Abstract
We demonstrate a compact and lightweight platform to conduct automated semen analysis using a lensfree on-chip microscope. This holographic on-chip imaging platform weighs ~46 g, measures ~4.2 × 4.2 × 5.8 cm, and does not require any lenses, lasers or other bulky optical components to achieve phase and amplitude imaging of sperms over ~24 mm2 field-of-view with an effective numerical aperture of ~0.2. Using this wide-field lensfree on-chip microscope, semen samples are imaged for ~10 s, capturing a total of ~20 holographic frames. Digital subtraction of these consecutive lens-free frames, followed by appropriate processing of the reconstructed images, enables automated quantification of the count, the speed and the dynamic trajectories of motile sperms, while summation of the same frames permits counting of immotile sperms. Such a compact and lightweight automated semen analysis platform running on a wide-field lensfree on-chip microscope could be especially important for fertility clinics, personal male fertility tests, as well as for field use in veterinary medicine such as in stud farming and animal breeding applications.
Semen analysis is an important routine that is extensively practiced in laboratories for evaluating male fertility1 and preparing artificial insemination.2 To determine the sperm concentration in semen, visual assessment by putting the sample into a counting chamber and then manually counting the sperms through an optical microscope is still the gold standard. Not only that this method is recommended by the World Health Organization (WHO), but also it is widely used in most laboratories that process semen.3 Due to the labor intensive nature of this manual method, several other optical approaches, including turbidimetry,4–11 laser Doppler velocimetry,12–17 and photon correlation spectroscopy,18–20 have also been proposed to automatically analyze semen. However, these approaches are still not widely adopted partially because they can only provide indirect estimations of the sperm concentration and motility.
Currently, computer-assisted semen analysis (CASA) systems,21–27 which utilize pattern analysis algorithms to automatically process the images recorded with a conventional optical microscope, is considered as one of the most promising technologies to replace the traditional manual semen analysis method. An important feature of CASA systems is their ability to provide quantitative information about sperm motility, such as the speed distribution of individual sperms, which has been proven to be rather important for predicting fertilization rate28,29 as well as for evaluation of the correlation between various drugs and sperm quality.30,31 However, despite the fact that state-of-the-art CASA systems are very efficient and versatile, their relatively large dimensions, high cost and maintenance needs partially hinder their widespread use in fertility clinics. For the same reason, application of this platform to field-use in veterinary medicine such as stud farming and animal breeding32–34 has also been significantly limited.
In addition to these, commercially available male fertility test kits for personal home use, such as FertilMARQ35 or Sperm-Check,36 also aim to indirectly quantify sperm concentration by a color change due to chemical staining or labeling of sperm-specific proteins. These tests, however, cannot quantify sperm motility or the concentration of motile sperms. Recently, an alternative semen analysis platform involving a compact microfluidic device that can measure electrical impedance changes due to sperm movement has also been reported.37 However, this lab-on-a-chip platform can only provide the total number of the sperms in the sample and cannot differentiate motile and immotile sperms from each other, which is an important limitation.
In this manuscript, as an alternative approach, we present a compact and lightweight platform to conduct automated semen analysis on a chip. At the core of our technology lies a lensfree holographic on-chip microscope (see Figure 1) which weighs ~46 g and measures ~4.2 × 4.2 × 5.8 cm providing an imaging field-of-view (FOV) of ~24 mm2 together with an effective numerical aperture (NA) of ~0.2.38 This imaging FOV is more than 20-fold larger than the FOV of a typical 10× objective-lens, and therefore provides a significant throughput advancement that permits automated monitoring of hundreds to thousands of sperms all in parallel. This lensfree on-chip microscope is based on digital in-line holography, and it utilizes an incoherent or partially coherent light source (such as a light emitting diode (LED)) that is filtered by a large aperture of ~0.1 mm to illuminate the sample of interest as illustrated in Figure 1. Over a short propagation distance of ~4 cm, this illumination light picks up partial spatial coherence, after which it scatters from each sperm to coherently interfere with the background light, forming lensfree holograms of the sperms over a large FOV (~24 mm2). These lensfree sperm holograms can then be rapidly processed (e.g., <1 s) using a Graphics Processing Unit (GPU) to reconstruct their microscopic images (both amplitude and phase38) as illustrated in Figure 2.
