Abstract
Background
20-hydroxyecdysone (20E) and its receptor complex ecdysone receptor (EcR) and ultraspiracle (USP) play a crucial role in controlling development, metamorphosis, reproduction and diapause. The ligand-receptor complex 20E-EcR/USP directly activates a small set of early-response genes and a much larger set of late-response genes. However, ecdysone-responsive genes have not been previously characterized in the context of insect chitin biosynthesis.
Principal Findings
Here, we show that injection-based RNA interference (RNAi) directed towards a common region of the two isoforms of SeEcR in a lepidopteron insect Spodoptera exigua was effective, with phenotypes including a high mortality prior to pupation and developmental defects. After gene specific RNAi, chitin contents in the cuticle of an abnormal larva significantly decreased. The expression levels of five genes in the chitin biosynthesis pathway, SeTre-1, SeG6PI, SeUAP, SeCHSA and SeCHSB, were significantly reduced, while there was no difference in the expression of SeTre-2 prior to 72 hr after injection of EcR dsRNA. Meanwhile, injection of 20E in vivo induced the expression of the five genes mentioned above. Moreover, the SeTre-1, SeG6PI, SeUAP and SeCHSB genes showed late responses to the hormone and the induction of SeTre-1, SeG6PI, SeUAP and SeCHSB genes by 20E were able to be inhibited by the protein synthesis inhibitor cycloheximide in vitro indicating these genes are 20E late-response genes.
Conclusions
We conclude that SeTre-1, SeG6PI, SeUAP and SeCHSB in the chitin biosynthesis pathway are 20E late-response genes and 20E and its specific receptors plays a key role in the regulation of chitin biosynthesis via inducing their expression.
Introduction
Throughout the insect life cycle, the steroid hormone 20-hydroxyecdysone (20E) coordinates multiple developmental events by eliciting a complex genetic program via a heterodimeric nuclear receptor composed of the ecdysone receptor (EcR) and ultraspiracle proteins (USP). Evidenced in a study of fruit fly Drosophila melanogaster by Ashburner et al. first revealed that a part of this developmental program consists of a genetic cascade in which the ligand-receptor complex 20E-EcR/USP directly activates the expression of a very small number of early-response genes. The products of the early-response genes in turn trigger the expression of a much larger set of late-response genes, so called secondary-response genes [1], [2], [3]. During the last decade, the EcR, USP and a large number of ecdysone-responsive genes from the two gene categories proposed in the Ashburner model have been characterized in D. melanogaster and several other insect species [4], [5], [6]. All of those studies published over the past decade have provided powerful evidence in support of the Ashburner model, meantime taken beyond that model which presented us with diverting new directions for our understanding of ecdysone signaling [7], [8], [9], [10].
Most of the previous studies are extensively emphasize DNA puffs induced by ecdysone, whereas a few specific DNA puff genes are studied for a better understanding of the mechanisms underlying the ecdysone response on molecular level. The roles and functions of many early-response genes (eg. E74, E75 and broad-complex) have been well studied [11], [12], while much less is known about the late-response genes (L63, L71 and L82) in insects. L63 has homology to the cyclin dependent kinase protein family and is required for Drosophila development [13]; L71 genes encode a set of polypeptides and provide an antimicrobial defense during metamorphosis [14]. L82 mutations displayed developmental delay and eclosion lethal phenotypes in Drosophila [15]. The late genes play direct or indirect roles and also a distinct role in controlling the appropriate biological response to hormone, including development, metamorphosis, reproduction and diapause. As outlined above, most of the studies regarding the ecdysone-responsive genes in insects have focused on the regulatory genes at the top of the ecdysone-elicit genetic hierarchy.
