Abstract
Since its discovery more than 50 years ago, the human Major Histocompatibility Complex (MHC) on chromosome 6p21.3 has been at the forefront of human genetic research. Here, we review from a historical perspective the major advances in our understanding of the nature and consequences of genetic variation which have involved the MHC, as well as highlighting likely future directions. As a consequence of its particular genomic structure, its remarkable polymorphism and its early implication in numerous diseases, the MHC has been considered as a model region for genomics, being the first substantial region to be sequenced and establishing fundamental concepts of linkage disequilibrium, haplotypic structure and meiotic recombination. Recently, the MHC became the first genomic region to be entirely re-sequenced for common haplotypes, while studies mapping gene expression phenotypes across the genome have strongly implicated variation in the MHC. This review shows how the MHC continues to provide new insights and remains in the vanguard of contemporary research in human genomics.
Keywords: Major Histocompatibility Complex, Human Leukocyte Antigen, polymorphism, haplotype, linkage disequilibrium, gene expression
The human major histocompatibility complex (MHC) was discovered more than 50 years ago [1]. It was initially known for its role in transplantation through histocompatibility antigens, hence the other commonly used nomenclature, ‘human leukocyte antigens’ (HLA) [2-5]. Several decades of intensive research has defined the remarkable genomic environment of the MHC and how genetic variation within this region plays a key role in susceptibility to autoimmune, infectious and other diseases. The MHC shows extreme levels of gene density and polymorphism. The nature, coinheritance and functional consequences of its genetic diversity have proved complex. Nonetheless, remarkable insights have been gained and this genomic region has become a paradigm for human genomics. In this review, rather than describing specific MHC associated diseases (for reviews see [6-10]), we recall the historical context and describe the genomic landscape of the MHC and some of the challenges which remain: an ongoing adventure in exploring human genetic diversity that keeps the MHC at the forefront of research in human genomics.
THE MHC: DISCOVERY AND BIOLOGICAL SIGNIFICANCE
Discovery of the MHC
The immunogenetics field was born in 1900 with the discovery of the ABO blood groups by Landsteiner, followed by the Rhesus system in 1940 (Figure 1). Peter Gorer was the first to describe in 1936 a histocompatibility system in mice from his observations of the agglutination of erythrocytes by rabbit immune sera [11]. This research was advanced by George Snell who established that graft rejection in mice was due to incompatibility at the level of certain antigens. The murine MHC was called ‘H2’ in honour of the antigen II discovered by Gorer [12].
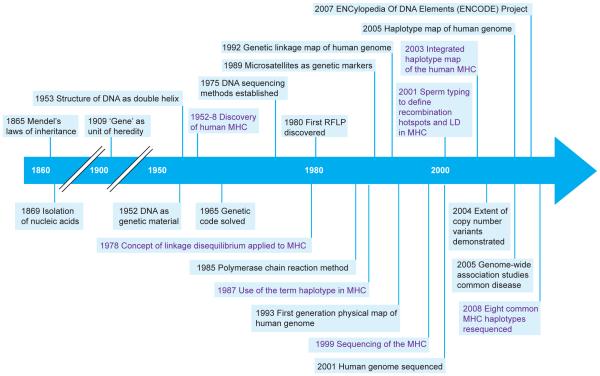
Figure 1.
Timeline of research in the MHC and human genome
As for humans, the history of the HLA complex began in 1952 with the princeps observation made by Jean Dausset. He hypothesised that a similar antigenic system to that seen in mouse erythrocytes could exist in humans on the surface of leucocytes, which he demonstrated by showing a massive leucoagglutination by the serum of a poly transfused patient. However, the firm discovery of the first human MHC antigen, MAC (HLA-A2), was made only in 1958 following a classical segregation population analysis using a serum reacting in only a subset of the studied sample [13]. This polymorphic system was then confirmed by the work of Jon van Rood, and of Rose Payne and Walter Bodmer, who defined respectively the antigens 4a and 4b (Bw4 and Bw6), and HLA-A2 and HLA-A3, through studies on multiparous women [2, 14]. An international research effort involving the International Histocompatibility Workshops (IHW) initiated in 1964, led progressively to the characterisation of a gene cluster on chromosome 6 with serologically defined alleles and including HLA-A, HLA-B and HLA-C. The complement system was also mapped to the same genetic region. In the 1970’s, HLA class II alleles were characterised through the identification of mixed lymphocyte reactions. Advances in molecular biology then allowed investigation of the HLA system directly at the level of the genes rather than of their products.
The role of MHC molecules is inherent to their polymorphism
The human MHC, on chromosome 6p21.3, is divided into the classical regions denoted class I and class II, and an intervening region dubbed class III (Figure 2). While the MHC was discovered based on its involvement in alloreactivity, its broader role in the immune response was established by Baruj Benacerraf who demonstrated that the MHC controlled the ability to mobilise an immune response against a particular antigen [15]. Indeed, HLA class I and II genes encode glycoprotein molecules expressed at the cell surface where they present antigenic peptides to CD8 positive and CD4 positive T cells respectively [16]. Their extraordinary genetic polymorphism guarantees the broadest diversity of recognized antigens and therefore reactivity against pathogens, conferring a selective advantage. The affinity between the HLA molecules and the antigenic epitopes is a pivotal feature of this mechanism, highlighted by elucidation of the structure of HLA molecules, established for the first time by Bjorkman in 1987 for HLA-A2 [17], and also for class II molecules [18]. Beyond this function of antigen presentation devoted to HLA molecules, MHC genes are involved in numerous aspects of the immune response and some have also non immune functions, such as olfaction [19, 20]. In particular, genes and polymorphisms in the class III region are involved in the regulation of the humoral immune response [21] and in the inflammatory reaction through genes encoding the tumour necrosis factor (TNF) [22], heat shock proteins or components of the complement cascade.
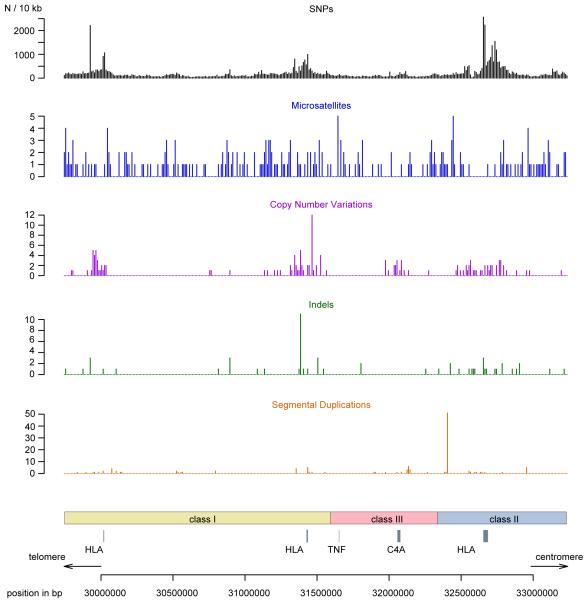
Figure 2. Genetic variation in the MHC.
Number of polymorphisms per 10 kb across the MHC. Polymorphisms were extracted from dbSNP build 129, microsatellites from dbMHC, copy number variants and indels from the Database of Genomic Variants version 6, and segmental duplications of more than 1 kb from the ucsc table browser [174].
THE GENOMIC LANDSCAPE OF THE MHC: EXTREME GENE DENSITY AND POLYMORPHISM
The first substantial human genomic region to be sequenced
As a consequence of its biological significance, defining the nucleotide sequence of the MHC was a priority and was achieved in 1999, providing important new insights and avenues for investigation. The entire 3.6 Mb was published through the combined efforts of the MHC Sequencing Consortium under the direction of Stephen Beck, Daniel Geraghty, Hidetoshi Inoko and Lee Rowen [23]. Covering about 0.12% of the human genome, this was the first substantial contiguous sequence to be determined, two years before the release of the whole genome draft sequence [24, 25]. Since it was derived from multiple individuals with different HLA types, this was a “mosaic” sequence, as is still the case for the rest of the reference genome assembly (build 36.3), although some complete diploid sequences are now becoming available through re-sequencing [26-29]).