Figure 1.
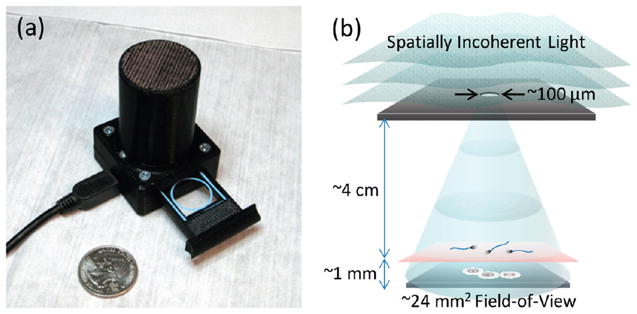
(a) A lensfree holographic on-chip microscope that measures 4.2 × 4.2 × 5.8 cm and weighs 46 g is shown. The embedded light source (an LED filtered by a pinhole) and the CMOS image sensor are both powered through a USB connection from the side. This compact on-chip microscope can provide both amplitude and phase images of the sperms (loaded within a sliding tray) over a field-of-view of 24 mm2 with an effective numerical aperture of ~0.2. (b) A schematic diagram of the lensfree holographic microscope shown in (a) depicts the relative positions of the light source, the semen sample and the sensor chip. This schematic diagram is not drawn to scale.
Figure 2.
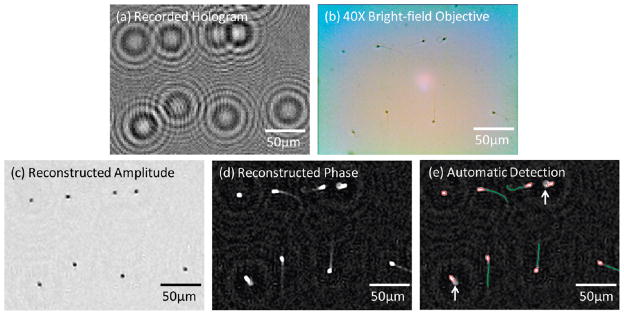
(a) A digitally cropped lensfree hologram of an immobilized semen sample (6.05 million sperms per mL) that is acquired with the unit in Figure 1(a) is shown. (b) For comparison purposes, a bright-field microscope image of the same FOV as in (a) is acquired with a 40× objective-lens (NA = 0.65). (c) The amplitude image reconstructed from the raw hologram shown in (a) for the same FOV indicates the locations of the heads of the sperms. (d) The phase image reconstructed from the raw hologram shown in (a) for the same FOV illustrates both the heads and the tails of the sperms. (e) Automatic characterization results that are generated based on the reconstructed phase image in (d) are illustrated. The elliptical areas corresponding to sperm heads are enclosed by red circles while the tails are labeled with green lines. Defective sperms with missing or unusually curved tails (marked with the white arrows in (e)) are not reported toward positive sperm counts.
To conduct automated semen analysis using this lensfree holographic microscope (Figure 1(a)) we capture ~20 holographic frames over ~10 s. Quite conveniently, digital summation of all these lensfree frames removes the moving sperms out of the picture and enables us to rapidly count only the immobile sperms based on the reconstructed phase images. On the other hand, digital subtraction of these consecutive holographic frames from each other this time removes the immobile sperms out of the picture, leaving behind only the moving ones, which permits automatic quantification of the individual speed and the trajectories of all the motile sperms within an FOV of ~24 mm2. We validated the performance of this platform for automated semen analysis by comparing our results against manual analysis of the same samples conducted using a conventional bright-field microscope.
We believe that such a compact, lightweight and cost-effective automated semen analysis platform running on a wide-field lensfree on-chip microscope would be especially useful for fertility clinics, personal male fertility tests, as well as for field-use in veterinary medicine.