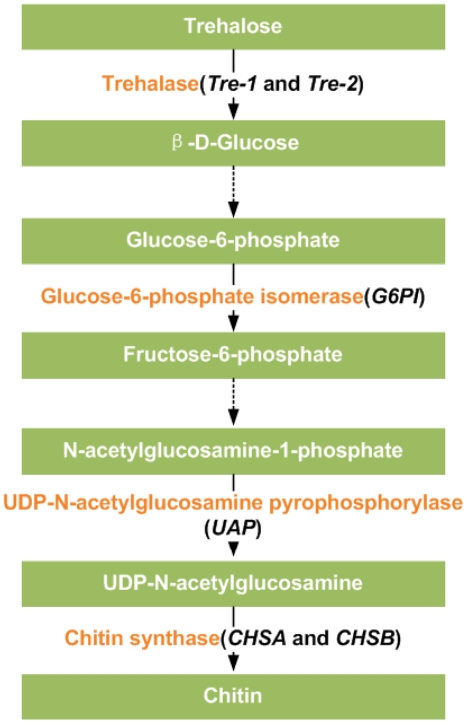
Chitin is the major polysaccharide layed in the cuticle, peritrophic matrix, tracheae and muscle attachment points as a characteristic constituent of insects and other arthropods. The chitin biosynthesis pathway begins with glycogen and trehalose, and consists of a patchwork of at least eight key enzymes (Fig. 1) [16]. Importantly, the process of chitin biosynthesis and degradation is strictly coordinated within the cycle of molts and behaves as an ecdysone-induced response [17], [18], [19]. Many studies have demonstrated the participation of 20E-EcR/USP complex in regulation of gene expression; however, the ecdysone-responsive genes involved in the process of chitin biosynthesis are still largely unclear.
Figure 1. A brief diagram of insect chitin biosynthesis pathway.
The black italics in parenthesis indicate six genes encoding the enzymes.
Here we cloned two EcR isoforms, SeEcR-A and SeEcR-B1, from the beet armyworm, Spodoptera exigua, a wide-spread, destructive and polyphagous noctuid lepidopteron pest. Consequently, we confirmed that the injection-based RNAi of SeEcR leads to a delay in developmental duration, reduced food intake, kinds of defect phenotypes in pupae formation and adults eclosion, and also chitin content reduction in the cuticle of abnormal larvae. The effects of RNAi on the target gene were proved to be gene-specific and effective, with the efficiency lasting for 108 hr. The results after the injection of dsRNA for EcR or 20E in vivo and the midgut culture experiments in vitro revealed that the regulation of EcR remarkably affects the expression of five genes (SeTre-1, SeG6PI, SeUAP, SeCHSA and SeCHSB) in the chitin biosynthesis pathway, and genes encoding SeTre-1, SeG6PI ,SeUAP and SeCHSB were identified as 20E late-response genes.
Results
Isolation and sequence analysis of EcR-A and EcR-B1 cDNAs
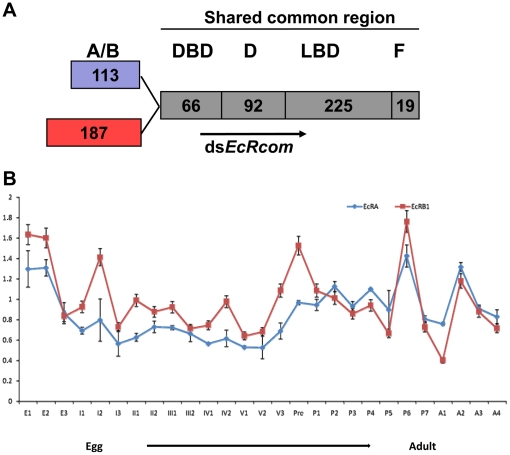
Cloning of EcR cDNAs from S. exigua was accomplished by RT-PCR using degenerate primers designed on the basis of conserved motifs in the DNA binding domain (DBD) and ligand binding domain (LBD) of insect EcR sequences. To subclone, cDNA from the first instar larvae was used as a template to obtain a 527-bp PCR fragment of the shared common domain, and two full-length cDNAs were obtained with a combination of 3′-RACE and 5′-RACE methodologies. Database BLAST search revealed that these cDNAs encoded S. exigua homologs of the nuclear receptor EcR, which contain a common carboxy-terminal region with a DNA-binding and ligand-binding domains but unique amino-termini of 113 and 187 amino acids (Fig. 2A). The two cDNAs were sequenced with an open reading frame of 1546 and 1767 nucleotides, encoding a protein of 514 and 588 amino acids with a calculated molecular weight of 57.4 and 66.0 kDa, respectively.
Figure 2. The domain structures and the developmental expression pattern of the two EcR isoforms in S. exigua.
(A) The domain structures of SeEcR-A and SeEcR-B1 and the common fragment used to generate the dsRNAs. The number of amino acids of each domain is indicated. (B) The developmental expression profiles of SeEcR-A and SeEcR-B1 mRNA. Total RNAs were prepared from individuals of S. exigua collected at all developmental stages from the first day of eggs to the last day of adults. The levels of the mRNA were detected using qRT-PCR. The mRNA level of β-Acitn was used as a control to normalize for loading. The data represent the mean values ± SE of three replicates. The age of the insects is indicated in: E1, the first day of eggs; I1, the first day of the first instar larvae; pre, prepupal period; P1, the first day of pupae; A1, the first day of adults.
