Abstract
MicroRNAs (miRNA) are small non-coding RNAs of ~22 nucleotides that regulate the translation and stability of mRNA to control different functions of the cell. Misexpression of miRNA has been linked to disruption of normal cellular functions, which results in various disorders including cancers such as leukemias. MicroRNA involvement in disease has been the subject of much attention and is increasing our current understanding of disease biology. Such linkages have been determined by high-throughput studies, which provide a framework for characterizing differential miRNA expression levels correlating to different cytogenetic abnormalities and their corresponding malignancies. In addition, functional studies of particular miRNAs have begun to define the effects of miRNA on predicted mRNA targets. It is clear that miRNAs can serve as molecular markers of leukemias and the hope is that they can also serve as new therapeutic targets. Studies are beginning to elucidate how to deliver therapeutic antagonists to attenuate overexpressed miRNAs and to replace underexpressed miRNAs. In this review, we: i) discuss the current understanding of miRNA function and expression in normal hematopoiesis, ii) provide examples of miRNAs that are misregulated in leukemias, and iii) evaluate the current status and potential future directions for the burgeoning field of antisense oligonucleotides and other therapeutic attempts to intervene in miRNA disregulation in leukemias.
Keywords: antagomir, diagnostic, hematopoiesis, leukemia, miR, microRNA, therapeutic
MiRNA Background
MicroRNAs (miRNA, miR, mir) are small noncoding RNAs of ~22 nucleotides (nt) that post-transcriptionally regulate mRNA expression levels. In 1993 the first miRNA was identified as the Caenorhabditis elegans lin-4 gene, which was found to be involved in the post-transcriptional regulation of lin-14[1]. In 2000, the let-7 miRNA was shown to control developmental timing in the larval-adult switch in C. elegans by repressing lin-41[2]. Let-7 was later found to be conserved in flies and humans [3]. Since then, more miRNAs have been identified in humans, and they have been implicated in a variety of cellular processes, including differentiation, apoptosis, and proliferation [4–6]. Currently, the Sanger Institute has listed 721 miRNAs in humans (www.mirbase.org). Many of the miRNAs are evolutionarily conserved, suggesting that they help control critical cellular processes.
MiRNAs are frequently located in polycistronic clusters. Approximately half of the known miRNAs are mapped to clusters averaging 2 to 3 miRNAs [7]. These clusters are often located in genomically fragile regions and are potentially subject to deletions, amplifications, and other genome-damaging events [7]. MiRNAs have sequence homology from which families with homologous miRNAs are recognized, often differing by a single nucleotide.
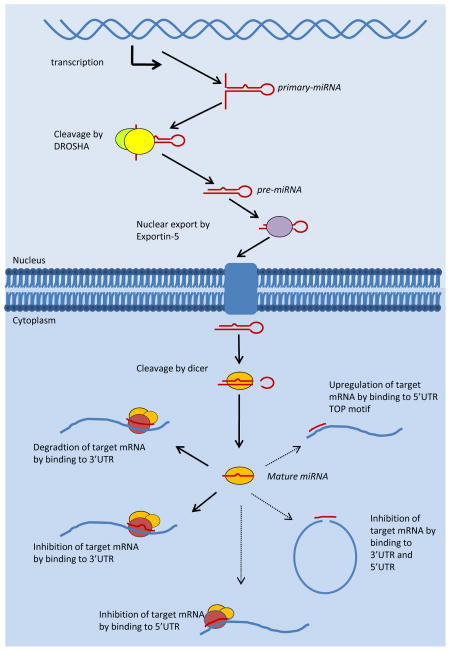
MiRNAs are transcribed by RNA Polymerase II as a primary miRNA transcript containing one or more miRNA sequences [8]. Transcription produces a primary microRNA transcript containing a 5’ cap and polyadenylated tail, as would be the case with other Polymerase II products (e.g., mRNA) [8]. This primary miRNA transcript forms a stable hairpin structure that is cleaved in the nucleus into pre-miRNA of approximately 80 nts by Drosha, a type III RNAse, with assistance from the double-stranded RNA-binding protein DGCR8/PASHA [9–11] (Figure 1). Pre-miRNA are then exported from the nucleus to the cytoplasm by exportin-5, a RAN-GTPase, and subsequently cleaved by Dicer, another type III RNAse, to produce a mature miRNA [12, 13] (Figure 1). Lewis et al. showed that miRNA binding depends on a 6–7 nt seed region that determines both the miRNA family and the targeting of the miRNA to the 3' untranslated region (UTR) of the target mRNA [14]. One strand of the mature miRNA associates with the RNA-induced silencing complex (RISC) as a guide strand whose seed region is projected outward to recognize and bind target mRNA [15] (Figure 1). Perfect and near-perfect binding of the full miRNA sequence to the 3'UTR of the target mRNA results in degradation of target mRNA, whereas imperfect binding results in translational inhibition [12, 16] (Figure 1). Putative miRNA targets with identical sequence matches to the miRNA should be easy to determine. However, when sequence mismatches to the target mRNA are tolerated, it is more difficult to determine miRNA targets because there are many more and a broader range of potential targets of miRNA regulation. This diversity of targets poses a huge problem for those trying to identify the critical targets of a specific miRNA. To predict targets of the miRNAs, several algorithms that differentially weigh various parameters have been developed, but more experimental validation will be required as understanding of miRNA increases.
Figure 1.
MicroRNA processing and function. MicroRNAs are transcribed in the nucleus by RNA Polymerase II and sequentially processed to the mature miRNA form which functions in the cytoplasm. Predominant mechanism of action is illustrated by a bold line. Recently discovered mechanisms are illustrated by thin lines.
Recent studies have increased our understanding of the processing and action of miRNAs on their target mRNA sequences. In addition to inhibition mediated by binding to the 3’UTR, new mechanisms of action for previously verified miRNAs have recently been discovered. Lytle et al. used reporter assays to show miRNA regulation of the 5’UTR of mRNA transcripts. They found that 5’UTR binding is equally able to inhibit translation as 3’UTR binding (Figure 1). Using a luciferase reporter system for the 5’UTR of Ce-lin-41 with an internal ribosome entry site (IRES), they showed repression and moderate levels of translation inhibition by 5’UTR binding of Ce-let-7 [17]. However, it is not yet known whether any mRNA is repressed primarily by this mechanism in vivo. Lee et al. recently proposed an alternative model for mRNA repression in which a miRNA simultaneously binds the 5’UTR and 3’UTR to prevent translational initiation [18] (Figure 1). Another mechanism of action proposed by Lund and Orom derives from the cancer-associated miRNA miR-10a. They showed that miR-10a can enhance the translation of terminal oligo pyrimidine motif– containing transcripts, which encode ribosomal proteins and elongation factors, by binding to the 5’UTR in an “atypical” manner where mismatches in the seed region are tolerated [13, 19] (Figure 1). While these new mechanisms of miRNA action present an additional level of complexity, the extent of their roles in gene regulation has not yet been clearly defined.