The most dense gene region of the human genome
The MHC Sequencing Consortium established that 224 gene loci were present in the human MHC [23], of which 42% were novel. Within the 1.9 Mb class I region, 18 HLA class I genes are known (the classical genes HLA-A, HLA-B and HLA-C, together with non-classical genes HLA-D, HLA-E, HLA-F, HLA-G and 12 pseudogenes) together with 7 MHC class I-chain related genes (MICA, MICB and 5 pseudogenes). Class I and MIC genes are organised in a three-fold repeated unit. The MHC class II region covers about 0.9 Mb and in the mosaic MHC sequence, 19 class II genes were counted including the classical genes HLA-DP, HLA-DQ and HLA-DR; the non-classical genes HLA-DM and HLA-DO; and 8 pseudogenes. Located between class I and class II regions, the class III is remarkably gene dense, including 62 expressed genes with more than 500 exons over 750 kb, many of which are involved in the immune response [30].
If pseudogenes are included, the average density of genes in the MHC is one gene per 16 kb, making this the most gene dense region of the human genome. There are numerous pseudogenes in the class I and II regions. Both these regions seem to have been duplicated several times, generating new gene family members which then diverged [31, 32]. It is believed that pseudogenes could have played a role in the generation of new alleles by genic conversion. In contrast, the class III region is unique in the near absence of pseudogenes except in the duplicated C4 region and contains one expressed gene at least every 15kb. In some instances, transcripts overlap as for example with TNXB and CYP21A2 genes [33].
Approximately 40% of the genes in the classical MHC are expressed in the immune system and they are physically clustered, reflecting their functional roles [2, 34, 35]. Further studies and sequencing of chromosome 6 [36, 37] revealed that flanking genomic regions showing evolutionary conservation now recognised as the extended class I and extended class II regions. The extended MHC spans 7.6Mb and includes 421 gene loci, of which 60% are thought to be expressed [38].
The most polymorphic human genomic region
Since the discovery of the first human biological polymorphism by Landsteiner, the remarkable nature of human genetic variation has been revealed, ranging from single nucleotide differences to large structural genomic variation spanning thousands of base pairs [39]. These include single nucleotide polymorphisms (SNPs), substitutions and deletion insertion polymorphisms (DIPs) as well as variation involving more than one base such as short tandem repeats (STRs) (microsatellites), large insertion deletions (INDELs), inversions and other repeats (Figure 2). As genetic markers, STRs and SNPs have proved particularly informative. While such variants have been identified in almost all genes, nowhere in the genome can yet compete with the extreme polymorphism found in HLA genes.
In 1980, for each of the four genes described by Jean Dausset in his Nobel lecture, HLA-A, -B, -C and –DR, 8 to 39 codominant alleles were known and the number of their haplotypic combinations was already reaching several millions. Other MHC-linked genes were assumed to also be extremely polymorphic, which led him to emphasise that virtually every human carries a unique combination [1]. By the year 2000, more than 15,000 HLA alleles were described [23]. With 86 SNPs per kb and 1,195 alleles, the HLA-B locus is the most polymorphic gene in the human genome, the number of alleles having doubled over the last four years (IMGT/HLA Database http://www.ebi.ac.uk/imgt/hla/ release 2.24.0) [36, 40]. The second most polymorphic is HLA-A with 733 alleles. A total of 3,371 alleles are now reported for HLA loci and 106 for non classical HLA loci (MICA, MICB, TAP1, TAP2).
Polymorphic sites in classical HLA genes are predominantly located in the second and third exons of the class I loci and in the second exon of HLA-DRB, -DQA, -DQB, -DPA and -DPB. Here, non synonymous single nucleotide substitutions lead to amino acid changes in the domain involved in the binding of antigenic peptides. The distribution of the HLA loci variants is different from the one observed in most functional genes where variants occur more often in introns.
Generation of such extreme polymorphism is attributable to point mutations, recombination and genic conversion [41]. In non coding sequences, variants are more frequent in regions flanking the most polymorphic genes [42] through “genetic hitch-hiking” [2, 43]. Polymorphism of class I and II genes is thought to be maintained by the selective advantage conferred by the heterozygous state allowing a more diverse immune response to infectious pathogens such as HIV-1 [44]. Polymorphism could also be selected because rare alleles provide an advantage on a population scale to escape from infections. Both mechanisms are likely to be overlapping. The extreme polymorphism in the MHC required a special nomenclature where the locus name is followed by an asterisk and 4 digits identifying the allele, the first two reflecting the previous serological nomenclature. Additional digits are added for synonymous changes or non coding variants. This system is applied for class I and class II genes, as well as for TAP or PSMB genes. Prior to the current era of genome-wide association studies and use of SNP microarrays as genetic markers, microsatellites were extensively studied to define disease associations. However there was no standardised nomenclature until the 13th IHW [45] which resolved aliases for 281 microsatellites with an average density of one per 45 kb. Since then, all microsatellites have been submitted to the dbMHC database (http://www.ncbi.nlm.nih.gov/gv/mhc) [46]. The most recent 14th IHW successfully mapped 664 microsatellite primer pairs in the current genome assembly [47].
Larger scale but submicroscopic structural genomic polymorphism is also increasingly recognised. It involves variants more than 1 kb in size, including copy number variants, segmental duplications, inversions and translocations. For example, within the MHC (specifically chromosome 6p21.3) a total of two inversions, 63 indels (100bp to 1kb in size) and 181 copy number variants are currently listed in the database of genomic variants (http://projects.tcag.ca/variation/) [48, 49], some being of potential functional importance such as CNV-507 or CNV-505 which overlap the susceptibility loci for sarcoidosis and psoriasis respectively [50]. Two main regions in the MHC are affected by segmental duplications, the HLA-DRB1 region and the complement region, although they are specific to certain allelic combinations or “haplotypes” [51].
A PARADIGM IN THE STUDY OF RECOMBINATION, LINKAGE DISEQUILIBRIUM AND HAPLOTYPIC STRUCTURE
Use of the term “haplotype”
The term “haplotype”, as a contraction of “haploid genotype”, refers to DNA sequence polymorphisms co-inherited at a minimum of two linked loci. It was first used in 1967 in the context of the MHC to describe “the combination of individual antigenic [MHC] determinants that are positively controlled by one allele” [36], although the concept was already applied to the MNS antigen system of human blood groups. It originated from the immunogeneticist Ruggero Ceppellini who observed familial genotype data that could be explained only by the joint inheritance of alleles at closely linked loci [52]. Ceppellini’s contribution to this field began even earlier through his studies of rhesus factor when he introduced the expectation maximization (EM) algorithm that is still used for haplotype reconstruction [53]. The earliest record of “haplotype” in PubMed is found one year later, again in relation to the MHC [54].
Numerous SNP studies have described the existence of DNA sequence segments in the human genome, ranging between 5 and 150 kb and maintained on haplotypes in the absence of genetic recombination. Within the MHC short blocks of less than 0.2 Mb involving regions such as the DR-DQ genes or the ‘complotype’ - a combination of alleles at the C4B, C4A, Bf and C2 complement genes – are described [55]. In addition, combinations of blocks forming single genetic units are found, highlighting the complexity of ‘genetic fixity’ in the MHC such that long conserved sequences representing combinations of multiple blocks are observed at relatively high frequencies within populations [56]. These “conserved extended haplotypes” or “ancestral haplotypes” can extend over 3.2 Mb, spanning the whole classical MHC [56-58].