Hence, on the basis of the similarity in their A/B domains when aligned with other insect EcR members, one cloned cDNA encodes an EcR-A isoform with a conserved 20 amino acid sequence (specific A-box) at the C-terminal of the A/B domain of all isoform-A receptors, and the other one is a SeEcR-B1 isoform. The deduced amino acid sequences of the two isoforms showed high similarity to the reported insect EcR sequences. SeEcR-A (GenBank accession number: GU296540) is 90% identical to SeEcR-B1 (GenBank accession number: EU426551).
By quantitative real-time PCR, the developmental expression patterns of SeEcR-A and SeEcR-B1 were investigated in all developmental stages including the first day of egg to the last day of adult. The results showed that both SeEcR-A and SeEcR-B1 were continuously expressed through the whole life cycle of S.exigua in correlation with the ecdysteroid pulse. (Fig. 2B).
The efficiency of RNAi towards the two EcR isoforms
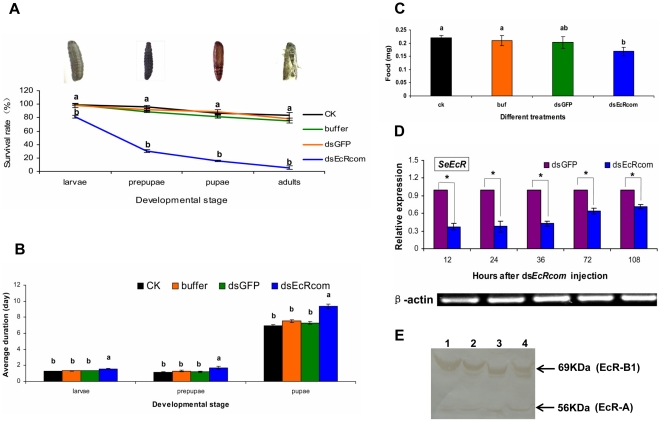
To analyze whether it was possible to reduce SeEcR levels by RNAi in vivo, a 546 bp fragment containing the coding region shared by the two EcR isoforms, named as dsEcRcom was used for dsRNA synthesis (Fig. 2A). qRT-PCR results showed that SeEcR mRNA levels clearly decreased in the dsEcRcom-injected insects as compared with those in the dsGFP-injected control (Fig. 3D). At 12 hours post-treatment, dsEcRcom injection resulted in an approximately 3-fold knock-down of the SeEcR transcript level. Moreover, in western blot analysis result, the protein level changes of the target genes showed that both SeEcR-A and SeEcR-B1 were not influenced at 12 hours after dsRNA injection, but decreased immune signals are detected in response to the dsEcRcom injection at 36 hours post-injection (Fig. 3E). Additionally, the silencing effect on the target gene lasted for as long as 108 hr, suggesting that the microinjection of dsRNA was effective at silencing the EcR genes in S. exigua.
Figure 3. The survival rates, developmental duration, food-intake and the changes of SeEcR mRNA and protein in S. exigua after SeEcR RNAi.
(A) The accumulated percentage of survival rates, (B) Developmental duration, (C) Food intake, (D) The relative expression level of SeEcR and (E) The SeEcR protein level of S.exigua larvae in different treatments. For (A, B, C), the following three controls were included: an equivalent amount of GFP dsRNA, the same volume of buffer group (DEPC water) and the CK control group (no treatment). Data are presented as mean ± SE from three independent experiments with 30 fifth-instar larvae in each group. The lowercase letter “a” at each developmental stage in Fig. 3A represents no significant difference among dsGFP, buffer and CK groups but the survival rates in all three control groups are significantly different from that in the dsEcRcom-injection group (labeled with a lowercase “b” at the same developmental stage). Different letters at the same developmental stage in Fig. 3B and Fig. 3C among different treatments indicate significant differences of either developmental duration or food-intake (p<0.05, Duncan multiple comparison test, SPSS). For (D), the expression level of SeEcR and Seβ-actin transcripts were detected using qRT-PCR. By definition, the mRNA level of the target gene was normalized to Seβ-actin in each group; and the relative expression level of the target gene was measured as the level in the dsEcRcom-injection group divided by the level in the dsGFP-injection group. Data are shown as mean ± SE from three independent experiments. An asterisk indicates significant differences in the expression level between the treated and control groups measured at the same time (p<0.05, T test). For (E), the protein levels of target genes were detected using Western blot. A total of 80 ug of proteins from each individual treated with dsGFP or dsEcRcom was applied to each lane and separated by 10% SDS-polyacrylamide gel electrophoresis. Lanes 1 and 3: 36 and 12 hours post-dsGFP injection; Lanes 2 and 4: 36 and 12 hours post- dsEcRcom injection, respectively. The results are representative of four replicates.