The miRNA biogenesis pathway itself (overall level of miRNA and disregulation of miRNA processing machinery) has also come under scrutiny as a potential target of misregulation in cancer. Schmittgen reviewed the efforts of several groups that have attempted to assess the overall level of miRNA in various malignancies to determine whether any global change was associated with cancer - but the results have been mixed. [20]. However, it remains to be seen how specific phases of miRNA biogenesis might be affected in different hematologic malignancies.
Hematopoiesis
Early miRNA research implicated miRNA in the developmental control of various tissues of lower organisms. Given the interrelation of developmentally controlled genes, differentiation, and cancer, it is important to determine the role of miRNAs in hematopoiesis to understand how miRNA contributes to hematologic malignancy.
Reasoning that conserved miRNAs might be critical for hematopoiesis in mammals, Chen et al. initially screened for miRNAs present in hematopoietic cells by cloning. Of the 100 miRNAs cloned, miR-181, miR-223, and miR-142 were identified as miRNAs involved in mammalian hematopoiesis because they were differentially expressed in hematopoietic cell types [4]. When miR-181 was ectopically expressed via retrovirus in lineage negative (Lin-) mouse hematopoietic progenitor cells, it induced B-cell fate lineages in vitro and in vivo in bone marrow replacement experiments. To a lesser extent, expression of miR-142 and miR-223 induced an increase in T-cell and myeloid fates in vitro [4].
Monticelli et al. used a murine model to determine how miRNA is involved in different lineages during hematopoiesis using an array analysis. After northern blotting for confirmation, they outlined the development of several lineages [21]. Spleen B-cell development was correlated with miR-150 upregulation, as well as miR-24, -142-5p, and -142-3p downregulation. Naïve T-cell development was characterized by upregulation of miR-150, followed by miR-150 downregulation during maturation. Th1 and Th2 cells showed differential expression of miR-146, with Th1 cells showing an increase and Th2 cells showing a decrease [21]. Careful consideration of the model system being used to investigate miRNAs in hematopoiesis is warranted because miRNA levels might be differently expressed between humans and mice. For example, Ramkissoon et al. observed that the expression level of miR-181 is restricted to expression in mouse B cells, but in humans miR-181 is also expressed in T cells, monocytes, and granulocytes [22].
Additional high-throughput studies have been undertaken to classify miRNA regulation in hematopoiesis. Georgantas et al. analyzed miRNA expression patterns of hematopoietic stem cells (HSCs) in conjunction with the mRNA expression patterns from bone marrow and peripheral blood stem cells (PBSC) to predict targets of miRNA regulation in the HSCs. Among 33 miRNAs determined to be hematopoietically expressed in both CD34+ and PBSC, the authors predicted the following three functional types of miRNAs: differentiation inhibitors of early hematopoiesis (miR-128a, -181a) differentiation-related miRNAs of multiple lineages (miR-146,-155, -24a, -17, -16, -103, -107), and lineage-specific miRNAs (miR-221, 22, 223) [23]. MiR-155 was predicted to block both myeloid and erythroid differentiation. To validate their results with functional studies, the authors virally introduced miR-155 into K562 cells. Consistent with their high-throughput data, expression of miR-155 inhibited both myeloid and erythroid differentiation [23]. Merkerova and colleagues correlated 13 miRNAs with specific cell lineages isolated from cell lines and healthy patients and then performed a clustering analysis to verify their results [24]. MiR-223 was upregulated in platelets, monocytes, and granulocytes, which is consistent with myeloid involvement of miR-223, as shown in other studies[4, 22]. MiR-150 was upregulated in both B- and T-cells, and miR-451 was upregulated in reticulocytes. They also observed global upregulation of miR-16 and miR-142-3p in all hematopoietic cells compared with non-hematopoietic cell lines [24].
In addition to high-throughput studies to determine the differential expression of miRNAs in hematopoietic lineages, functional studies have experimentally verifed miRNA targets and assigned functional roles to miRNAs in hematopoiesis. Such studies are critical in both verifying correlation of miRNA regulation to hematopoietic lineage with greater sensitivity and understanding the causal roles miRNA disregulation plays in hematopoiesis. Fazi et al. showed that miR-223 is induced during granulopoiesis by C/EBPα-dependent activation in a retinoic acid-dependent manner. MiR-223 acts in a positive feedback mechanism by inhibiting NFI-A, a competitor for C/EBPα on the miR-223 promoter [25]. Fontana et al. studied the regulation of monocytopoiesis by miR-17-5p, -20a, and -106a[26]. MiR-17-5p and -20a are members of the miR-17-92 cluster, while miR-106a is a homolog of miR-17. This study revealed an miRNA circuit where the miRNAs repress translation of the transcription factor, AML1, which negatively regulates the miRNAs in a negative feedback loop[26]. Additionally, AML1 positively regulates M-CSFR to promote monocyte differentiation. In this way, miRNA levels control monocytopoiesis through inhibition of differentiation. Velu et al. found that miR-21 and -196b are regulated by the transcription factor Gfi1 and direct lineage negative bone marrow cells away from granulopoiesis, while miR-21 also promoted monocytopoiesis [27]. Erythropoiesis is regulated by the miRNA cluster of miR-221/222 [28], which downregulates c-kit expression in CD43+ hematopoietic progenitor cells.
As data are compiled and consensus is formed, the role of miRNA regulation throughout the process of hematopoiesis is beginning to be defined. These data have the potential to help explain defects that shape the development of leukemic cells.
MicroRNA involvement in Leukemia
MiRNA involvement in leukemia provides an additional layer of complexity to understanding the development and progression of the disease state. Just as miRNA involvement has been investigated in developmental processes including normal hematopoiesis, the roles of disregulated miRNAs in the development and progression of leukemias has been the subject of multiple studies. As with hematopoiesis, investigators have approached this task using the dual strategies of high-throughput analysis and functional analysis of specific disregulated miRNAs. Collectively, the goals of these types of studies are two-fold: 1) to determine differentially expressed miRNAs that might prove useful diagnostically, and 2) as starting points for developing novel therapies. While high throughput methods may provide clear signatures of miRNA disregulation correlating to karyotypic abnormalities and/or disease prognosis, functional analysis of individual miRNAs is necessary to understand the altered cellular processes.