Haplotypes differ between populations with greater diversity in African populations than in any other ethnic group. The 12th IHW in 1996 highlighted the marked differences in MHC haplotype frequencies seen both between and within population groups. Among those of Asian ancestry particular haplotypes were very common in Japan such as HLA-A2-B52-DR15-DQ1 but of very different frequency in Singapore and Thailand where HLA-A2-Cw11-B46-DR9/DQ3 was common; in African populations the most common haplotype seen in South Africa HLA-A30-B42-DR1-DQ1 for example, is not found in neighbouring Zimbabwe where HLA-A30-B45-DR1-DQ1 is commonly found. Whether these population differences in haplotype frequencies correlate with variable prevalence of HLA-associated diseases has been little investigated at present. However, it has been suggested that the strikingly lower prevalence of type 1 diabetes in Asians compared with Caucasians might result from a parallel variation of HLA susceptibility allele frequencies in both of these ethnic groups [59, 60].
The first region for detailed study of linkage disequilibrium and recombination hotspots
The concept of linkage disequilibrium (LD) was described for the first time in the MHC by Bodmer, Piazza and Dausset [61]. LD refers to an association or correlation between alleles at two or more linked loci. Geneticists working on susceptibility to autoimmune diseases quickly became familiar with the issue as associations between autoimmune diseases and particular HLA loci are often the consequence of strong LD within the MHC. This facilitated the initial mapping of diseases but has made fine mapping of specific variants very difficult [62].
Studies on the H-2 complex in mice suggested different haplotypes could affect the frequency and location of recombination spots [63]. The analysis was more difficult in humans until high density microsatellite and SNP genotyping data was available and sperm typing became possible. Systematic studies showed that recombination and LD patterns across the MHC are not uniform [64, 65]. Two regions are poor in recombination events and show high LD: between HLA-B and HLA-C, and between HLA-DQA1 and HLA-DRB1. The recombination rate in class II and class III regions of 0.74 and 0.94cM/Mb is consistent with the average value of 0.9 cM/Mb, whereas it is only 0.31% in the class I region between HLA-A and HLA-B [66]. This is consistent with the existence of recombination hotspots [67, 68]. Indeed, random association of alleles in TAP1 and TAP2 genes suggested the existence of a recombination hotspot in this 15kb region [69], which was then formally confirmed in 40 families [64].
Using SSCP (single strand conformation polymorphism) analysis, the TAP2 hotspot was fine mapped to the second intron of the TAP2 gene [70], becoming at that time the most precisely defined region of recombination in the human genome. A combination of microsatellite typing and SSCP analysis, allowed to identify two other hotspots in the same region [68]. A first LD map covering 1.5 Mb, between BRD2 and HLA-B genes, was established in a Swedish population using STRs [71]. Analysis using SNPs and haploid genomes (sperm typing) then allowed high resolution mapping of the TAP2 recombination hotspot [72]. Its size of 1.2kb appeared similar to hotspots outside the MHC. It was more active in female than in male meioses and characterized by reciprocal crossovers. It is one of the best known hotspots in the human genome but interestingly it was not found in Chimpanzee genome [73].
Applied to the entire class II region, sperm typing using SNPs revealed three extended regions with high LD, dramatically decreasing around HLA-DOA, DMB and TAP2 [74]. The first two recombination hotspots could actually be resolved into three (DOA) and two (DMB) hotspots of size ranging from 1 to 2 kb and of activity varying between 0.4 and 140 cM/Mb. Nonetheless, all show a remarkably symmetrical distribution of recombination frequencies. These recombination hotspots hold almost all the recombination occurring within the MHC. LD block sizes of 60-90 kb are comparable to the ones described at the same time on chromosome 5 [75] or in the yeast genome [76], suggesting conservation of the mechanisms involved in hotspot distribution. This research culminated in 2002 with the publication of a high resolution meiotic recombination map covering 3.3 Mb of the MHC, with genotyping of 20,031 single sperm from 12 unrelated donors [77]. Six hotspots were found, accounting for 94% of the recombination events. They were separated by “coldspots” covering 87% of the MHC and matching LD blocks. Recombination rates, estimated at 0.49 cM/Mb in the MHC versus 0.92 cM/Mb in the whole genome, may significantly vary between non MHC identical individuals [78], and the recombination pattern is similar between sexes [77]. Hotspots are present at least every 0.8 Mb but are not necessarily evenly spaced. Between these hotspots, low levels of recombination are regularly observed every 100 kb, suggesting the presence of “warm-spots” delimitating the LD blocks. Sperm typing for the whole genome only became possible in 2008 and confirmed the existence of 1-2 kb hotspot regions with recombination rate heterogeneity between individuals [79]. However, recent research has shown the relationship between recombination hotspots and LD blocks is more complex. Recombination hotspots are in general found flanking LD blocks but this need not be the case.
Genomic architecture in the MHC compared to the rest of the genome
How does this detailed characterisation of genetic variation and LD in the MHC relate to the whole genome? The first integrated haplotype map of the MHC revealed that LD extends over larger physical distances in the MHC than in the rest of the genome, the average length being 31.1 kb versus 22.3 kb for the genome [58, 80]. Nonetheless, although physically larger, these blocks are actually shorter in terms of genetic distances (recombination rate of 0.49 cM/Mb versus 0.81 cM/Mb), leading to similar sizes of 0.012 cM versus 0.017 cM. Haplotypic diversity in MHC appears comparable to the rest of the genome except for classical HLA genes.
Higher resolution maps were then established, initially in population of European ancestry [57], then for HapMap populations of European, African, Chinese and Japanese ancestry by the MHC Consortium [81]. In addition to bringing new insights into the evolutionary dynamics of ancestral haplotypes, these two studies identified haplotype tagging SNPs which can be used as surrogates to genotype the classical HLA loci. These informative tagging SNPs are also used in the forthcoming IMAGEN (International Major Histocompatibility Complex and Autoimmunity Genetics Network) project aimed at refining MHC associations in several autoimmune diseases [9].
Maintenance of LD in the MHC
Little is known about the mechanisms maintaining LD in the MHC. Again, work in this area should provide clues to the rest of the genome. Three mechanisms may be involved. First, some allelic associations may have existed in a population of limited size and would have been present after successive generations without yet reaching equilibrium. Second, preferential allelic associations could involve epistatic mechanisms [82] and confer a selective advantage in a given environment. Third, haplotypes may be subject to differential recombination rates [83]. These mechanisms may co-exist and are overlapping in nature. We know that MHC alleles are subject to selective pressures although some suggest that recombination hotspots could be more significant for the LD pattern than population history, with less sharing of haplotypes between populations than in other genomic regions [83]. Numerous studies have suggested that LD and recombination rate of the same genomic region are variable between haplotypes. Observations in MHC are consistent with this hypothesis [84-87].
Potential mechanisms maintaining LD in the MHC are nicely illustrated by the ‘8.1 haplotype’ (or HLA-A1-B8-DR3 haplotype) which consistently shows the highest level with a half-length of LD above 3.5 Mb [87]. With a frequency of about 6%, this is the most common extended haplotype in Caucasian populations [88]. It may be maintained by positive selection of one particular allele, epistatic selection of a combination of alleles, or the existence of a mechanism inhibiting recombination in this specific haplotype.
The first genomic region entirely re-sequenced for common haplotypes
Sequencing single haplotypes has been a dream for geneticists, with attempts to separate parental haplotypes or to perform long-range allele-specific PCRs [89-91]. The latter is now dramatically improved by the next generation DNA sequencing technologies, with the pyrosequencing methods currently generating up to 450 nucleotide reads [92]. However, theoretical determination of haplotype sequence requires a massive coverage. The application of such technologies is more challenging in the MHC due to the extreme polymorphism of the region, notably gene duplication and structural variation. When whole genome re-sequencing studies have been undertaken, the reconstruction of the sequence MHC region remains one of the most difficult.
