Alexa-Fluor® 555, chicken anti-mouse IgG AlexaFluor® 647 and goat anti-mouse IgG-AlexaFluor® 488 were from Molecular Probes (Eugene, OR).
Cell constructs and flow cytometry
CD4+ naïve T cells from C57BL/6 WT, Lag3−/− and OT II TCR transgenic mice were negatively purified by MACS separation (AutoMACS, Miltenyi Biotec, Auburn, CA). Briefly, the single cell suspension from spleens and lymph nodes of mice was prepared by homogenization of tissue using a cell strainer followed by red blood cell lysis with Gey’s solution. After washing the cells with labeling buffer (PBS containing 2 mM EDTA), cells were incubated with 10% normal mouse serum on ice for 5 min. Subsequently, cells were stained with biotinylated anti-B220, anti-Gr-1, anti-CD8, anti-TER119, anti-pan NK, anti-CD25, anti-CD11b, anti-CD11c and anti-CD19 antibodies on ice for 15 min. The stained cells were washed twice with labeling buffer and incubated with streptavidin-conjugated magnetic beads (Miltenyi Biotec) at 4°C for 15 min. After incubation, CD4+ naïve T cells were negatively purified by MACS separation. Purity was 96–98% evaluated by flow cytometry. The isolated CD4+ naïve T cell were resuspended in RPMI medium (Mediatech, Manassas, VA) supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and distributed into 6-well plates (5 × 106/well), which were precoated with anti-CD3 and anti-CD28 Ab (2 μg/mL) (eBioscience). For surface and intracellular LAG-3 staining, the cells were harvested 72 h after activation, distributed in 96-well V-bottom plates and washed twice with FACS buffer (PBS plus 5% FBS and 0.05% NaN3). LAG-3 mAb (4-10-C9) AlexaFluor 647 or isotype control was added and the cells incubated for 20 min on ice. The stained cells were washed twice with FACS buffer and analyzed using a FACSCalibur (Becton Dickinson). For intracellular staining of LAG-3, activated T cells were fixed with 4% formaldehyde (polysciences, Warrington, PA) at room temperature (RT) for 15 min and permeabilized with 0.2% Triton X-100 at RT for 5 min. The fixed cells were washed with FACS buffer, stained with the anti-LAG-3 mAb and analyzed as described previously.
LAG-3 and CD4 constructs were produced using recombinant PCR as previously described [2, 26]. All LAG-3 and CD4 constructs were cloned into a murine stem cell virus-based retroviral vector, MSCV-IRES-GFP (pMIG). Details of primers and strategy will be provided on request (vignali.lab@stjude.org). The CD4+ 3A9 T-cell hybridoma (hen egg lysozyme 48–63-specific; H-2Ak-restricted) [27] and a CD4 loss variant (3A9.CD4−) [28] T-cell hybridoma were transduced as described previously [10]. Cells were sorted on a MoFlow (Cytomation, Ft. Collins, CO) for uniform GFP expression.
Surface biotinylation and crosslinking
Biotinylation of cell surface proteins was performed as described previously. In brief, all cells (5 × 106 for T-cell hybridoma and 107 for normal T cells) were washed three times in HBSS (Mediatech, Holly Hill, FL) and then treated with 1 mg/mL NHS-SS-biotin (Pierce, Rockford, IL) for 30 min on ice. Lysine/HBSS (25 mM) was used to quench excess biotin. Cells were then washed three times with HBSS before lysis in 1% NP40 (Sigma-Aldrich, St. Louis, MO).
Immunoprecipitation and immunoblotting
Cells were lysed on ice for 30 min with lysis buffer containing 1% NP40 (50 mM Tris, 150 mM NaCl, 1% NP40, 10 μg/mL leupeptin, 10 μg/mL pepstatin, 10 μg/mL aprotinin, 2 mM pefabloc, pH 7.4). Whole cell lysate was centrifuged at 15 000 × g for 10 min. Supernatant was collected and immunoprecipitated with the Ab indicated. Lysates or eluted proteins from immunoprecipitates were resolved by SDS-PAGE (Invitrogen, Carlsbad, CA) and blots probed as detailed. Blots were developed using ECL (Amersham, Piscataway, NJ) and autoradiography.