Chronic Lymphocytic Leukemias
More than half of all chronic lymphocytic leukemias (CLL) have some type of clonal abnormality. However, there is no single, well-characterized cytogenetic abnormality that defines the disease. Deletion of chromosome 13 band q14 (13q14) is one of the most common cytogenetic abnormalities in CLL, occurring in approximately 10–35% of cases as observed cytogenetically, but with 70% frequency when more sensitive fluorescence in situ hybridization (FISH) probes are used [29].
Clinically, CLLs are divided into two forms without regard to cytogenetic abnormalities— an indolent form that does not require immediate treatment and an aggressive form characterized by disease progression that requires therapeutic intervention. Prognosis is correlated with several different markers, i.e., poor prognosis is correlated with loss of ZAP-70 and unmutated IgVH [29]. Calin et al. found that over half of all CLLs contain a limited deletion at 13q14 encompassing the polycistronic miRNA cluster, miR-15a/16-1 [30]. This deletion was the first evidence of miRNA involvement in cancer. MiR-15a/16-1 deletion is associated with the indolent form of the disease and is involved with repression of Bcl-2, a pro-survival protein that antagonizes intrinsic apoptosis[6]. Additional loss of miR-29 and miR-181 has been correlated with the more aggressive form of the disease [31]. MiR-29b and miR-181b regulate Tcl1, which is implicated as an oncogene in an early aggressive form of B-lineage CLL in an AP-1- dependent and NF-κB-dependent manner [32]. In transgenic mice, Tcl1 overexpression under an IgVH promoter resulted in a disease state closely resembling CLL [33, 34]. Chen et al. found that overexpression of miR-181 led to increased B cell differentiation in vitro and in vivo [4). MiR-181a and -181b have also been implicated in repression of the homeotic gene, HOXA11, in myoblasts, with a greater effect observed for miR-181a [35]. In a high-throughput study to clarify miRNA signatures in CLL, two main clusters were found in unsupervised analysis, which correlated with the status of the negative prognostic markers ZAP-70 and unmutated IgVH (Table 1)[36].
TABLE 1.
Differentially expressed miRNA signatures among leukemias.
| Subject | Compared | Upregulated | Downregulated | Reference |
|---|---|---|---|---|
| CLL (ZAP70 high, IgVH unmutated) | CLL (no ZAP70, IgVH mutated) | miR-15a, -195, -221, -23b, -155, -24-1, -146, -16-1, -16-2 | miR-223, -29a-2, -29b-2, -29c | Calin, et al. [36] |
| CLL | CD19+ normal | miR-331, -29a, -195, -34a, -29c | miR-135b, -199s, -142-5p, -185, -181c | Zanette, et al. [44] |
| CML (imatinib resistant) | CML (imatinib responsive) | miR-191 | miR-7, -23a,-26a, -29a, -29c, -30b, -30c, - 100, -126*, -134, -141, -183 ,-196b, -199, - 224, -326, -422b, -520a | San Jose Enirez, et al. [43] |
| ALL | CD19+ normal | miR-128b, -204, -218, -331, -181b | miR-135b, -132, -199, -139, -150 | Zanette, et al. [44] |
| ALL | normal CD34+ | miR-128a, -142-3p, 142-5p, -150, -151-5p, - 181a, -181b, -181c, -193, -30e-5p, -34b, -365, -582, -708 | miR-100, -125b, -99a, -196b, let-7e | Schotte, et al. [48] |
| t(11q23) ALL | AML | miR-128a, - 128b,-130, -151, -210, j-miR-1 | miR-223, -125a, -221, -222, -23a, -23b, -24, - 27a, -27b, -199b, -26a, -335, -21, -22, -424, - 451, let-7a, let-7b, let-7c, let-7e | Mi, et al. [46] |
| t(11q23) ALL | other ALL | miR-196b | miR-193, -151-5p, -30e-5p, -34b, -582, -708, let-7e | Schotte, et al. [48] |
| AML | normal CD34+ | NA | miR-126, -130a, -135, -93, -146, -106b, -224, 125a, -92, -106a, -95, -155, -25, -96, -124a, - 18, -20, -7d, -26a, -222, -101, -338, -371, - 199b, -29b, -301 | Garzon, et al.[49] |
| t(8;21), inv(16), t(16;16) AML | other AMLs | miR-126/126*, -130a | miR-196b, -17-5p, -17-3p, -18a, -19a, 19b, 20a, 92 | Li, et al. [51] |
| inv(16) | other AMLs | miR-424, -199b, -365, -335, -511, -193a | miR-192, -296, -155, -148a, -218, -135b, -196b, -196a, -432, -135a, -10a, -10b, -127, - let-7b | Jongen-Lavrencic, et al. [50] |
| t(8;21) AML | other AMLs | miR-126* | miR -19a, -221, -107, -188, -338, -342, -20b,-187, -501, -339, -210, -502, -182, -500, -152,-135a, -148a, -125b, -100, -99a, -1, -133a, - 133b, -224, -9, -10a, - 10b, -196a, -196b, let- 7b, let-7c | Jongen- Lavrencic, et al. [50] |
| t(15;17) AML | other AMLs | miR-130a, -130b, -335, -148a, -222, -146a, - 181d, -193a, -450, -213, -199, -409-5p, - 181b, -496, -181a, -424, -497, -154, -125b, - 365, -369-5p, -99a, -203, -433, -323, -494, - 100, -370, -432, -224, -127, -452, -299-5p, - 376a, -134, -485-5p, -382, -379, -193b | miR-196a, -196b, -151,-10b, let-7c | Jongen- Lavrencic, et al. [50] |
| t(15;17) AML | other AMLs | miR-181a, -181b, -181c, -181d, -100, -125b, -224, -368, -382, -424 | miR-126/126*, -422b, -10a, -150, -124a, -17- 5p, -20a, j-miR-2 | Li, et al. [51] |
| t(11q23) AML | other AMLs | miR-9, -429 | miR-213, -146a | Jongen- Lavrencic, et al. [50] |
| t(11q23) AML | other AMLs | miR-326, -219, -194, -301, -324, -339, -99b, 328 | miR-34b, -15a, -29c, -372, -30a, -29b, -30e, - 196a, let-7f, -102, -331, -229, -193 | Garzon, et al.[49] |
| t(11q23) AML | other AMLs | miR-10a, -10b, -124a, -196b, -17-5p, -17-3p, -18a, -19a, -19b, -20a, -92, j-miR-2 | miR-126/126*, -130a, -146a, -181a, -181b, - 181c, -181d, -224, -368, -382, -424 | Li, et al.[51] |
| NPM1 mutated AML | other AMLs | miR-10a, -10b, -135a, -196b, -196a, -152, let-7b | miR-99b, 323, -143, -146a, -497, -320 -511, - 450, -151, -494, -193b, -365, -203, -335, - 130a, -485-5p, -126*, -299-5p, -433, -451, - 134, -370, -379, -432, -224, -382, -376a, - 424, -127 | Jongen- Lavrencic, et al. [50] |