EXPERIMENTAL METHODS
Lensfree Holographic on-Chip Microscope
A self-contained on-chip microscope (see Figure 1(a)) is designed to record the holographic images of the semen samples over a field-of-view of ~24 mm2 with an effective NA of ~0.2 without utilizing any lenses or scanning mechanical components.38 This entire lensfree on-chip microscope assembly (Figure 1(a)) weighs ~46 g and measures ~4.2 × 4.2 × 5.8 cm. Inside this on-chip microscope, a simple light-emitting diode (LS E67B, OSRAM Opto Semiconductors, center wavelength: 645 nm, bandwidth: ~15 nm) is filtered by a 0.1 mm pinhole to provide partially coherent illumination over the semen sample that is placed at a distance of ~4 cm from the source (Figure 1(b)). Lensfree holograms of the sperms are recorded by a monochrome CMOS image sensor (MT9P031STM, Aptina Imaging, 2.2 μm pixel size, 5 megapixel, 24 mm2 active area). This entire imaging system, including the light source and the sensor-chip, is powered and controlled by a laptop computer through a USB 2.0 connection. A smart-phone or a personal digital assistant (PDA) could also be used for the same purpose. The sample of interest is loaded into this microscope using a sliding tray (see Figure 1(a)) and held at ~1 mm above the active area of the CMOS image sensor.
Sample Preparation
Frozen semen specimens were obtained from California Cyrobank and were stored in liquid nitrogen. Before use, the specimen vials were thawed in 37 °C water bath for 10 min to revive the sperms. A sperm washing medium (9983, Irvine Scientific), premixed with a small number of 20 μm polystyrene microspheres (4220A, Thermo Scientific, ~40 beads/μL), was used to dilute the semen sample to a desired concentration. The diluted semen was pipetted into a glass sperm counting chamber (DRM-600, Millennium Sciences, chamber depth: 20 μm) and then enclosed with a no. 1 glass coverslip (12-548A, Fisher Scientific) to suppress liquid evaporation. The added microspheres act as mechanical spacers to ensure a constant chamber height and a uniform sperm distribution. For preparing semen samples with immobilized sperms, the diluent was replaced by a bicarbonate-formalin diluting fluid (6710-4, Ricca Chemical) while the rest of the procedures remained the same.
Hologram Recording and Automated Sperm Analysis
Twenty consecutive lensfree holographic frames for each semen sample of interest were recorded using the on-chip microscope shown in Figure 1(a) at a frame rate of two frames per second (FPS) and an integration time of ~35 ms per frame. Two different processing approaches, digital summation and subtraction of these lensfree holographic frames, were applied to separately identify and quantify the immotile and the motile sperms in the semen sample.
For identification and quantification of the immotile sperms, all the individual holographic images were normalized to their own mean intensity and then were summed up digitally. This summation operation not only increases the digital signal-to-noise ratio (SNR) of the immotile sperms’ holograms, but also smears out the lensfree holograms of the motile sperms. As a result, this step creates sufficient contrast and SNR to observe the faint images of the sperms’ tails, which enables automated identification of their signatures from the background (see, e.g., Figures 2(d and e)).
Next, an iterative holographic reconstruction algorithm38 was used to process this summation hologram and reconstruct the microscopic images (containing both amplitude and phase information, see, e.g., Figures 2(c and d), respectively) of the immotile sperms. Toward automated counting of immotile sperms, candidate objects were initially screened by their distinct and bright elliptical heads in the reconstructed phase images (see, e.g., Figure 2(d)). These immotile sperm candidates were isolated from the background through a threshold operation, where pixels above a certain intensity value were grouped together. After this thresholding step, several digital properties of each connected region were calculated such as its pixel area, orientation, and a coefficient indicating the object circularity.39 Thresholded regions of invalid size or shape were discarded, and a line was fitted to match the orientation and the length of the fitted ellipse to each sperm head (shown with red colored lines in Supporting Information (SI) Figure 1(b)). Following this initial screening step, the tail of each sperm must also be identified and matched to its corresponding sperm head orientation. The condition for a viable sperm is the presence of a healthy tail (in accordance with the WHO laboratory manual3); and therefore for a positive count, the tail must have an adequate length that is also aligned with the orientation of the sperm head. Toward this end, for digital enhancement of the contrast of each tail in the reconstructed phase image, a determinant-of-Hessian filter40 was applied to extract the ridge-like features of each individual sperm tail (see SI Figure 1(c)). A threshold was then applied to the resulting image after which the tails were identified by searching for the remaining ridges in the neighborhood of the end points of each head orientation line (i.e., by matching the green and red lines as shown in SI Figure 1(d)). If a ridge is absent from this sperm head neighborhood, or if the tail is not long enough, or has an abnormal shape it was discarded. All the valid sperms were then automatically counted and marked in the image for further comparison, if necessary.