Confocal microscopy
CD4+ T cells were incubated in anti-CD3/anti-CD28 coated plates for 72 h, harvested and purified by gradient density centrifugation using Ficoll (Lymphocyte Separation Medium, MP Biomedicals, Solon, OH). Purified CD4+ T cells were fixed with 4% formaldehyde and permeabilized with 0.2% Triton-X-100. Fixed cells were placed on coverglass (Microscope Cover Glass, Fisher Scientific, Pittsburgh, PA) or in glass slide chambers (Lab-Tek® II Chamber Slide™ Syatem, Nunc, Naperville, IL), which were precoated with 0.1% polyethyleneimine solution (Sigma-Aldrich) and allowed to adhere to the slide for 1 h. The attached cells were washed twice with PBS and Image-iT® FX signal enhancer (Invitrogen, Eugene, OR) was added and incubated at RT for 30 min. After washing the cells twice with PBS, 2% non-fat dry milk (Bio-Rad Lab, Hercules, CA) solution was added and incubated at RT for 30 min. Primary Abs in 2% milk solution were added and incubated at RT for 1 h. The slide was washed extensively with PBS and fluorochrome-labeled secondary Abs diluted in 2% milk solution were added and incubated at RT for 1 h. After washing the chamber four times with PBS, the stained cells were mounted using Prolong® Gold antifade reagent with DAPI (Invitrogen, Eugene, OR) and cover slides. Images of the stained cells were taken using a Zeiss Axiovert 200 M confocal microscope (Thornwood, NY) and were analyzed using SlideBook 5.0 software (3i, Santa Monica, CA).
Supplementary Material
Acknowledgements
We are very grateful to Cliff Guy for help with image analysis, Richard Cross, Greig Lennon and Stephanie Morgan for FACS, to the staff of the St. Jude Flow Cytometry core for MACS sorting, to the staff of the Hartwell Center for oligo synthesis and DNA sequencing and especially to Lingqing Zhang, Jennifer Peters and Samuel Connell of the Cell and Tissue Imaging Center for assistance with confocal microscopy analysis. We also wish to thank Klaus Karjalainen, Yueh-hsiu Chien, Christophe Benoist, Diane Mathis, Steve Schoenberger and Bill Heath for reagents, and the Vignali lab for constructive discussion. This work was supported by the National Institutes of Health (NIH) (AI-39480), a Cancer Center Support CORE grant (CA-21765) and the American Lebanese Syrian Associated Charities (ALSAC) (to D.A.A.V).
Abbreviations
- CTLA-4
cytotoxic T-Lymphocyte antigen 4
- EEA1
early endosomal antigen 1
- LAG-3
lymphocyte activation gene-3 (CD223)
- LCMV
lymphocytic choriomeningitis virus
- MTOC
microtubule organizing center
- RT
room temperature
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Supporting Information available online
References
- 1.Triebel F, Jitsukawa S, Baixeras E, Roman-Roman S, Genevee C, Viegas-Pequignot E, Hercend T. LAG-3, a novel lymphocyte activation gene closely related to CD4. J. Exp. Med. 1990;171:1393–1405. doi: 10.1084/jem.171.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Workman CJ, Dugger KJ, Vignali DA. Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J. Immunol. 2002;169:5392–5395. doi: 10.4049/jimmunol.169.10.5392. [DOI] [PubMed] [Google Scholar]
- 3.Workman CJ, Vignali DA. Negative regulation of T cell homeostasis by lymphocyte activation gene-3 (CD223) J. Immunol. 2005;174:688–695. doi: 10.4049/jimmunol.174.2.688. [DOI] [PubMed] [Google Scholar]
- 4.Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, Hipkiss EL, et al. Role of LAG-3 in regulatory T cells. Immunity. 2004;21:503–513. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, et al. Coregulation of CD8+T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosso JF, Kelleher CC, Harris TJ, Maris CH, Hipkiss EL, De Marzo A, Anders R, et al. LAG-3 regulates CD8+T cell accumulation and effector function in murine self- and tumor-tolerance systems. J. Clin. Invest. 2007;117:3383–3392. doi: 10.1172/JCI31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruniquel D, Borie N, Triebel F. Genomic organization of the human LAG-3/CD4 locus. Immunogenetics. 1997;47:96–98. doi: 10.1007/s002510050332. [DOI] [PubMed] [Google Scholar]
- 8.Huard B, Gaulard P, Faure F, Hercend T, Triebel F. Cellular expression and tissue distribution of the human LAG-3-encoded protein, an MHC class II ligand. Immunogenetics. 1994;39:213–217. doi: 10.1007/BF00241263. [DOI] [PubMed] [Google Scholar]