| NPM1 mutated AML | NPM1 unmutated AML | miR-10a, -10b, -100, -21, -16a, -16b, -19b, - 18a, -29c, -29a, -16-1, -29b, -24, -20, -17, - 369, - 19a, -106, -16-2, -195, -102, -152, -9, - 142, -378, -98, -374, -15a, 155, let-7a-3, let- 7f, let-7c, let-7a-2, let-7a-1, let-7g, let-7d | miR-22, -192, -128a, -383, -373, -324, -127, - 373, -324, -127, -373*, -139, -193b, -145, - 498, -135a, -299, -429, -493, -326, -204, - 198, 486 | Garzon, et al. [62] |
| FLT3 mutated AML | FLT3 wt | miR-155, -10a, -10b | NA | Garzon, et al.[49] |
| FLT3 mutated AML | FLT3 wt | miR-155, -302a, -133a | NA | Garzon, et al. [62] |
| FLT3 mutated AML | other AMLs | miR-511, -155, -10b, -135a | miR-30a-3p, -203, -130a, -214, -338, -143, - 145, -182 | Jongen- Lavrencic, et al. [50] |
Bold miRNAs indicate miRNAs whose disregulation is verified by multiple studies on this table.
Chronic Myelogenous Leukemia
Chronic myelogenous leukemia (CML) is overwhelmingly caused by a common cytogenetic abnormality. Over 95% of cases have a balanced translocation t(9;22)(q34;q11) known as the Philadelphia chromosome, which produces a chimeric BCR-ABL protein with tyrosine kinase activity. Currently, treatment consists of imatinib mesylate to inhibit the kinase activity of BCR-ABL. Although that strategy often successfully treats CML, complete eradication appears difficult, and resistance to imatinib develops. Recent studies have identified therapeutic candidates for imatinib-resistant CMLs, namely, two proteins from the Src kinase family, Fyn and Lyn [37, 38].
CML is associated with upregulation of the MYC-regulated miRNA cluster miR-17-92 through an indirect and unknown mechanism. This polycistronic cluster of 7 miRNAs (17-5p, 17-3p, 18a, 19a, 20a, 19b-1, and 92-1) has been implicated in cell-cycle control by inhibiting E2F1 [5]. MiR-17 and miR-20a exert an anti-apoptotic effect in rapidly dividing cells by downregulating E2F1 and preventing cell-cycle arrest as a result of a G1 checkpoint caused by an accumulation of double strand breaks [39]. MiR-17-5p mediates cell-cycle progression through G1 by inhibiting the tumor suppressor p21 in neuroblastoma, which might be similar in CML [40]. Additionally, CMLs express lower levels of miR-10a, miR-150, and miR-151 and elevated levels of miR-96 [41]. Bueno et al. showed that miR-203, which directly targets BCR-ABL for downregulation, is lost in many cases of BCR-ABL-related leukemias by genomic instability and CpG methylation, which potentially disables a key negative feedback pathway [42]. San Jose-Eneriz found a distinct miRNA signature in analyses of 8 patients with imatinib-resistant CML, with 18 downregulated and 1 upregulated miRNA (Table 1)[43].
Acute Lymphocytic Leukemia
Acute lymphocytic leukemias (ALL), including T- or B-lineage ALL, are characterized by several different cytogenetic abnormalities, including t(12;21)TEL-AML1; the t(8;14), t(2;8) and t(8;22) MYC-related translocations; TAL1; t(1;19) E2A-PBX; 11q23 translocations (MLL); t(9;22) translocations (BCR-ABL), and others. Zanette et al. first characterized miRNA expression abnormalities in ALL by pooling samples of 7 patients and grouping the abnormalities according to the cell lineage compared with CD19+ normal cells [44]. ALL showed multiple disregulated miRNAs highlighted by 5 upregulated miRNAs and 5 downregulated miRNAs (Table 1). The miR-17-92 cluster was upregulated to a lesser extent and was shown to repress E2F1 in T-lineage ALL[44, 45]. Mi et al. performed a high-throughput bead-based profile comparing B-lineage ALL to AML and normal bone marrow cells (mononuclear and CD15+)[46]. In that study, the authors compared the miRNA signatures of 18 ALL samples, all with 11q23 translocations, against 54 AML samples that had a broad spectrum of cytogenetic abnormalities. They found differential expression of 27 miRNAs in ALL, with 6 upregulated and 21 downregulated (Table 1) [46]. These findings confirmed that miR-128a/b is upregulated, which they attributed to epigenetic misregulation of CpG island methylation. The authors also found let-7b and miR-223 to be downregulated compared with AML leukemias. They proposed that the disregulation of these 4 miRNAs (miR-128a, miR-128b, let-7b, and miR-223) serve as the minimal criteria to distinguish ALL and AML leukemias, with miR-128a and miR-128b indicating ALL and let-7b and miR-223 indicating AML [46].
More recently, Fulci et al. attempted to discriminate between T-cell lineage and B-cell lineage in ALL [47]. They performed a clustering analysis of several adult ALL subgroups (B- or T-ALL without known molecular abnormalities or those involving E2A, BCR-ABL, or MLL-AF4). Unsupervised cluster analysis showed distinct expression patterns for each subgroup, whereas a supervised cluster analysis showed that several genes were differentially regulated between T-ALL and B-ALL. T-ALL was characterized by miR-148, -151, and -424 overexpression, whereas B-ALL was characterized by miR-425-5p, -191, -146, -128, -629, and -126 overexpression. It is also possible to discriminate between MLL-related ALL and other ALL subtypes. MLL-related ALLs have an additional definitive subset of disregulated miRNAs independent of differentiation status, including upregulation of miR-196b and downregulation of miR-708 with respect to other ALL samples [48]. Taken together, these results begin to identify miRNA signatures that are distinctly expressed in different ALL subtypes. However, further clarification is needed, as only some subtypes have been characterized by multiple studies, and few have concretely shown an impact on translational regulation of target genes. The relevance of these classifications to prognosis also remains to be determined.