Quantification of the motile sperms in semen is a relatively easier task for our technique since the only moving objects within the sample are the motile sperms. For this purpose, consecutive lensfree holographic frames were digitally subtracted from each other to create new digital holograms (see, e.g., Figure 3(a)) that represent differential motion of the sperms. The same iterative algorithm38 discussed above was then used to reconstruct the differential motion of the motile sperms as shown in Figure 3(b). In these reconstructed holographic images, the motion of each sperm generates one negative (dark) and one positive (bright) spot, which indicate the start and the end positions of the sperm’s motion, respectively. Therefore, by quantifying the relative distance between these bright and dark spots we can simultaneously infer the speed and the trajectories of all the motile sperms within our imaging field-of-view (~24 mm2). For this purpose, the locations of these spots were identified by simple thresholding followed by calculation of their centroid positions.41 As a result of this, the displacements of individual sperms between consecutive holographic frames can be easily calculated by using the shortest distance between the centroid positions of each dark-bright spot pair (see, e.g., Figure 3(b)). These displacements of motile sperms in all consecutive frames were then linked to each other to plot the dynamic trajectories of the sperms within our FOV (see, e.g., Figure 4(a)). The average speed of each motile sperm was calculated by summing up the magnitudes of its displacements in all frames and dividing this sum with the total duration of the frame acquisition.
Figure 3.
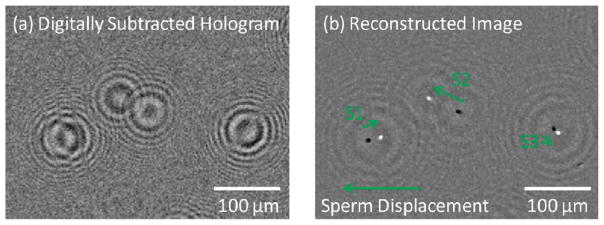
(a) A digitally subtracted lensfree hologram of three moving sperms is generated from two successive frames (500 ms apart) (b) A microscopic image, that is digitally reconstructed from the lensfree differential hologram shown in (a), illustrates the positions of three sperms in two successive frames (white spots show the sperms’ end positions and black spots show their starting positions). The displacement vectors of these sperms are labeled as S1, S2, and S3.
Figure 4.
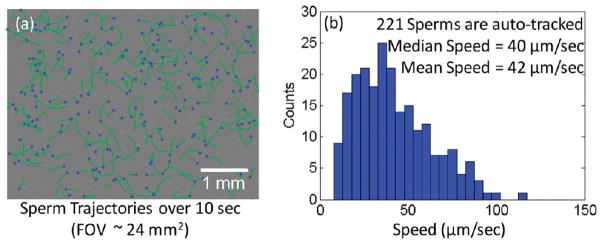
(a) Dynamic trajectories of 221 sperms within a field-of-view of ~24 mm2 are automatically tracked over a time-span of 10 s. The blue spots mark the end positions of the tracked sperms, while the green lines refer to their trajectories. (b) The speed histogram of these motile sperms is calculated using the information in (a) by summing the sperm displacements from all the consecutive frames and then dividing this sum by the total image acquisition duration.
Digital processing of the acquired lensfree holographic images, including both immotile sperm detection and motile sperm tracking, were implemented in Matlab. The computation time for each semen sample was ~15 min for ~24 mm2 FOV using a PC with an Intel Core2Duo E8400 3.00 GHz CPU. By moving these image processing routines to a GPU (e.g., NVIDIA GeForce GTX 285), the time required to analyze each semen samples can be significantly reduced (by, e.g., >10× ).