Fortunately, the MHC has already been the object of a unique re-sequencing approach performed at the haplotypic level by the MHC Haplotype Consortium led by Stephan Beck, Stephen Sawcer, John Todd and John Trowsdale [93]. The MHC Haplotype Project made use of DNA from individuals entirely homozygous for the MHC, as a consequence of consanguinity. Selected haplotypes were frequent in Caucasian populations and associated with common autoimmune diseases [94-96]. Sequences for the ‘8.1 haplotype’, the ‘18.2 haplotype’ [HLA-A26-B18-Cw5-DR3-DQ2] and the ‘7.1 haplotype’ [HLA-A3-B7-Cw7-DR15] were prioritised with the latter replacing the “mosaic” sequence from 1999 as the reference sequence for the MHC. The sequence of the remaining 5 haplotypes of this project was released in 2008 [96], microsatellites have been mapped to the alternate haplotypes [47] and all annotations are available on the Vega database (http://vega.sanger.ac.uk/).
The project revealed different gene content and polymorphisms on each haplotype, with genetic variation between haplotypes mainly concentrated in the classical class I and class II HLA loci and major sequence differences in the complement region (RCCX module) and HLA-DRB. The approach has been extended to 46 other haplotypes [97]. It provided a better understanding of the 8.1 haplotype structure with an estimated age of 35,500 years matching the population migration to Europe. It also revealed new rare SNPs, notably for the class III region, and provided evidence in favour of the rare-SNP hypothesis for MHC associated diseases. An ongoing project sequencing 200 MHC haplotypes using next generation sequencing technology should further advance our knowledge in this area (www.molgen.mpg.de/~genetic-variation/Projects.html).
THE MHC AND GENETIC SUSCEPTIBILITY TO DISEASE: INSIGHTS FOR THE REST OF THE GENOME
The pre-eminent genomic region associated with disease
Point mutations, deletions and other variants within the MHC are responsible for monogenic diseases showing Mendelian inheritance such as haemochromatosis [98] or congenital adrenal hyperplasia [99]. More significant however was the role played by this region of the human genome in many common multifactorial diseases. Serological typing established clear and dramatic evidence of association by the early 1970s between specific HLA antigens and autoimmune diseases such as ankylosing spondylitis [100-102], psoriasis [103] and celiac disease [104, 105]. The importance of the MHC was subsequently robustly demonstrated using microsatellites, single nucleotide polymorphisms (SNPs) and other genetic markers for a range of diseases, and continues today in the era of genome-wide association studies [106]. Mainly autoimmune diseases are associated with genetic variation in the MHC, including the major risk loci for type 1 diabetes [107-109], rheumatoid arthritis [110-112] and multiple sclerosis [113-115] while other disease associations include with infectious and inflammatory diseases [44, 116] as well as cancer [117-119], smoking behaviour [19] or mating preferences [20].
For the majority of disease associations in the MHC, it is the haplotype which is implicated, as opposed to specific variants. Notably, common ancestral haplotypes may be associated with several diseases [120]. Among them, the 8.1 haplotype is the most remarkable because of its association with a very large number of immune phenotypes [121] and diseases [122], such as type 1 diabetes [123], myasthenia gravis [21], rheumatoid arthritis [124], systemic lupus erythematosus [125], celiac disease [126] or IgA deficiency [127]. Fine mapping such haplotypic associations has proved difficult because of the extent of LD which hampers the identification of the causative variants
How the HLA indirectly contributed to create the first physical and genetic map of the human genome
Thanks to the shared Nobel Prize in 1980, Jean Dausset received in 1984 for his laboratory a huge legacy by an art collector, Helène Anavi. With this unexpected gift, he created in collaboration with Howard Cann and Daniel Cohen, the Centre d’Etude du Polymoprhisme Humain (CEPH) [128], which soon after became Foundation Jean Dausset-CEPH. Inspired by the MHC association studies, Dausset and Cohen decided to look at polymorphism across the whole genome to generate markers that would segregate in families to investigate linkage in multiple diseases. The collection included several large families from numerous collaborators, including the large Utah pedigrees studied by Ray White [129]. This CEPH consortium led to the publication of the first microsatellite genetic and physical maps of the human genome [130-132], an essential prelude to enabling the Human Genome Project [133]. This invaluable resource opened a new area in genetic studies, allowing the identification of hundreds of disease susceptibility genes. Today, the CEPH samples are still intensively studied and played a key role in recent genomic advances such as the International HapMap project [134] or the ongoing ‘1000 genomes’ sequencing project (http://www.1000genomes.org).
GENETICAL GENOMICS AND MODULATION OF GENE EXPRESSION IN THE MHC
It has been recently demonstrated that variation in gene expression in humans is heritable [135-138] and genetic determinants can be defined by linkage or association analysis [139]. Approaches developed in model organisms are now applied in humans to map expression quantitative trait loci (eQTLs) by so-called “genetical genomics” [140]. This is an important tool to identify functional variants in disease susceptibility genes [141], particularly when combined with genome wide disease association studies, as illustrated by recent work on asthma [142] or high-density lipoprotein cholesterol [143]. Given its many disease associations and remarkable genetic polymorphism, as might be expected, the human MHC has been shown to be a major site of association for eQTLs (summarized in Table 1) [137, 138, 143-151].
Table 1.
eQTL mapping studies showing strong evidence of linkage or association with the MHC
| Study | Description | Expression phenotypes at MHC loci |
|---|---|---|
| Cheung et al. 2003 |
|
|
| Pastinen et al. 2004 |
|
|
| Monks et al. 2004 |
|
|
| Cheung et al. 2005 |
|
|
| Spielman et al. 2007 |
|
|
| Stranger et al. 2007 |
|
|
| Dixon et al. 2007 |
|
|
| Göring et al. 2007 |
|
|
| Kwan et al. 2008 |
|
|
| Emilsson et al. 2008 |
|
|
These associations are striking but care in interpreting such results in the context of the MHC is appropriate for a number of reasons, notably relating to the technology used in analyses of gene expression. First, the commercial expression microarrays used are designed against the reference sequence of the human genome, so do not take into account genetic and haplotypic diversity which is particularly significant in the MHC. As a consequence, individuals carrying haplotypes other than the reference haplotype (for the MHC, HLA-A3-B7-DR15) may not show expression of certain genes because the designed probes do not match the gene sequence of their particular haplotypic background [152]. This could skew the distribution of the expressed gene and may lead to false positive eQTL associations. The second issue is that the arrays used in almost all human eQTL studies to date only target the 3′ end of the transcripts, hence miss a large number of alternatively spliced gene isoforms. Alternative splicing is common across the genome, including the MHC, and is itself modulated by DNA sequence diversity, further compounding these difficulties [146, 153-157]. Methods quantifying accurately, both qualitatively and quantitatively, all transcripts in a haplotypic specific manner - the “haplo-spliceo-transcriptome” [158] - are required to investigate the genetic basis of gene expression in the human MHC. Further molecular and cellular approaches are needed to identify regulatory variants modulating gene expression within haplotypes. Such efforts are facilitated by current projects to define the regulatory landscape of the human genome such as the ENCODE (ENCyclopedia Of DNA Elements) Project [159] and a variety of approaches based on animal models, assays of chromatin accessibility and modifications, protein-DNA interactions and gene expression [160]. For example, at the TNF gene locus, we recently defined novel regulatory elements based on quantitative chromatin profiling [161] while earlier work resolved specific promoter SNPs modulating affinity of transcription factor binding [162, 163]. A further approach is to consider allele-specific gene expression which occurs relatively commonly and is heritable: this approach has been extended from analysis based on relative transcript abundance to considering RNA polymerase loading, allowing resolution of regulatory variants [144, 162, 164]. For example, using the haplotype specific chromatin immunoprecipitation (haploChIP) approach, differences in gene expression were resolved to LTA rather than TNF within an LD block spanning these two genes [164].