Acute Myeloid Leukemias
Acute myeloid leukemias (AML) display a number of distinct cytogenetic abnormalities that define the disease phenotypes. In several studies, each phenotype was found to have a distinct miRNA expression pattern. In fact, the most extensive effort to characterize miRNA expression patterns was done for AML; several studies correlated the miRNA expression patterns of the different AML subtypes with the different cytogenetic abnormalities[49-51]. Here, we will focus on the most common subtypes that have been investigated in multiple, independent studies, although there are several other cytogenetic abnormalities, as well as cytogenetically normal AMLs (which comprise approximately half of all AMLs).
Several high-throughput studies have classified AMLs according to cytogenetic abnormality [49–51]. Overlap in the focus of these studies is present for Core Binding Factor (CBF), PML-RARα, and MLL AMLs. Jongen-Lavrencic et al. analyzed 215 AML samples by multiplex real-time PCR for 206 miRNAs [50]. In unsupervised analysis of miRNA signatures, they distinguished clustering of miRNAs between different cytogenetic subgroups and molecular aberrations. Supervised analysis revealed broad signature patterns for cytogentic abnormalities and common mutations (Table 1). Li et al. performed a high-throughput bead-based screen of miRNA expression levels in 52 AML patient samples to characterize the differential expression of miRNAs among AMLs with common translocations (Table 1)[51]. Garzon et al. used microarrays to analyze 122 pre-treatment AML patients and validated their results with an additional 60 patients[49]. Signatures were also developed for abnormal karyotype and normal karyotype AML(Table 1). Overall survival and event free survival were shown to negatively correlate with high levels of miR-199a and -191[49].
Some of the most common chromosomal abnormalities in AML affect subunits of the heterodimeric transcription factor comprised of AML1 (RUNX1) and CBFB. The balanced translocation t(8;21)(q22;q22) results in a chimeric protein AML1-ETO (RUNX1-RUNX1T1), which affects AML1, the alpha subunit of the CBF protein complex. Additionally, abnormalities of chromosome 16 affect the core binding factor beta subunit(CBFB), either as inv(16)(p13;q22) or t(16;16)(p13;q22) creating the chimeric protein CBFB-MYH11. Jongen-Lavrencic et al. characterized the predictive marker for t(8;21) and inv(16) CBF-related AMLs. Each abnormality clustered separately and provided a unique signature(Table 1). Let-7b and let-7c were downregulated in both abnormalities; however, the authors were unable to verify any effect on the known let-7 target mRNAs, Ras and HMGA at the mRNA level [50]. In CBF-related AMLs, Li et al. found a distinct signature of for CBF AMLs involving t(t8;21), inv(16), or t(16;16) (Table 1). MiR-126/126* were upregulated and indicated to be part of a minimal identification signature; these miRNAs were found to inhibit apoptosis and enhance proliferation in validation experiments in the same study [51]. Additional studies have shown that miR-126 regulates HOXA9 through binding to the mRNA sequence encoding the homeobox [52].
PML-RARα is another common molecular abnormality in AML, specifically acute promyelocytic leukemia (APL) as a result of the t(15;17)(q22;q12) [53]. Currently, AML with PML-RARα fusions are treated with all-trans-retinoic acid and arsenic trioxide to induce differentiation by dissociating PML-RARα from repressive histone deacetylase (HDAC) complexes and promoting PML-RARα degradation [53]. Jongen-Lavrencic et al. described a miRNA signature for t(15;17) characterized by upregulation of miR-382, -134, -376a, -127, -299-5p, and -323(Table 1) [50]. Li et al. characterized this leukemia subset as having multiple miRNAs upregulated, including miR-181a/b/c/d, which are involved in B-cell differentiation and HOX gene regulation[4, 35, 51]. MiR-224, -368, -384 were also upregulated; hence they have been proposed as criteria for the diagnosis of this subtype. Several miRNAs, including miR-422b, miR-10a, miR-150, j-miR-2, and miR-124a, are downregulated in PML-RARα AMLs(Table 1)[51].
Translocations between 11q23 and a number of fusion partners create chimeric proteins that fuse the amino-terminal portion of the large multi-domain protein Mixed Lineage Leukemia (MLL) to a C-terminal portion of a partner protein. MLL regulates expression of many target genes, including the developmentally important HOX genes [54]. Jongen-Lavrencic observed upregulation of miR-9 and -429 as well as downregulation of miR-213 and miR-146a(Table 1)[50]. Garzon et al. characterized 11q23 translocations as having upregulation of 8 miRNAs and downregulation of 14 miRNAs (Table 1)[49]. Importantly, this study indicated a downregulation of miR-29a, -29b, -29c in AMLs with 11q23 translocations. The involvement of the miR-29 family members in AML and CLL implies a potential functional importance to this disregulation. In a follow-up study they investigated the function of miR-29b in AML [55]. Intriguingly, administration of synthetic miR-29b resulted in increased apoptosis in vitro and greatly decreased tumor volume and mass in a xenograft model. Specific anti-apoptotic targets, including the Bcl-2 family member, Mcl-1, were found to be regulated by miR-29b in primary patient samples[55]. Li et al. showed that 11q23 translocations result in the upregulation of the miR-17-92 cluster as well as miR-196b and miR-10a. Additionally, miR-181a/b/c/d are downregulated in 11q23-translocated AMLs(Table 1) [51]. In a later study characterizing gene misregulation of MLL-ENL and MLL-ELL leukemias, Li et al. confirmed that the miR-17-92 cluster is upregulated in mice and human leukemias of this variety [56]. MiR-196b is located in the HOXA cluster within the HOXA9 locus, which is regulated by CpG island methylation protection by MLL [57, 58], and targets HOXB8 for degradation [59]. Popovic et al. demonstrated that miR-196b contributes to the transformation of MLL leukemias. In that study, in vitro treatment to antagonize miR-196b inhibited the replating potential of MLL-AF9-transformed bone marrow progenitors in an in vitro colony assay [60].