Microscope Comparisons to Validate Lensfree on-Chip Imaging
To verify our automatic characterization results made with this holographic on-chip microscope, videos of the same semen samples were also recorded using a conventional bright-field microscope (ME300TZ-2 L-5M, AmScope). For each semen sample, these videos were recorded across ≥9 adjacent FOVs (~0.20 mm2 each) using a 20× objective-lens (NT38-339, Edmund Optics, NA = 0.4). Motile and immotile sperms were then separately counted on the recorded video by visual inspection, following the WHO guidelines.3 For samples that had less than 0.5 million motile sperms per mL, videos were also recorded with a 10× objective-lens (NA = 0.25) that had a relatively larger field-of-view than a 20× objective-lens. However, these lower resolution videos were used for counting of motile sperms only, and were not used to quantify immotile sperm densities. These conventional bright-field microscope videos were recorded right before the same semen samples were imaged by our lensfree holographic on-chip microscope. The delay between these two imaging experiments was kept below 30 s to minimize the error from the change of semen status.
RESULTS AND DISCUSSION
To demonstrate the capabilities of our platform shown in Figure 1(a) for automated detection of sperms from their lensfree holograms, we initially worked with semen samples that had immobilized sperms. In these experiments, lensfree holograms of the immobilized sperms were digitally reconstructed as detailed in the Experimental Methods section to yield both an amplitude and a phase image of each sperm over an imaging FOV covering ~24 mm2. As an example, Figure 2 and Supplementary Figure 2 digitally focus on smaller regions of this FOV to illustrate some of these raw lensfree holograms, and their corresponding reconstruction results.
An interesting observation based on these images is that the sperm tails can only be observed in the reconstructed phase images, while the heads of the sperms are clearly visible in both the amplitude and the phase images (see, e.g., Figures 2(c and d) or SI Figures 2(b and c)). The main reason behind this behavior is that the tail of a sperm is a submicrometer structure which generates a rather weak scattering signal. Therefore, with the limited NA and SNR of our wide-field lensfree microscope, such a small feature does not contribute enough scattering signal to be reconstructed in the amplitude image. On the other hand, the refractive index difference between the tail and the surrounding medium still creates a sufficient contrast in the reconstructed phase image, permitting observation and automated detection of the sperm tails as described in the Experimental Methods section (see, e.g., Figures 2(d and e) and SI Figures 1–2). In these results presented in Figure 2 and SI Figures 1–2, the tail of each sperm was automatically identified and matched to its corresponding sperm head orientation. In this process, some of the sperms were disqualified from a positive count because of unusually curved tails (refer to the Experimental Methods section for details). To further validate our analysis, these automatic detection results were also compared against a regular microscope image taken on the same field-of-view as shown in Figure 2(b, d, and e).
To demonstrate the performance of our platform on quantification of motile sperms, we characterized several semen samples with varying sperm concentrations. Figures 3 and 4 and SI Figure 3 show the differential imaging and automated tracking results of some of these semen samples. As detailed in the Experimental Methods section, digital subtraction of consecutive frames from each other removes all the stationary holograms and leaves behind only the differential holograms of the motile sperms, as also illustrated in Figure 3(a) or SI Figure 3(a). Therefore, each motile sperm has a distinctive signature on the new differential hologram that represents the magnitude and the direction of the sperm displacement between these two consecutive frames. Performing holographic reconstruction on these differential holograms reveal two spots for each motile sperm: one dark spot indicating its start position and another bright one indicating its end position (see, e.g., Figure 3(b) or SI Figure 3(b)). Based on these reconstructed differential images, we have quantified the dynamic trajectories of the motile sperms over our entire imaging FOV (~24 mm2) as illustrated in Figure 4(a) or SI Figure 3(c). For further quantification of the sperms shown in Figure 4(a), the displacements of all these motile sperms were linked across all the 20 lensfree frames acquired over ~10 s, which enabled us to determine the speed histogram of the sperms as shown in Figure 4(b). Similar results were also obtained at different sperm concentrations as illustrated in SI Figure 3 for 2.36 million sperms per mL. This average speed histogram calculated by our system is essentially equivalent to the distribution of straight-line velocity (also known as VSL) of the sperms, which is reported to highly correlate with fertilization rate,28 suggesting that the average sperm speed provided by our platform could be an effective indicator of male fertility.