Other studies have combined ChIP with RNA-FISH and chromosomal capture conformation at the HLA-DRB1 locus to reveal a new mechanism of regulation of MHC class II genes involving the insulator CTCF binding factor and a particular chromatin long-distance loop configuration [165]. Some specific associations of chromosome territories with nuclear proteins were invoked [166]. An open chromatin configuration has been described in relation to the timing of DNA replication for the MHC which occurred early in S-phase [167]. The generation of a 2 kb resolution tiling path MHC array [168] has also allowed chromatin profiling and identification of cell-specific genomic anchors which show dynamic changes on transcriptional activation [169]. This MHC array has also allowed analysis of methylation across the MHC to define 90 differentially methylated regions using DNA from liver, placenta, CD+ lymphocytes and sperm. This builds on data in which the MHC was studied as the pilot region for the Epigenome project [170-173], which had revealed that 10% of MHC loci had a cell-specific methylation pattern although the relationship of methylation to haplotypic background currently remains unclear.
CONCLUSIONS
The MHC has intrigued, enthralled and challenged several generations of researchers. It is a remarkable region of the human genome which has indeed proved a paradigm for many different aspects of human genetic research. In the genetics Olympics, the MHC would hold many records notably in terms of extreme genetic variation, gene density, biological significance and disease association. Rightly, much research effort has been focused on trying to dissect the complexities of the MHC and many general lessons have been learnt, notably relating to the nature and inheritance of genetic polymorphism. We have seen how this was the first substantial genomic region to be sequenced and the first to be clearly delineated in terms of its genomic landscape, pioneering our understanding of linkage disequilibrium, recombination and haplotypic structure. Despite this, progress in fine mapping disease associations and resolving specific functional variants remains difficult. It is now clear that both structural and regulatory variants are important and may indeed be operating in tandem. Advances in functional genomics to map and characterise polymorphisms modulating gene expression should help advance this field. However the complexities are many-layered, from sequence level diversity to gene regulation, epigenetic and environmental factors. Effects will be context specific, relating to particular cell or tissue types and indeed developmental stages, as well as particular environmental stimuli or other conditions. The MHC will undoubtedly continue to challenge us and has many secrets yet to be revealed which will be of relevance to the rest of the genome. However an integrated approach, building on current successes, should ensure research into the MHC remains at the avant-garde of human genomics.
Key points.
The MHC is the most polymorphic and gene dense region of the human genome
Genetic variation in the MHC is associated with more diseases than any other genomic region
The MHC has been a model to study linkage disequilibrium and haplotypic structure
This is the first region of the genome to be sequenced for common haplotypes
Current efforts to map expression quantitative trait loci highlight the importance of genetic variation in the MHC
Acknowledgements
We are grateful to friends and colleagues within the Wellcome Trust Centre for Human Genetics and elsewhere for many stimulating discussions relating to genetic variation and the biology of the MHC.
Funding: This work was supported by the Wellcome Trust [074318 to J.C.K].
Footnotes
Publisher's Disclaimer: This is a pre-copyediting, author-produced pdf of an article accepted for publication in Briefings in Functional Genomics and Proteomics following peer review. The definitive publisher-authenticated version Vandiedonck C & Knight JC Brief Funct Genomic Proteomic. 2009 8(5):379-94 is available at http://bfgp.oxfordjournals.org/cgi/content/full/8/5/379.
References
- 1.Dausset J. The major histocompatibility complex in man. Science. 1981;213:1469–1474. doi: 10.1126/science.6792704. [DOI] [PubMed] [Google Scholar]
- 2.Bodmer WF. Evolutionary significance of the HL-A system. Nature. 1972;237:139–145. doi: 10.1038/237139a0. passim. [DOI] [PubMed] [Google Scholar]
- 3.Dausset J, Le Brun A, Sasportes M. Lymphocyte mixed culture reaction (LMC) between parents and children with the same HL-A phenotype. Hypothesis of a genetic recognition system. C R Acad Sci Hebd Seances Acad Sci D. 1972;275:2279–2282. [PubMed] [Google Scholar]
- 4.Snell GD. Methods for the study of histocompatibility genes. J Genet. 1948;49:87–108. doi: 10.1007/BF02986826. [DOI] [PubMed] [Google Scholar]
- 5.Snell GD. The H-2 locus of the mouse: observations and speculations concerning its comparative genetics and its polymorphism. Folia Biol (Praha) 1968;14:335–358. [PubMed] [Google Scholar]
- 6.Ryder LP, Svejgaard A, Dausset J. Genetics of HLA disease association. Annu Rev Genet. 1981;15:169–187. doi: 10.1146/annurev.ge.15.120181.001125. [DOI] [PubMed] [Google Scholar]
- 7.Lie BA, Thorsby E. Several genes in the extended human MHC contribute to predisposition to autoimmune diseases. Curr Opin Immunol. 2005;17:526–531. doi: 10.1016/j.coi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Trowsdale J. HLA genomics in the third millennium. Curr Opin Immunol. 2005;17:498–504. doi: 10.1016/j.coi.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Fernando MM, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen CE, Alper CA. The genetics of HLA-associated disease. Curr Opin Immunol. 2004;16:660–667. doi: 10.1016/j.coi.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Gorer PA. The genetic and antigenic basis of tumour transplantation. J Pathol Bacteriol. 1937;44:691–697. [Google Scholar]
- 12.Snell GD. Some recollections of Peter Gorer and his work on this fiftieth anniversary of his discovery of H-2. Immunogenetics. 1986;24:339–340. doi: 10.1007/BF00377948. [DOI] [PubMed] [Google Scholar]
- 13.Dausset J. Iso-leuko-antibodies. Acta Haematol. 1958;20:156–166. doi: 10.1159/000205478. [DOI] [PubMed] [Google Scholar]
- 14.Payne R, Tripp M, Weigle J, et al. A New Leukocyte Isoantigen System in Man. Cold Spring Harb Symp Quant Biol. 1964;29:285–295. doi: 10.1101/sqb.1964.029.01.031. [DOI] [PubMed] [Google Scholar]
- 15.McDevitt HO, Benacerraf B. Genetic control of specific immune responses. Adv Immunol. 1969;11:31–74. doi: 10.1016/s0065-2776(08)60477-0. [DOI] [PubMed] [Google Scholar]
- 16.Benacerraf B. Role of MHC gene products in immune regulation. Science. 1981;212:1229–1238. doi: 10.1126/science.6165083. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkman PJ, Saper MA, Samraoui B, et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 18.Jones EY, Fugger L, Strominger JL, et al. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 19.Fust G, Arason GJ, Kramer J, et al. Genetic basis of tobacco smoking: strong association of a specific major histocompatibility complex haplotype on chromosome 6 with smoking behavior. Int Immunol. 2004;16:1507–1514. doi: 10.1093/intimm/dxh152. [DOI] [PubMed] [Google Scholar]
- 20.Jacob S, McClintock MK, Zelano B, et al. Paternally inherited HLA alleles are associated with women’s choice of male odor. Nat Genet. 2002;30:175–179. doi: 10.1038/ng830. [DOI] [PubMed] [Google Scholar]
- 21.Vandiedonck C, Beaurain G, Giraud M, et al. Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia. Proc Natl Acad Sci U S A. 2004;101:15464–15469. doi: 10.1073/pnas.0406756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayley JP, Ottenhoff TH, Verweij CL. Is there a future for TNF promoter polymorphisms? Genes Immun. 2004;5:315–329. doi: 10.1038/sj.gene.6364055. [DOI] [PubMed] [Google Scholar]