Although cytogentically abnormal AMLs occur in a sizable percentage of cases, approximately half of all AMLs are cytogenetically normal. Common mutations that affect both cytogenetically normal and abnormal AMLs add an additional layer of complexity. Garzon et al. examined normal karyotype AML (NK-AML) against abnormal karyotype AML to discern a signature that is common among NK-AMLs but does not distinguish between NK-AML and AMLs with defined cytogenetic abnormalities(Table 1)[49]. Marcucci et al. examined the miRNA expression profiles of high risk cytogenetically normal AMLs with FLT3-ITD mutations and/or NPM1 mutations. In their study, patient samples were divided into a training group, which provided the initial signature, and a validation group of similarly characterized patient samples. MiR-181a/b correlated with positive outcomes; whereas miR-124, -128-1, 194, 219-5p, 220a, and 320 correlated with more negative outcomes [61]. Garzon et al. investigated the miRNA signature of patients with NPM1 and FLT3-ITD mutations. They found that miR-155 was upregulated in FLT3-ITD AMLs, which corresponds to findings from earlier studies(Table 1) [49, 50]. NPM1 AMLs had a signature of miRNA disregulation, including upregulation of miR-10a, -10b, and -29a/b/c (Table 1). Additionally, NPM1 AMLs were characterized by a decrease in miR-204, and -128a, which were functionally verified to repress HOXA10and MEIS1 [62]. The upregulation of miR-10a, and -10b corresponds to previous data from Jongen-Lavrencic which identified miR-10a, -10b, -196a, and -196b as upregulated miRNAs in NPM1 mutated AMLs[50].
Current Trends in MiRNA Therapeutic Technology
The emerging portrait of miRNA involvement in leukemia has provided both signatures of consistently deregulated miRNAs and specific functional information of the processes controlled by miRNAs. While the signatures themselves may prove valuable in diagnosis, functionally critical miRNA may prove useful as targets for therapeutics. The ability to target miRNA for downregulation and for replacement in cancer treatment will depend on the ability to address key points, mainly delivery systems and the biochemical nature of oligonucleotide modifications. Therapeutic treatments to downregulate and to replace miRNA share some common elements with current therapeutic designs for small interfering RNA (siRNA) with regard to oligonucleotide modifications and delivery systems.
Oligonucleotide Design
Antisense oligonucleotides (oligos) act via multiple independent pathways to silence miRNA or their targets [63]. In addition to RISC-dependent silencing of target mRNA, oligos may promote RNAse H degradation of target miRNA by forming duplexes with target miRNA, inhibiting activity by binding to the target miRNA, or acting as a ribozyme to catalyze cleavage of the target miRNA[63]. Although ribozyme oligonucleotides are appealing for their potential to degrade target miRNA or mRNA, they are not yet applicable for oligo therapy. However, silencing of miRNA via an inhibitory or degradative mechanism has been developed for in vitro and in vivo study as well as clinical trials. Several early studies found that oligo therapy has the potential problem of evoking an immune response mediated by interferon signaling via Toll-like receptors. Oligos are also vulnerable to phagocytosis and exonuclease degradation [64]. Any oligo-based therapeutic intervention, however, must also consider the endogenous small RNA processing pathways. Grimm et al. showed that persistent high-level treatment with short hairpin RNA (shRNA) resulted in high mortality rates in mice attributed to overwhelming the endogenous processing machinery[65].
Antisense oligos can escape the pitfall of degradation through the use of chemical modifications. Kurreck reviewed several generations of antisense oligos in extensive detail [63]. The first generation of modifications consisted of a phosphorothioate modification on the phosphodiester backbone, which increased nuclease resistance at the expense of a lower binding affinity and a potential increase in toxicity [63]. Phosphorothioate inclusion also increases the bioavailability of oligos to the blood, keeping them in circulation longer and out of tissues until eventual accumulation and clearance by the kidneys [66]. In second-generation oligos, a methyl or a methoxy ethyl is added to the 2’-hydroxyl group of ribose. 2’-O-methyl modifications have lower toxicity than phosphorothioate linkages, and they increase the affinity for target miRNA but cannot induce target cleavage by RNAse H [67]. Minimally, incorporating 20% of nucleotides with this modification can abrogate immune system responses to oligo treatment (e.g., inflammation, oligo phagocytosis) [68].
Krutzfeldt et al. used the hepatic miRNA miR-122 as a model to develop and characterize the in vivo activity of a specific motif of modifications directed against miRNA, which they termed “antagomirs” [69, 70]. In those studies, phosphorothioate linkages on the phosphates of the four 5’ nucleotides and two 3’ nucleotides were combined with 2’O-Me modifications on all ribose moieties. Antagomirs also have cholesterol linkages to the 3’ end to facilitate delivery. The subject of delivery will be covered in greater detail later in this review. The resulting oligos downregulate the expression of target miRNA in a sequence-dependent manner, independent of the RISC complex and exonucleases [69]. Studies have shown that similar modifications can also be used to target mRNA and act through the RISC complex to promote mRNA degradation in mice and nonhuman primates [71, 72]. By using modifications similar to those described by Krutzfeld et al., John et al. demonstrated that using siRNA to target apolipoprotein B in the liver can silence mRNA without the deleterious effects on miRNA processing previously seen with shRNA[73]. Taken together, these studies suggest that the same modifications and treatments might be used to target mRNA or replace lost or otherwise misregulated miRNA.
Locked nucleic acids (LNA) comprise another therapeutic technology to enhance miRNA efficiency. LNAs improve the binding of complementary sequences through increased affinity between the modified nucleic acid and the target [74, 75] as well as through increased serum stability [76]. LNA modifications consist of a 2’-O,4’-C-methylene-β-D-ribofuranosyl nucleotide (methyl bridge linkage of the 4’C to the 2’ hydroxyl group) on a single nucleotide or several nucleotides [77, 78]. Several groups have described additional modifications using the same basic structure and mechanism as LNA, but altering the methyl bridge to generate amino-LNAs, thio-LNAs, α-L-ribo-LNAs, and β-dxylo-LNAs [77]. To mechanically increase binding affinity to the target sequence, it is necessary to lock ribose into a C3’endo-conformation [77], which increases binding affinity in a sequence-dependent manner that is determined by adjacent nucleotides [79, 80]. LNA-modified oligos can be involved in any of the aforementioned degradation pathways (e.g., miRNA silencing via binding, mRNA silencing via binding, mRNA silencing via RISC complex) that depend on sequence. Importantly, LNA modifications represented throughout the oligo allow for a “mixmer” and are fully compatible with RISC activity [76]. However, it is necessary to incorporate a 7-10 nt gap, excluding LNAs, to allow RNAse H cleavage in a “gap-mer.” [78] Despite the context-dependent nature of LNA oligos, the flexibility afforded by LNA incorporation might prove valuable for developing more complex oligo treatments. Recently, for example, LNA modifications were used to construct an oligo that can target multiple distinct miRNAs [81]. Development of such multitarget oligos could help to direct treatments against miRNA clusters or functionally redundant miRNAs.