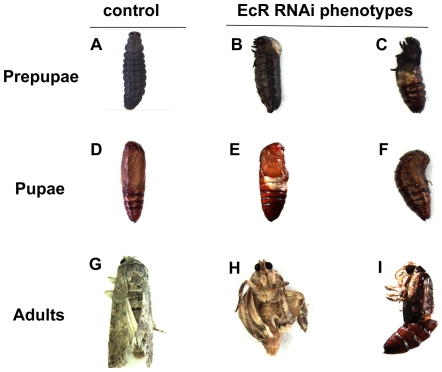
We observed severe developmental abnormity and phenotypic defects caused by the dramatic changes in the mRNA level of SeEcR by SeEcR-specific RNAi in S. exigua. Quite remarkably, following dsEcRcom injection, the average survival rates were 81.11%, 30.00%, 15.56%, and 5.56% in the fifth-instar larvae, prepupae, pupae and adults, respectively, and were significantly lower than those in the three controls (Fig. 3A). A sharp decline in survival rate was observed between larva-prepupa metamorphosis after RNAi. In the dsEcRcom-injected insect, pupation was abnormal and the pupa–adult metamorphasis could not complete, with the pupal exocuticle partially shelled, the adult in pupal cuticle arrested entrapped or the wing brokenly extended. Although the severity of the phenotype defects varied somewhat, two typical severe defect phenotypes, “half pupation” larvae and “abnormal eclosion” adults, were observed (Fig. 4B, C, H, I). These “half pupation” larvae, which were not observed in the controls, always failed to complete pupation and died within one or two days. Moreover, the dsEcRcom-injected group also had evidently longer developmental durations in the larva, prepupa and pupa phase (Fig. 3B). In contrast, no significant lethality differences were found among the three controls. In addition, many individuals injected with dsEcRcom shrank in body size and had reduced food-intake (Fig. 3C).
Figure 4. Lethal phenotypes caused by dsEcRcom injection into fifth-instar larvae.
(A, D, G) Normal prepupa, pupa and adult (B, C) Abnormal prepupae (E, F) Severe misshaped pupae, including typical “half pupation” pupa (E) and “Abnormal eclosion” adults (H, I).
Chitin levels in the epidermis and midgut/peritrophic membrane after RNAi
To address the impact of lower SeEcR transcript levels on the chitin metabolism, the chitin contents in the epidermis and the midgut/peritrophic membrane (PM) were determined. From 12 hr to 36 hr post injection of dsRNA, the chitin contents in both the midgut/PM and the epidermis decreased in both treatments of dsGFP and dsEcRcom (Table 1). At 12 hr post injection, no significant difference on chitin levels was found between treatments of dsGFP and dsEcRcom. At 36 hr post injection, however, the chitin content of the midgut/PM in the dsGFP treatment group was relatively lower than that in the dsEcRcom treatment group. Since chitin degradation starts in the midgut/PM in this stage [18], [20], the reduction in SeEcR levels may result in slow chitin degradation carried out in the midgut/PM. While in the epidermis at 36 hr post injection, both degradation of larvae epidermis chitin and generation of pupae cuticle chitin happened in prepupal stage. Lower levels of SeECR may have different effects on the degradation and the generation of the chitin, which may result in relatively higher chitin content in the dsGFP treatment. These results were also in correlation with the longer developmental duration observed in this study.
Table 1. The effect of injecting EcR dsRNAs on chitin content of different tissues in S. exigua larvae.
| Tissues | Group | Mean (µg chitin/insect) ± SE | |
| 12 hr | 36 hr | ||
| Midgut/peritrophic membrane | dsGFP | 5.7219±0.7580 a | 0.7094±0.0665 b |
| dsEcRcom | 5.6726±0.4949 a | 1.9533±0.3530 a | |
| Epidermis | dsGFP | 39.7933±2.1478 a | 33.1551±1.8894 a |
| dsEcRcom | 39.6455±2.9804 a | 29.1587±1.5632 b | |
Different letters between treatments of dsGFP and dsEcRcom in the same column indicate significant difference in the chitin contents in either the midgut or the epidermis (p<0.05, T test, n = 8).