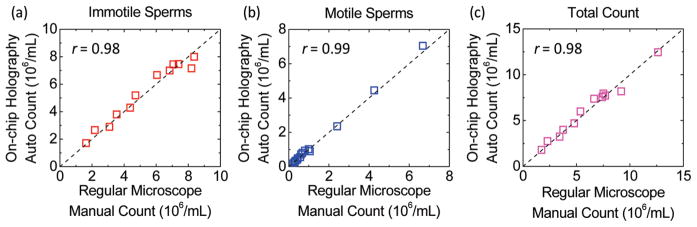
Finally, using our on-chip imaging platform, we characterized 12 semen samples containing both immotile and motile sperms at varying concentrations to validate the automated counting accuracy of our system. In Figure 5, our automatic counting results were compared against manual counting results achieved using a bright-field microscope (see the Experimental Methods section). This comparison confirmed that our automated analysis can accurately quantify the concentration of both immotile and motile sperms up to a density of 8.3 and 6.8 Million/mL, respectively. Such a large dynamic range allowed our system to reliably analyze human semen samples that were diluted by ~1–10×. In addition to this, Figure 5 also revealed our automated platform’s superior ability to analyze semen samples with very low sperm concentrations. For example, the lowest concentration reported in Figure 5(b), 87 500 sperms/mL, corresponds to ~42 sperms tracked by our system over its 24 mm2 field-of-view. To achieve the same level of statistical characterization accuracy for such a low density semen sample, a conventional microscope equipped with a 20× objective-lens would need to look at close to 100 different field-of-views, making it quite inconvenient. Therefore, the large field-of-view of our lensfree on-chip imaging platform is also quite valuable for providing quantitative results at extremely low sperm densities such as observed in postvasectomy.42,43
Figure 5.
Counting accuracy of the presented automated semen analysis platform for (a) immotile sperms; (b) motile sperms; and (c) both the motile and immotile sperms is illustrated at various sperm concentrations up to 12.5 × 106/mL. The x-axes are the sperm concentrations that are manually counted using a conventional bright-field microscope. The y-axes are the sperm concentrations that are automatically counted for the same semen samples using lensfree holographic images acquired with the on-chip microscope shown in Figure 1(a). The total counts in (c) are the summed up concentrations of the immotile sperms in (a) and the motile sperms in (b). Correlation coefficients (r) of these characterization results shown in (a), (b), and (c) (0.98, 0.99, and 0.98, respectively) further validate the accuracy of this compact and lightweight holographic lensfree microscope as a semen analysis platform.
Potential application areas of this semen analysis platform include automated sperm counting in fertility clinics, semen quality evaluation in sperm banks, personal male fertility tests at home, postvasectomy tests either at home or in clinic, stud performance assessment at animal breeding centers, and stud health monitoring in farms. In addition to these, by using a larger-area image sensor, this platform can also be transformed into a significantly higher throughput analysis platform that can measure sperms’ response to different drugs or chemicals of various concentrations all in parallel. Furthermore, this optical technology with its automated analysis capability can be utilized for characterization of cells within bodily fluids such as whole blood samples or for detection of bacteria or parasites in drinking water. The compact, lightweight and cost-effective design of this imaging platform and its connectivity to PDAs or smart-phones may allow the users to perform whole blood count or water quality screening in field settings with minimal resources.
CONCLUSION
In conclusion, we have demonstrated a compact and lightweight platform to conduct automated semen analysis using a lensfree on-chip microscope. This automated semen analysis platform weighs ~46 g and measures ~4.2 × 4.2 × 5.8 cm, and therefore it fills up an important gap between simple qualitative male fertility test kits and sophisticated quantitative characterization systems based on, for example, CASA. In a very compact and lightweight format, this platform can provide quantitative semen analysis including the measurements of the absolute concentrations of both motile and immotile sperms, as well as the trajectories and the speed distributions of motile sperms. Since several different factors other than the sperm concentration are now being used to predict fertility,44 such a compact and versatile semen analysis tool could be very useful for fertility clinics, personal male fertility test kits, as well as for field-use in veterinary medicine.