Delivery Systems
MiRNA-based therapies are currently undergoing a transformation shaped by the development of appropriate delivery vehicles. The mechanism of delivery determines the cell specificity and biological availability through prevention of renal clearance, and plays a critical role in avoiding immune stimulation/providing tolerance [64, 82].
The most basic delivery mechanism currently in use is the conjugation of the therapeutic oligo to a chemical modification that facilitates serum stability and cellular uptake. Conjugation to cholesterol was used in the landmark studies by Krutzfeld et al. and has proven valuable for widespread delivery of antagomirs in vivo[69, 70]. However, cholesterol conjugation only provides a limited increase in bioavailability and could be problematic if the chosen targets play critical roles in non-target cell-types or if they direct hematopoietic differentiation toward a particular cell lineage.
Viral vectors have been used with some success in vitro and in vivo to deliver genes that can be transcribed into pri-miRNA or decoy mRNA targets [83-86]. This therapeutic model has a significant drawback in potential immune activation that must be counterbalanced against the potential to direct therapeutic miRNAs and miRNA antagonists to target cells [83]. Furthermore, virally produced miRNA must go through the processing steps of miRNA biogenesis; thus they present a challenge in ensuring that cellular toxicity does not occur as a result of overwhelming the miRNA biogenesis machinery. However, the requirement that they be transcribed might allow for the advantages in miRNA antagonism. Viral delivery systems provides opportunity to produce mRNAs designed to more efficiently; the addition of repeated miRNA binding sites to an abundantly produced mRNA transcript might function as a “miRNA sponge”[87].
Viral therapeutic strategies rely on different virus types, including adenoviruses (which are nonintegrating but possess a higher immunogenicity as nonenveloped particles) and lentiviruses (which integrate into dividng and nondividing cells and are less immunogenic than adenoviral systems as lentiviruses are enveloped particles) [88]. Recently, miRNA replacement using an adenoviral delivery system was shown to suppress development of hepatocellular carcinomas in vivo [89]. Kota et al. utilized an adenoviral delivery system to suppress hepatocellular carcinomas. In this system, miR-26a, which had been downregulated in hepatocellular carcinomas, was effectively replaced and shown to functionally repress the miR-26a trargets cyclin D2 and cyclin E2[85]. Scherr et al. utilized a lentiviral system for antagonism of the miR-17-92 polycistron[86]. Using this system, restoration of mi-17-92 cluster targets was observed in an in vitro model utilizing the BCR-ABL cell line K562[86]. Advances have also been made in targeting by incorporating antibodies into lentiviral surfaces to provide target cell specificity [84]. Yang et al. developed a system to target B-cells in vitro and in vivo for infection using an immunoconjugate directed at CD20 presented alongside a viral fusogen[84].
Recent studies have examined the utility of additional synthetic vehicles for antagomir delivery. Liposomes and other lipid-based particles consisting of a lipid coating over a therapeutic load have been developed to improve bioavailability. This extensive field of study has been reviewed by Whitehead et al., Alexis et al., Kawakami, et al., and Akhtar et al.[64, 82, 90, 91]. Liposomal particles increase size of the siRNA/carrier complex to prevent renal clearance and direct delivery to specific organs or cell types through modifications added to the liposomal surface [64]. Such particles can have a variety of compositions and surface modifications, including PEG-ylation, RGD peptides to direct the antagomir to tumor vasculature, cationic modifications to aid in membrane interaction and repel serum proteins, and specific targeting proteins (often composed of antibodies or antibody fragments)[82]. Of particular interest for hematologic malignancies is the development of immunoconjugates such as gemtuzumab ozogamicin, a targeting immunoconjugate directed at CD33 in AML treatment [82]. Zimmerman et al. introduced siRNA with second-generation modifications using a SNALP (stable nucleic acid lipid particles) system in nonhuman primates with only limited toxicity [72]. However, the addition of complex particles as a drug vehicle has the potential for toxicity and immunogenicity over prolonged exposure.
When a particular inhibitory oligo modification or delivery method is used, clinicians will need to consider the desired mechanism of inactivation and the context of the therapy itself. It is tempting to speculate that low-level immune system activation might be acceptable in specific cases. IFNα induces division of quiescent stem cells in mice via JAK/STAT1 signaling, and, when coupled with a toxic agent, it eradicates bone marrow cells [92]. If a miRNA is necessary for the survival of a specific malignancy but unessential or redundant in other cell lineages, an IFNα-mediated immune response could potentially be useful for eliminating leukemic stem cell reservoirs harboring mutations.
Future Perspectives/Concluding remarks
Since their discovery in 1993, the importance of miRNAs as a regulatory element has continuously increased. Coupled with the discovery of RNAi by Fire and colleagues[93] 1998, miRNA and their exogenous counterpart siRNA are gaining attention as potentially useful therapeutic tools. Although there is much focus on siRNA against upregulated oncogenic mRNA, the need to correct misregulation of miRNA could prove to be equally valuable as our understanding of miRNA involvement in hematologic malignancies and other illnesses improves. Although we have paid particular attention to high-throughput efforts to categorize miRNA misregulations and provide an outline of miRNA involvement in hematopoiesis and cancer, it is important to acknowledge the need to further explore the functional activity of each miRNA and the limitations of high-throughput methods in detecting changes in miRNA expressed at low levels. The combinatorial approach of outlining expression levels followed by in-depth verification of activity will be necessary to discriminate between miRNAs that are misregulated as a consequence of disease state and miRNAs whose misregulation contributes to a disease state.
Therapeutic use of miRNA either to replace underexpressed miRNA or to inhibit overexpressed miRNA has not yet begun. However, there are several ongoing clinical trials testing similar technology in siRNA. The selection of targets is central to the use of miRNA as therapeutic agents, and the development of targets to be used in therapeutic models will depend on the outcome of many ongoing studies.
Acknowledgments
The authors thank Noah Birch for help with editing, and NIH HL087188 (NJZ-L) for funding.