Effects of SeEcR RNAi on the expression of key genes in the chitin biosynthesis pathway
The chitin analysis results (Table 1) suggested that SeEcR expression affected the chitin contents in the epidermis and the peritrophic membrane. Thus, six genes involved in the chitin biosynthesis pathway were examined to ascertain their responsiveness to ecdysone receptor expression using qRT-PCR.
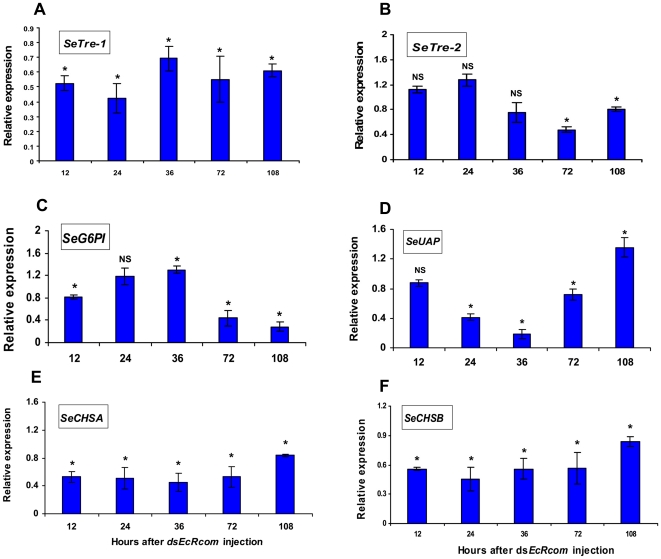
Concurrently, at 12 hr after dsEcRcom injection, the expressions of SeTre-1 (EU427311), SeCHSA (DQ062153) and SeCHSB (EU622827) were dramatically down-regulated in the dsEcRcom-injected larvae and reduced to approximately half of the mRNA levels in the control insects. Coinciding with the low expression level of SeEcR, the significantly suppressed effect on these three genes lasted for 108 hr (Fig. 5A, E and F). The SeCHSA and SeCHSB genes seem to share a similar down-regulation pattern. Meanwhile, SeTre-2, SeG6PI and SeUAP had extraordinary responses to SeEcR RNAi. The transcriptional abundance of SeG6PI (unpublished) was repressed at 12 hr after dsEcRcom injection, with a small rise at 36 hr, and then remained low until 108 hr post-injection of dsEcRcom (Fig. 5C). Quite differently, SeUAP (FJ380203) expression was slightly affected at the first testing point, and the repressed transcription level of SeUAP was only observed at 24 hr and 72 hr post-injection with a detectable but subtle increase in expression at 108 hr after dsEcRcom injection (Fig. 5D). Conversely, hardly any expressional variation of SeTre-2 (EU106080) was observed before 72hr post-injection of dsEcRcom (Fig. 5B). The qRT-PCR results revealed different transcript level changes in the six genes involved in chitin biosynthesis, which suggests that the effects of EcR on the six genes varied somewhat.
Figure 5. Changes in mRNA expression levels for key genes in the pathway following SeEcR RNAi.
The expression levels of six transcripts (SeTre-1, SeTre-2, SeG6PI, SeUAP, SeCHSA and SeCHSB) were detected by qRT-PCR at five detection points after dsEcRcom injection. Data are represented as mean ± SE from three independent experiments, which were analyzed in the same way as in Fig. 3. *: a significant difference between the dsEcRcom injection group and the dsGFP injection group (p<0.05, T test); NS: no significant difference between the two groups.