Supplementary Material
Acknowledgments
A.O. gratefully acknowledges the support of NSF (CAREER Award on BioPhotonics), the Office of Naval Research (ONR) under the Young Investigator Award 2009 and the NIH Director’s New Innovator Award, Award No. DP2OD006427 from the Office of The Director, National Institutes of Health. We also acknowledge the support of the Okawa Foundation, Vodafone Americas Foundation, DARPA DSO (under 56556-MS-DRP), NSF BISH program (under Award Nos. 0754880 and 0930501), NIH (under 1R21EB009222-01), and AFOSR (under Project No. 08NE255).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supplementary figures as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Björndahl L, Mortimer D, Barratt CLR, Castilla JA, Menkveld R, Kvist U, Alvarez JG, Haugen TB. A Practical Guide to Basic Laboratory Andrology. 1. Cambridge University Press; New York: 2010. [Google Scholar]
- 2.Foote RH. J Anim Sci. 2002;80:1–10. [Google Scholar]
- 3.World Health Organization (WHO) Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction. 4. Cambridge University Press; New York: 1999. [Google Scholar]
- 4.Sokoloski JE, Blasco L, Storey BT, Wolf DP. Fertil Steril. 1977;28:1337–1341. doi: 10.1016/s0015-0282(16)42980-8. [DOI] [PubMed] [Google Scholar]
- 5.Atherton RW, Radany EW, Polakoski KL. Biol Reprod. 1978;18:624–628. doi: 10.1095/biolreprod18.4.624. [DOI] [PubMed] [Google Scholar]
- 6.Atherton RW, Jackson FL, Bond G, Radany EW, Kitchin RM, Polakoski K. Arch Androl. 1979;3:301–308. doi: 10.3109/01485017908988420. [DOI] [PubMed] [Google Scholar]
- 7.Levin RM, Shofer J, Greenberg SH. Fertil Steril. 1980;33:631–635. doi: 10.1016/s0015-0282(16)44777-1. [DOI] [PubMed] [Google Scholar]
- 8.Levin RM, Greenberg SH, Wein AJ. Fertil Steril. 1981;35:332–336. doi: 10.1016/s0015-0282(16)45382-3. [DOI] [PubMed] [Google Scholar]
- 9.Morton BE, Sagadraca R. Arch Androl. 1981;7:219–227. doi: 10.3109/01485018108999310. [DOI] [PubMed] [Google Scholar]
- 10.Levin RM, Hypolite JA, Wein AJ. Andrologia. 1984;16:434–438. doi: 10.1111/j.1439-0272.1984.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 11.Halangk W, Bohnensack R. Biomed Biochim Acta. 1986;45:331–341. [PubMed] [Google Scholar]
- 12.Jouannet P, Volochine B, Deguent P, Serres C, David G. Andrologia. 1977;9:36–49. doi: 10.1111/j.1439-0272.1977.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 13.Naylor GP, Martin JS, Chantler EN. Med Biol Eng Comput. 1982;20:207–214. doi: 10.1007/BF02441357. [DOI] [PubMed] [Google Scholar]
- 14.Craig T, Hallett FR, Nickel B. Biophys J. 1982;38:63–70. doi: 10.1016/S0006-3495(82)84531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolford MW, Harvey JD. Biophys J. 1982;40:7–16. doi: 10.1016/S0006-3495(82)84452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigler R, Thyberg P. Cytometry. 1984;5:327–332. doi: 10.1002/cyto.990050407. [DOI] [PubMed] [Google Scholar]
- 17.Earnshaw JC, Munroe G, Thompson W, Traub AI. Med Biol Eng Comput. 1985;23:263–268. doi: 10.1007/BF02446869. [DOI] [PubMed] [Google Scholar]
- 18.Frost J, Cummins H. Science. 1981;212:1520–1522. doi: 10.1126/science.7233239. [DOI] [PubMed] [Google Scholar]