- 9.Baixeras E, Huard B, Miossec C, Jitsukawa S, Martin M, Hercend T, Auffray C, et al. Characterization of the lymphocyte activation gene 3-encoded protein. A new ligand for human leukocyte antigen class II antigens. J. Exp. Med. 1992;176:327–337. doi: 10.1084/jem.176.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Workman CJ, Rice DS, Dugger KJ, Kurschner C, Vignali DA. Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3) Eur. J. Immunol. 2002;32:2255–2263. doi: 10.1002/1521-4141(200208)32:8<2255::AID-IMMU2255>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 11.Li N, Wang Y, Forbes K, Vignali KM, Heale BS, Saftig P, Hartmann D, et al. Metalloproteases regulate T-cell proliferation and effector function via LAG-3. EMBO J. 2007;26:494–504. doi: 10.1038/sj.emboj.7601520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alegre ML, Noel PJ, Eisfelder BJ, Chuang E, Clark MR, Reiner SL, Thompson CB. Regulation of surface and intracellular expression of CTLA4 on mouse T cells. J. Immunol. 1996;157:4762–4770. [PubMed] [Google Scholar]
- 13.Linsley PS, Bradshaw J, Greene J, Peach R, Bennett KL, Mittler RS. Intracellular trafficking of CTLA-4 and focal localization towards sites of TCR engagement. Immunity. 1996;4:535–543. doi: 10.1016/s1074-7613(00)80480-x. [DOI] [PubMed] [Google Scholar]
- 14.Leung HT, Bradshaw J, Cleaveland JS, Linsley PS. Cytotoxic T lymphocyte-associated molecule-4, a high-avidity receptor for CD80 and CD86, contains an intracellular localization motif in its cytoplasmic tail. J. Biol. Chem. 1995;270:25107–25114. doi: 10.1074/jbc.270.42.25107. [DOI] [PubMed] [Google Scholar]
- 15.Valk E, Leung R, Kang H, Kaneko K, Rudd CE, Schneider H. T cell receptor-interacting molecule acts as a chaperone to modulate surface expression of the CTLA-4 coreceptor. Immunity. 2006;25:807–821. doi: 10.1016/j.immuni.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 16.Mead KI, Zheng Y, Manzotti CN, Perry LC, Liu MK, Burke F, Powner DJ, et al. Exocytosis of CTLA-4 is dependent on phospholipase D and ADP ribosylation factor-1 and stimulated during activation of regulatory T cells. J. Immunol. 2005;174:4803–4811. doi: 10.4049/jimmunol.174.8.4803. [DOI] [PubMed] [Google Scholar]
- 17.Iida T, Ohno H, Nakaseko C, Sakuma M, Takeda-Ezaki M, Arase H, Kominami E, et al. Regulation of cell surface expression of CTLA-4 by secretion of CTLA-4-containing lysosomes upon activation of CD4+T cells. J. Immunol. 2000;165:5062–5068. doi: 10.4049/jimmunol.165.9.5062. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Workman CJ, Martin SM, Vignali DA. Biochemical analysis of the regulatory T cell protein lymphocyte activation gene-3 (LAG-3; CD223) J. Immunol. 2004;173:6806–6812. doi: 10.4049/jimmunol.173.11.6806. [DOI] [PubMed] [Google Scholar]
- 19.Marsh M, Pelchen-Matthews A. Endocytic and exocytic regulation of CD4 expression and function. Curr. Top. Microbiol. Immunol. 1996;205:107–135. doi: 10.1007/978-3-642-79798-9_6. [DOI] [PubMed] [Google Scholar]
- 20.Pelchen-Matthews A, Armes JE, Griffiths G, Marsh M. Differential endocytosis of CD4 in lymphocytic and nonlymphocytic cells. J. Exp. Med. 1991;173:575–587. doi: 10.1084/jem.173.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelchen-Matthews A, Boulet I, Littman DR, Fagard R, Marsh M. The protein tyrosine kinase p56lck inhibits CD4 endocytosis by preventing entry of CD4 into coated pits. J. Cell Biol. 1992;117:279–290. doi: 10.1083/jcb.117.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelchen-Matthews A, Parsons IJ, Marsh M. Phorbol ester-induced downregulation of CD4 is a multistep process involving dissociation from p56lck, increased association with clathrin-coated pits, and altered endosomal sorting. J. Exp. Med. 1993;178:1209–1222. doi: 10.1084/jem.178.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kupfer A, Dennert G. Reorientation of the microtubule-organizing center and the Golgi apparatus in cloned cytotoxic lymphocytes triggered by binding to lysable target cells. J. Immunol. 1984;133:2762–2766. [PubMed] [Google Scholar]
- 24.Miyazaki T, Dierich A, Benoist C, Mathis D. Independent modes of natural killing distinguished in mice lacking Lag3. Science. 1996;272:405–408. doi: 10.1126/science.272.5260.405. [DOI] [PubMed] [Google Scholar]
- 25.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 26.Workman CJ, Vignali DA. The CD4-related molecule, LAG-3 (CD223), regulates the expansion of activated T cells. Eur. J. Immunol. 2003;33:970–979. doi: 10.1002/eji.200323382. [DOI] [PubMed] [Google Scholar]
- 27.Allen PM, McKean DJ, Beck BN, Sheffield J, Glimcher LH. Direct evidence that a class II molecule and a simple globular protein generate multiple determinants. J. Exp. Med. 1985;162:1264–1274. doi: 10.1084/jem.162.4.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vignali DA, Strominger JL. Amino acid residues that flank core peptide epitopes and the extracellular domains of CD4 modulate differential signaling through the T cell receptor. J. Exp. Med. 1994;179:1945–1956. doi: 10.1084/jem.179.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.