- 23.The MHC sequencing consortium Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 24.McPherson JD, Marra M, Hillier L, et al. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 25.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 26.Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Wang W, Li R, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie T, Rowen L, Aguado B, et al. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 2003;13:2621–2636. doi: 10.1101/gr.1736803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawkins R, Leelayuwat C, Gaudieri S, et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 32.Shiina T, Tamiya G, Oka A, et al. Molecular dynamics of MHC genesis unraveled by sequence analysis of the 1,796,938-bp HLA class I region. Proc Natl Acad Sci U S A. 1999;96:13282–13287. doi: 10.1073/pnas.96.23.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bristow J, Tee MK, Gitelman SE, et al. Tenascin-X: a novel extracellular matrix protein encoded by the human XB gene overlapping P450c21B. J Cell Biol. 1993;122:265–278. doi: 10.1083/jcb.122.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howard JC. Restrictions on the use of antigenic peptides by the immune system. Proc Natl Acad Sci U S A. 1993;90:3777–3779. doi: 10.1073/pnas.90.9.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trowsdale J. Genomic structure and function in the MHC. Trends Genet. 1993;9:117–122. doi: 10.1016/0168-9525(93)90205-v. [DOI] [PubMed] [Google Scholar]
- 36.Mungall AJ, Palmer SA, Sims SK, et al. The DNA sequence and analysis of human chromosome 6. Nature. 2003;425:805–811. doi: 10.1038/nature02055. [DOI] [PubMed] [Google Scholar]
- 37.Stephens R, Horton R, Humphray S, et al. Gene organisation, sequence variation and isochore structure at the centromeric boundary of the human MHC. J Mol Biol. 1999;291:789–799. doi: 10.1006/jmbi.1999.3004. [DOI] [PubMed] [Google Scholar]
- 38.Horton R, Wilming L, Rand V, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 39.Scherer SW, Lee C, Birney E, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson J, Waller MJ, Parham P, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Little AM, Parham P. Polymorphism and evolution of HLA class I and II genes and molecules. Rev Immunogenet. 1999;1:105–123. [PubMed] [Google Scholar]
- 42.Horton R, Niblett D, Milne S, et al. Large-scale sequence comparisons reveal unusually high levels of variation in the HLA-DQB1 locus in the class II region of the human MHC. J Mol Biol. 1998;282:71–97. doi: 10.1006/jmbi.1998.2018. [DOI] [PubMed] [Google Scholar]
- 43.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 44.Carrington M, Nelson GW, Martin MP, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 45.Gourraud PA, Cambon-Thomsen A, Dauber EM, et al. Nomenclature for HLA microsatellites. Tissue Antigens. 2007;69(Suppl 1):210–213. doi: 10.1111/j.1399-0039.2006.00771.x. [DOI] [PubMed] [Google Scholar]
- 46.Gourraud PA, Feolo M, Hoffman D, et al. The dbMHC microsatellite portal: a public resource for the storage and display of MHC microsatellite information. Tissue Antigens. 2006;67:395–401. doi: 10.1111/j.1399-0039.2006.00600.x. [DOI] [PubMed] [Google Scholar]
- 47.Gourraud PA, Hoffman D, Cambon-Thomsen A, et al. 14th International HLA and Immunogenetics Workshop: report on mapping microsatellite markers in the major histocompatibility complex region. Tissue Antigens. 2007;69(Suppl 1):206–209. doi: 10.1111/j.1399-0039.2006.00770.x. [DOI] [PubMed] [Google Scholar]
- 48.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 49.Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–192. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ceppellini R, Curtoni E, Mattiuz P, et al. Genetics of Leukocyte Antigens: A Family Study of Segregation and Linkage. In: Curtoni E, Mattiuz P, Tosi R, editors. Histocompatibility Testing. Munksgaard; Copenhagen: 1967. [Google Scholar]
- 53.Ceppellini R, Siniscalco M, Smith CA. The estimation of gene frequencies in a random-mating population. Ann Hum Genet. 1955;20:97–115. doi: 10.1111/j.1469-1809.1955.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 54.Amos B, Ward FE, Zmijewski CM, et al. Graft donor selection based upon single locus (haplotype) analysis within families. Transplantation. 1968;6:524–534. doi: 10.1097/00007890-196807000-00003. [DOI] [PubMed] [Google Scholar]
- 55.Alper CA, Raum D, Karp S, et al. Serum complement ‘supergenes’ of the major histocompatibility complex in man (complotypes) Vox Sang. 1983;45:62–67. doi: 10.1111/j.1423-0410.1983.tb04124.x. [DOI] [PubMed] [Google Scholar]
- 56.Yunis EJ, Larsen CE, Fernandez-Vina M, et al. Inheritable variable sizes of DNA stretches in the human MHC: conserved extended haplotypes and their fragments or blocks. Tissue Antigens. 2003;62:1–20. doi: 10.1034/j.1399-0039.2003.00098.x. [DOI] [PubMed] [Google Scholar]
- 57.Miretti MM, Walsh EC, Ke X, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh EC, Mather KA, Schaffner SF, et al. An integrated haplotype map of the human major histocompatibility complex. Am J Hum Genet. 2003;73:580–590. doi: 10.1086/378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Todd JA, Mijovic C, Fletcher J, et al. Identification of susceptibility loci for insulin-dependent diabetes mellitus by trans-racial gene mapping. Nature. 1989;338:587–589. doi: 10.1038/338587a0. [DOI] [PubMed] [Google Scholar]
- 60.Park Y, Eisenbarth GS. Genetic susceptibility factors of Type 1 diabetes in Asians. Diabetes Metab Res Rev. 2001;17:2–11. doi: 10.1002/1520-7560(2000)9999:9999<::aid-dmrr164>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 61.Dausset J, Legrand L, Lepage V, et al. A haplotype study of HLA complex with special reference to the HLA-DR series and to Bf. C2 and glyoxalase I polymorphisms. Tissue Antigens. 1978;12:297–307. doi: 10.1111/j.1399-0039.1978.tb01337.x. [DOI] [PubMed] [Google Scholar]
- 62.Bell JI. Single nucleotide polymorphisms and disease gene mapping. Arthritis Res. 2002;4(Suppl 3):S273–278. doi: 10.1186/ar555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinmetz M, Stephan D, Fischer Lindahl K. Gene organization and recombinational hotspots in the murine major histocompatibility complex. Cell. 1986;44:895–904. doi: 10.1016/0092-8674(86)90012-7. [DOI] [PubMed] [Google Scholar]
- 64.Carrington M, Stephens JC, Klitz W, et al. Major histocompatibility complex class II haplotypes and linkage disequilibrium values observed in the CEPH families. Hum Immunol. 1994;41:234–240. doi: 10.1016/0198-8859(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez-Mazas A, Djoulah S, Busson M, et al. A linkage disequilibrium map of the MHC region based on the analysis of 14 loci haplotypes in 50 French families. Eur J Hum Genet. 2000;8:33–41. doi: 10.1038/sj.ejhg.5200391. [DOI] [PubMed] [Google Scholar]
- 66.Martin M, Mann D, Carrington M. Recombination rates across the HLA complex: use of microsatellites as a rapid screen for recombinant chromosomes. Hum Mol Genet. 1995;4:423–428. doi: 10.1093/hmg/4.3.423. [DOI] [PubMed] [Google Scholar]
- 67.Carrington M. Recombination within the human MHC. Immunol Rev. 1999;167:245–256. doi: 10.1111/j.1600-065x.1999.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 68.Cullen M, Noble J, Erlich H, et al. Characterization of recombination in the HLA class II region. Am J Hum Genet. 1997;60:397–407. [PMC free article] [PubMed] [Google Scholar]