- 19.Wilson MC, Harvey JD. Biophys J. 1983;41:13–21. doi: 10.1016/S0006-3495(83)84400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson M, Harvey J, Shannon P. Biophys J. 1987;51:509–512. doi: 10.1016/S0006-3495(87)83373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortimer ST. J Androl. 2000;21:515–524. [PubMed] [Google Scholar]
- 22.Mortimer S. Hum Reprod Update. 1997;3:403–439. doi: 10.1093/humupd/3.5.403. [DOI] [PubMed] [Google Scholar]
- 23.Agarwal A, Sharma RK. Fertil Steril. 2007;87:156–162. doi: 10.1016/j.fertnstert.2006.05.083. [DOI] [PubMed] [Google Scholar]
- 24.Tomlinson MJ, Pooley K, Simpson T, Newton T, Hopkisson J, Jayaprakasan K, Jayaprakasan R, Naeem A, Pridmore T. Fertil Steril. 2010;93:1911–1920. doi: 10.1016/j.fertnstert.2008.12.064. [DOI] [PubMed] [Google Scholar]
- 25.Ramió L, Rivera M, Ramírez A, Concha I, Peña A, Rigau T, Rodríguez-Gil J. Theriogenology. 2008;69:501–512. doi: 10.1016/j.theriogenology.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Maree L, du Plessis S, Menkveld R, van der Horst G. Hum Reprod. 2010;25:1369–1382. doi: 10.1093/humrep/deq075. [DOI] [PubMed] [Google Scholar]
- 27.Coetzee K, de Villiers A, Kruger TF, Lombard CJ. Fertil Steril. 1999;71:222–225. doi: 10.1016/s0015-0282(98)00465-8. [DOI] [PubMed] [Google Scholar]
- 28.Liu DY, Clarke GN, Baker HW. J Androl. 1991;12:231–239. [PubMed] [Google Scholar]
- 29.Barratt CL, Tomlinson MJ, Cooke ID. Fertil Steril. 1993;60:520–525. [PubMed] [Google Scholar]
- 30.Kuo YL, Tzeng WL, Chiang HK, Ni RF, Lee TC, Young ST. Arch Androl. 1998;41:127–133. doi: 10.3109/01485019808987954. [DOI] [PubMed] [Google Scholar]
- 31.Glenn DR, McVicar CM, McClure N, Lewis SE. Fertil Steril. 2007;87:1064–1070. doi: 10.1016/j.fertnstert.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 32.McCurnin DM, Bassert JM. Clinical Textbook for Veterinary Technicians. 5. W.B. Saunders Company; Philadelphia, PA: 2002. [Google Scholar]
- 33.Clifford FS. Swine Health Prod. 1999;7:117. [Google Scholar]
- 34.Turner RM. Clin Tech Equine Pract. 2005;4:257–268. [Google Scholar]
- 35.The BabyStart Male Infertility Test (FertilMARQ) [Accessed on 07/10/2010]; http://www.embryotech.com/babystart/bs-male/index.htm.
- 36.ContraVac, SpermCheck Fertility. [Accessed on 07/10/2010]; http://www.contravac.com/products/spermcheck/fertility.php.
- 37.Segerink LI, Sprenkels AJ, Braak PMT, Vermes I, Berg AVD. Lab Chip. 2010;10:1018–1024. doi: 10.1039/b923970g. [DOI] [PubMed] [Google Scholar]
- 38.Mudanyali O, Tseng D, Oh C, Isikman SO, Sencan I, Bishara W, Oztoprak C, Seo S, Khademhosseini B, Ozcan A. Lab Chip. 2010;10:1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo S, Su T, Tseng DK, Erlinger A, Ozcan A. Lab Chip. 2009;9:777–787. doi: 10.1039/b813943a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindeberg T. Int J Comput Vision. 1998;30:117–156. [Google Scholar]
- 41.Su T, Isikman SO, Bishara W, Tseng D, Erlinger A, Ozcan A. Opt Express. 2010;18:9690–9711. doi: 10.1364/OE.18.009690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chawla A. Urology. 2004;64:1212–1215. doi: 10.1016/j.urology.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Dhar NB, Bhatt A, Jones JS. BJU Int. 2006;97:773–776. doi: 10.1111/j.1464-410X.2006.06107.x. [DOI] [PubMed] [Google Scholar]
- 44.Lewis SEM. Reproduction. 2007;134:31–40. doi: 10.1530/REP-07-0152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.