Reference List
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 4.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 5.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 6.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 12.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 13.Lund AH. miR-10 in development and cancer. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee I, Ajay SS, Yook JI, et al. New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen TD. Regulation of microRNA processing in development, differentiation and cancer. J Cell Mol Med. 2008;12:1811–1819. doi: 10.1111/j.1582-4934.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monticelli S, Ansel KM, Xiao C, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramkissoon SH, Mainwaring LA, Ogasawara Y, et al. Hematopoietic-specific microRNA expression in human cells. Leuk Res. 2006;30:643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 25.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 26.Fontana L, Pelosi E, Greco P, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 27.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Felli N, Fontana L, Pelosi E, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Codony C, Crespo M, Abrisqueta P, Montserrat E, Bosch F. Gene expression profiling in chronic lymphocytic leukaemia. Best practice \& research. Clinical haematology. 2009;22:211–222. doi: 10.1016/j.beha.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calin GA, Pekarsky Y, Croce CM. The role of microRNA and other non-coding RNA in the pathogenesis of chronic lymphocytic leukemia. Best Pract Res Clin Haematol. 2007;20:425–437. doi: 10.1016/j.beha.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Pekarsky Y, Santanam U, Cimmino A, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 33.Bichi R, Shinton SA, Martin ES, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicoloso MS, Kipps TJ, Croce CM, Calin GA. MicroRNAs in the pathogeny of chronic lymphocytic leukaemia. Br J Haematol. 2007;139:709–716. doi: 10.1111/j.1365-2141.2007.06868.x. [DOI] [PubMed] [Google Scholar]
- 35.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 36.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Meng F, Kong LY, et al. Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst. 2008;100:926–939. doi: 10.1093/jnci/djn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grosso S, Puissant A, Dufies M, et al. Gene expression profiling of imatinib and PD166326-resistant CML cell lines identifies Fyn as a gene associated with resistance to BCR-ABL inhibitors. Mol Cancer Ther. 2009;8:1924–1933. doi: 10.1158/1535-7163.MCT-09-0168. [DOI] [PubMed] [Google Scholar]
- 39.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–145. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontana L, Fiori ME, Albini S, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6:1830–1840. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- 42.Bueno MJ, Perez de Castro I, Gomez de Cedron M, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 43.San Jose-Eneriz E, Roman-Gomez J, Jimenez-Velasco A, et al. MicroRNA expression profiling in Imatinib-resistant Chronic Myeloid Leukemia patients without clinically significant ABL1-mutations. Mol Cancer. 2009;8:69. doi: 10.1186/1476-4598-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanette DL, Rivadavia F, Molfetta GA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 45.Nagel S, Venturini L, Przybylski GK, et al. Activation of miR-17-92 by NK-like homeodomain proteins suppresses apoptosis via reduction of E2F1 in T-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2009;50:101–108. doi: 10.1080/10428190802626632. [DOI] [PubMed] [Google Scholar]
- 46.Mi S, Lu J, Sun M, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fulci V, Colombo T, Chiaretti S, et al. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48:1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- 48.Schotte D, Chau JC, Sylvester G, et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–322. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 49.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 51.Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen W, Hu Y, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mrozek K, Bloomfield CD. Clinical significance of the most common chromosome translocations in adult acute myeloid leukemia. J Natl Cancer Inst Monogr. 2008;(39):52–57. doi: 10.1093/jncimonographs/lgn003. [DOI] [PubMed] [Google Scholar]
- 54.Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? J Cell Biochem. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 55.Garzon R, Heaphy CE, Havelange V, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z, Luo RT, Mi S, et al. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. 2009;69:1109–1116. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popovic R, Erfurth F, Zeleznik-Le N. Transcriptional complexity of the HOXA9 locus. Blood Cells Mol Dis. 2008;40:156–159. doi: 10.1016/j.bcmd.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erfurth FE, Popovic R, Grembecka J, et al. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc Natl Acad Sci U S A. 2008;105:7517–7522. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 60.Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 62.Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 64.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 66.Braasch DA, Paroo Z, Constantinescu A, et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg Med Chem Lett. 2004;14:1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 67.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 68.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 69.Krutzfeldt J, Kuwajima S, Braich R, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 71.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 72.Zimmermann TS, Lee AC, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 73.John M, Constien R, Akinc A, et al. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007;449:745–747. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koshkin AA, Singh SK, Nielsen P, et al. LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine,5-methylcytosine, thymine, and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998:3607–3630. [Google Scholar]
- 75.Obika S, Nanbu D, Hari Y, et al. Stability and structural features of the duplexes containing nucleoside analogues with fixed N-type conformation, 2'-O,4'C'methyleneribonucleosides. Tetrahedron Lett. 1998:5401–5404. [Google Scholar]
- 76.Elmen J, Thonberg H, Ljungberg K, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frieden M, Orum H. Locked nucleic acid holds promise in the treatment of cancer. Curr Pharm Des. 2008;14:1138–1142. doi: 10.2174/138161208784246234. [DOI] [PubMed] [Google Scholar]
- 78.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 79.McTigue PM, Peterson RJ, Kahn JD. Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)-DNA duplex formation. Biochemistry. 2004;43:5388–5405. doi: 10.1021/bi035976d. [DOI] [PubMed] [Google Scholar]
- 80.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Y, Xiao J, Lin H, et al. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res. 2009;37:e24. doi: 10.1093/nar/gkn1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raty JK, Pikkarainen JT, Wirth T, Yla-Herttuala S. Gene therapy: the first approved gene-based medicines, molecular mechanisms and clinical indications. Curr Mol Pharmacol. 2008;1:13–23. doi: 10.2174/1874467210801010013. [DOI] [PubMed] [Google Scholar]
- 84.Yang L, Bailey L, Baltimore D, Wang P. Targeting lentiviral vectors to specific cell types in vivo. Proc Natl Acad Sci U S A. 2006;103:11479–11484. doi: 10.1073/pnas.0604993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scherr M, Venturini L, Battmer K, et al. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007;35:e149. doi: 10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Venturini L, Eder M, Scherr M. RNA-Mediated Gene Silencing in Hematopoietic Cells. J Biomed Biotechnol. 2006;2006:87340. doi: 10.1155/JBB/2006/87340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawakami S, Hashida M. Targeted delivery systems of small interfering RNA by systemic administration. Drug Metab Pharmacokinet. 2007;22:142–151. doi: 10.2133/dmpk.22.142. [DOI] [PubMed] [Google Scholar]
- 92.Essers MA, Offner S, Blanco-Bose WE, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 93.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]