- 69.van Endert PM, Lopez MT, Patel SD, et al. Genomic polymorphism, recombination, and linkage disequilibrium in human major histocompatibility complex-encoded antigen-processing genes. Proc Natl Acad Sci U S A. 1992;89:11594–11597. doi: 10.1073/pnas.89.23.11594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cullen M, Erlich H, Klitz W, et al. Molecular mapping of a recombination hotspot located in the second intron of the human TAP2 locus. Am J Hum Genet. 1995;56:1350–1358. [PMC free article] [PubMed] [Google Scholar]
- 71.Vorechovsky I, Kralovicova J, Laycock MD, et al. Short tandem repeat (STR) haplotypes in HLA: an integrated 50-kb STR/linkage disequilibrium/gene map between the RING3 and HLA-B genes and identification of STR haplotype diversification in the class III region. Eur J Hum Genet. 2001;9:590–598. doi: 10.1038/sj.ejhg.5200688. [DOI] [PubMed] [Google Scholar]
- 72.Jeffreys AJ, Ritchie A, Neumann R. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet. 2000;9:725–733. doi: 10.1093/hmg/9.5.725. [DOI] [PubMed] [Google Scholar]
- 73.Ptak SE, Roeder AD, Stephens M, et al. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2004;2:e155. doi: 10.1371/journal.pbio.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 75.Daly MJ, Rioux JD, Schaffner SF, et al. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 76.Gerton JL, DeRisi J, Shroff R, et al. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cullen M, Perfetto SP, Klitz W, et al. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet. 2002;71:759–776. doi: 10.1086/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Broman KW, Murray JC, Sheffield VC, et al. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coop G, Wen X, Ober C, et al. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 80.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 81.de Bakker PI, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gregersen JW, Kranc KR, Ke X, et al. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature. 2006;443:574–577. doi: 10.1038/nature05133. [DOI] [PubMed] [Google Scholar]
- 83.Kauppi L, Sajantila A, Jeffreys AJ. Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum Mol Genet. 2003;12:33–40. doi: 10.1093/hmg/ddg008. [DOI] [PubMed] [Google Scholar]
- 84.Thomsen M, Neugebauer M, Arnaud J, et al. Recombination fractions in the HLA system based on the data set ‘provinces Francaises’: indications of haplotype-specific recombination rates. Eur J Immunogenet. 1994;21:33–43. doi: 10.1111/j.1744-313x.1994.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 85.Levo A, Westman P, Partanen J. An approach to mapping haplotype-specific recombination sites in human MHC class III. Immunogenetics. 1996;43:136–140. doi: 10.1007/BF00176674. [DOI] [PubMed] [Google Scholar]
- 86.D’Alfonso S, Borelli I, Dall’Omo A, et al. The natural history of an HLA haplotype and its recombinants. Immunogenetics. 1998;48:8–15. doi: 10.1007/s002510050394. [DOI] [PubMed] [Google Scholar]
- 87.Ahmad T, Neville M, Marshall SE, et al. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet. 2003;12:647–656. [PubMed] [Google Scholar]
- 88.Candore G, Modica MA, Lio D, et al. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: a genetically determined defect of C4 influences immunological parameters of healthy carriers of the haplotype. Biomed Pharmacother. 2003;57:274–277. doi: 10.1016/s0753-3322(03)00079-9. [DOI] [PubMed] [Google Scholar]
- 89.Kidd JM, Cheng Z, Graves T, et al. Haplotype sorting using human fosmid clone end-sequence pairs. Genome Res. 2008;18:2016–2023. doi: 10.1101/gr.081786.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Turner DJ, Tyler-Smith C, Hurles ME. Long-range, high-throughput haplotype determination via haplotype-fusion PCR and ligation haplotyping. Nucleic Acids Res. 2008;36:e82. doi: 10.1093/nar/gkn373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dapprich J, Ferriola D, Magira EE, et al. SNP-specific extraction of haplotype-resolved targeted genomic regions. Nucleic Acids Res. 2008;36:e94. doi: 10.1093/nar/gkn345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 93.Allcock RJ, Atrazhev AM, Beck S, et al. The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens. 2002;59:520–521. doi: 10.1034/j.1399-0039.2002.590609.x. [DOI] [PubMed] [Google Scholar]
- 94.Stewart CA, Horton R, Allcock RJ, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Traherne JA, Horton R, Roberts AN, et al. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horton R, Gibson R, Coggill P, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith WP, Vu Q, Li SS, et al. Toward understanding MHC disease associations: partial resequencing of 46 distinct HLA haplotypes. Genomics. 2006;87:561–571. doi: 10.1016/j.ygeno.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 98.Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 99.Miller WL, Morel Y. The molecular genetics of 21-hydroxylase deficiency. Annu Rev Genet. 1989;23:371–393. doi: 10.1146/annurev.ge.23.120189.002103. [DOI] [PubMed] [Google Scholar]
- 100.Brewerton DA, Hart FD, Nicholls A, et al. Ankylosing spondylitis and HL-A 27. Lancet. 1973;1:904–907. doi: 10.1016/s0140-6736(73)91360-3. [DOI] [PubMed] [Google Scholar]
- 101.Caffrey MF, James DC. Human lymphocyte antigen association in ankylosing spondylitis. Nature. 1973;242:121. doi: 10.1038/242121a0. [DOI] [PubMed] [Google Scholar]
- 102.Schlosstein L, Terasaki PI, Bluestone R, et al. High association of an HL-A antigen, W27, with ankylosing spondylitis. N Engl J Med. 1973;288:704–706. doi: 10.1056/NEJM197304052881403. [DOI] [PubMed] [Google Scholar]
- 103.Russell TJ, Schultes LM, Kuban DJ. Histocompatibility (HL-A) antigens associated with psoriasis. N Engl J Med. 1972;287:738–740. doi: 10.1056/NEJM197210122871503. [DOI] [PubMed] [Google Scholar]
- 104.Falchuk ZM, Rogentine GN, Strober W. Predominance of histocompatibility antigen HL-A8 in patients with gluten-sensitive enteropathy. J Clin Invest. 1972;51:1602–1605. doi: 10.1172/JCI106958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stokes PL, Asquith P, Holmes GK, et al. Histocompatibility antigens associated with adult coeliac disease. Lancet. 1972;2:162–164. doi: 10.1016/s0140-6736(72)91330-x. [DOI] [PubMed] [Google Scholar]
- 106.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cudworth AG, Woodrow JC. Evidence for HL-A-linked genes in “juvenile” diabetes mellitus. Br Med J. 1975;3:133–135. doi: 10.1136/bmj.3.5976.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nerup J, Platz P, Andersen OO, et al. HL-A antigens and diabetes mellitus. Lancet. 1974;2:864–866. doi: 10.1016/s0140-6736(74)91201-x. [DOI] [PubMed] [Google Scholar]
- 109.Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 110.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 111.Stastny P. Mixed lymphocyte cultures in rheumatoid arthritis. J Clin Invest. 1976;57:1148–1157. doi: 10.1172/JCI108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wordsworth BP, Lanchbury JS, Sakkas LI, et al. HLA-DR4 subtype frequencies in rheumatoid arthritis indicate that DRB1 is the major susceptibility locus within the HLA class II region. Proc Natl Acad Sci U S A. 1989;86:10049–10053. doi: 10.1073/pnas.86.24.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jersild C, Fog T, Hansen GS, et al. Histocompatibility determinants in multiple sclerosis, with special reference to clinical course. Lancet. 1973;2:1221–1225. doi: 10.1016/s0140-6736(73)90970-7. [DOI] [PubMed] [Google Scholar]
- 114.Winchester R, Ebers G, Fu SM, et al. B-cell alloantigen Ag 7a in multiple sclerosis. Lancet. 1975;2:814. doi: 10.1016/s0140-6736(75)80033-x. [DOI] [PubMed] [Google Scholar]
- 115.Fogdell A, Hillert J, Sachs C, et al. The multiple sclerosis- and narcolepsy-associated HLA class II haplotype includes the DRB5*0101 allele. Tissue Antigens. 1995;46:333–336. doi: 10.1111/j.1399-0039.1995.tb02503.x. [DOI] [PubMed] [Google Scholar]
- 116.Hill AV. Aspects of genetic susceptibility to human infectious diseases. Annu Rev Genet. 2006;40:469–486. doi: 10.1146/annurev.genet.40.110405.090546. [DOI] [PubMed] [Google Scholar]
- 117.Browning M, Dunnion D. HLA and cancer: implications for cancer immunotherapy and vaccination. Eur J Immunogenet. 1997;24:293–312. doi: 10.1111/j.1365-2370.1997.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 118.Howell WM, Calder PC, Grimble RF. Gene polymorphisms, inflammatory diseases and cancer. Proc Nutr Soc. 2002;61:447–456. doi: 10.1079/pns2002186. [DOI] [PubMed] [Google Scholar]
- 119.Bateman AC, Howell WM. Human leukocyte antigens and cancer: is it in our genes? J Pathol. 1999;188:231–236. doi: 10.1002/(SICI)1096-9896(199907)188:3<231::AID-PATH325>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 120.Dawkins RL, Christiansen FT, Kay PH, et al. Disease associations with complotypes, supratypes and haplotypes. Immunol Rev. 1983;70:1–22. doi: 10.1111/j.1600-065x.1983.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 121.Price P, Witt C, Allcock R, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 122.Candore G, Lio D, Colonna Romano G, et al. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev. 2002;1:29–35. doi: 10.1016/s1568-9972(01)00004-0. [DOI] [PubMed] [Google Scholar]
- 123.Degli-Esposti MA, Abraham LJ, McCann V, et al. Ancestral haplotypes reveal the role of the central MHC in the immunogenetics of IDDM. Immunogenetics. 1992;36:345–356. doi: 10.1007/BF00218041. [DOI] [PubMed] [Google Scholar]
- 124.Jawaheer D, Li W, Graham RR, et al. Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am J Hum Genet. 2002;71:585–594. doi: 10.1086/342407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Christiansen FT, Dawkins RL, Uko G, et al. Complement allotyping in SLE: association with C4A null. Aust N Z J Med. 1983;13:483–488. doi: 10.1111/j.1445-5994.1983.tb02699.x. [DOI] [PubMed] [Google Scholar]
- 126.Shidrawi RG, Parnell ND, Ciclitira PJ, et al. Binding of gluten-derived peptides to the HLA-DQ2 (alpha1*0501, beta1*0201) molecule, assessed in a cellular assay. Clin Exp Immunol. 1998;111:158–165. doi: 10.1046/j.1365-2249.1998.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schroeder HW, Jr., Zhu ZB, March RE, et al. Susceptibility locus for IgA deficiency and common variable immunodeficiency in the HLA-DR3, -B8, -A1 haplotypes. Mol Med. 1998;4:72–86. [PMC free article] [PubMed] [Google Scholar]
- 128.Dausset J, Cann H, Cohen D, et al. Centre d’etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 129.Wyman AR, White R. A highly polymorphic locus in human DNA. Proc Natl Acad Sci U S A. 1980;77:6754–6758. doi: 10.1073/pnas.77.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weissenbach J, Gyapay G, Dib C, et al. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- 131.NIH/CEPH Collaborative Mapping Group A comprehensive genetic linkage map of the human genome. Science. 1992;258:67–86. [PubMed] [Google Scholar]
- 132.Cohen D, Chumakov I, Weissenbach J. A first-generation physical map of the human genome. Nature. 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- 133.Murray JC, Buetow KH, Weber JL, et al. Cooperative Human Linkage Center (CHLC) A comprehensive human linkage map with centimorgan density. Science. 1994;265:2049–2054. doi: 10.1126/science.8091227. [DOI] [PubMed] [Google Scholar]
- 134.The International HapMap Project Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 135.Yan H, Yuan W, Velculescu VE, et al. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 136.Schadt EE, Monks SA, Drake TA, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 137.Monks SA, Leonardson A, Zhu H, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cheung VG, Conlin LK, Weber TM, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 139.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 140.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 141.Cookson W, Liang L, Abecasis G, et al. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 143.Goring HH, Curran JE, Johnson MP, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 144.Cheung VG, Spielman RS, Ewens KG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 146.Kwan T, Benovoy D, Dias C, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 147.Pastinen T, Sladek R, Gurd S, et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- 148.Spielman RS, Bastone LA, Burdick JT, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 150.Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang W, Duan S, Kistner EO, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Johnson JM, Castle J, Garrett-Engele P, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 154.Modrek B, Resch A, Grasso C, et al. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kwan T, Benovoy D, Dias C, et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hull J, Campino S, Rowlands K, et al. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 2007;3:e99. doi: 10.1371/journal.pgen.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 158.Graveley BR. The haplo-spliceo-transcriptome: common variations in alternative splicing in the human population. Trends Genet. 2008;24:5–7. doi: 10.1016/j.tig.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Consortium TEP The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 160.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Taylor JM, Wicks K, Vandiedonck C, et al. Chromatin profiling across the human tumour necrosis factor gene locus reveals a complex, cell type-specific landscape with novel regulatory elements. Nucleic Acids Res. 2008;36:4845–4862. doi: 10.1093/nar/gkn444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Knight JC, Keating BJ, Kwiatkowski DP. Allele-specific repression of lymphotoxin-alpha by activated B cell factor-1. Nat Genet. 2004;36:394–399. doi: 10.1038/ng1331. [DOI] [PubMed] [Google Scholar]
- 163.Knight JC, Udalova I, Hill AV, et al. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat Genet. 1999;22:145–150. doi: 10.1038/9649. [DOI] [PubMed] [Google Scholar]
- 164.Knight JC, Keating BJ, Rockett KA, et al. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 165.Majumder P, Gomez JA, Chadwick BP, et al. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shiels C, Islam SA, Vatcheva R, et al. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- 167.Takousis P, Johonnett P, Williamson J, et al. Replication timing profile reflects the distinct functional and genomic features of the MHC class II region. Cell Cycle. 2007;6:2393–2398. doi: 10.4161/cc.6.19.4762. [DOI] [PubMed] [Google Scholar]
- 168.Tomazou EM, Rakyan VK, Lefebvre G, et al. Generation of a genomic tiling array of the human Major Histocompatibility Complex (MHC) and its application for DNA methylation analysis. BMC Med Genomics. 2008;1:19. doi: 10.1186/1755-8794-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ottaviani D, Lever E, Mitter R, et al. Reconfiguration of genomic anchors upon transcriptional activation of the human major histocompatibility complex. Genome Res. 2008;18:1778–1786. doi: 10.1101/gr.082313.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bradbury J. Human epigenome project--up and running. PLoS Biol. 2003;1:E82. doi: 10.1371/journal.pbio.0000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Eckhardt F, Beck S, Gut IG, et al. Future potential of the Human Epigenome Project. Expert Rev Mol Diagn. 2004;4:609–618. doi: 10.1586/14737159.4.5.609. [DOI] [PubMed] [Google Scholar]
- 173.Rakyan VK, Hildmann T, Novik KL, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bailey JA, Yavor AM, Massa HF, et al. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]