Influence of 20E on expression of SeEcR and key genes in the pathway
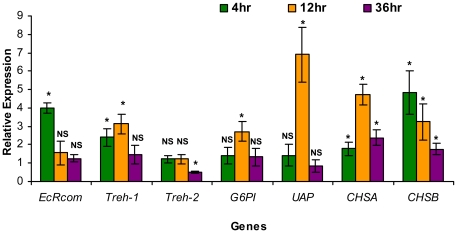
After 20E injection, the relative mRNA expressions of SeEcR and the six genes in the various groups of S. exigua larvae were assayed using qRT-PCR. The results showed that there was a burst in SeEcR transcription that increased nearly 4-fold when compared to the mRNA level in control insects at 4 hr after 20E injection, while there was no difference in the relative abundance at 12 hr and 36 hr after 20E injection as compared with the control (Fig. 6). At 12 hr post-injection of 20E, the mRNA expressions of SeTre-1, SeG6PI and SeCHSB showed about 3-fold up-regulation, SeCHSA transcript was up-regulated over 4-fold and SeUAP mRNA level was increased nearly 7-fold. Thereafter, a significantly stimulated up-regulation of the SeCHSA and SeCHSB genes were observed at 36 hr after 20E injection, synchronously coinciding with distinct expression of the SeTre-1, SeG6PI and SeUAP genes whose variation of transcript abundance were barely detectable at this detection point. On the contrary, treatment with 20E resulted in a delay in changes of the SeTre-2 expression before 36 hr post-injection of 20E. Hence, the expression of the five genes (excluding SeTre-2) continued to increase in the 20E injection group, suggesting that these five genes are probably among the ecdysone-responsive genes.
Figure 6. The influence of 20E on expression of SeEcR and key genes in the pathway.
The mRNA expression levels of SeEcR and the six chitin biosynthesis pathway genes were detected using qRT-PCR at 4 hr, 12 hr and 36 hr after injection of 20E. Data are represented as mean ± SE from Six independent experiments which were analyzed in the same way as Fig. 3. *: significant difference between the dsEcRcom injection group and the dsGFP injection group (p<0.05, T test); NS: no significant difference between the two groups.
Response of genes in the pathway to ecdysone or cycloheximide in vitro
To further determine the hierarchy position in the EcR/USP-20E-mediated genetic regulatory network, the transcript accumulation patterns of all genes (except SeCHSA, a non-midgut gene) in the pathway were investigated in cultured midguts in vitro. Incubation of the midguts in the presence of both 10−6 M 20E and 10−3 M cycloheximide (Chx,the protein synthesis inhibitor) in the medium slightly or hardly induced the following four genes: SeTre-1, SeG6PI, SeUAP and SeCHSB, a pattern quite similar to that in incubation with the same dose of Chx in the hormone-free medium as determined at p<0.05 using T test (Data not shown). As opposed to two treatments mentioned above, the four mRNA induction profiles in the cultured tissues incubated with 10−6 M 20E were siginificantly different at various times points (Fig. 7). When analyzing larval midguts cultured in the presence of 20E, the induced expression peaks of SeTre-1 and SeCHSB were detected 4 hr later, and those of SeG6PI and SeUAP could be sensed 8 hr later (Fig. 7A, B, C, D). In this in vivo midgut culture experiment, the data demonstrated that the expression of these four genes could be induced by 20E but inhibited by Chx.
Figure 7. The effect of 20E and cycloheximide on the mRNA expression level of the key genes in midgut in vitro culture.
The midguts were cultured with10−6 M 20E (20E), 10−3 M cycloheximide in the hormone-free medium (Chx), in the continuous presence of both 10−6 M 20E and10−3 M Chx (20E+Chx), or with no addition in the medium (control). The mRNA expression levels of four genes, including SeTre-1 (A), SeG6PI (B), SeUAP (C) and SeCHSB (D), were determined using qRT- PCR after incubating for 0 hr, 4 hr, 8 hr and 12 hr. The expression level of each target gene was normalized to the Seβ-actin at each time point, then the mRNA levels in the three treatment groups were normalized to the control group at each time point. The relative expression level of each target gene (y-axis) was calculated as the mRNA level at each time divided by the level at 0 hr in each group. Data are represented as mean ± SE from three independent experiments. Different letters on the broken line (20E incubation group) indicate significant differences among different culture time points in this group. (p<0.05, Duncan multiple comparison test, SPSS).
Discussion
Characteristics of S. exigua EcR isoforms and their role in developmental processes
We cloned and characterized two EcR isoforms (Fig. 2). Analysis of the deduced amino acid sequences indicated that they were two isoforms of EcR, a member of the steroid hormone receptor superfamily, homologous to the Drosophila EcR (Fig. 8). The fact that the two isoforms have distinctive N-terminal A/B domain and share a highly conserved amino acid sequence in all other domains at the C-termini suggests they are splice variants generated from the same EcR gene transcript. Ever since the Drosophila EcR gene was cloned and the three EcR isoforms (EcR-A, EcR-B1 and EcR-B2) were characterized [21], two isoforms of EcR (EcR-A and EcR-B1) were only reported in a few lepidopteron species, including Bombyx mori, Choristoneura fumiferana and Chilo suppressalis [22], [23], [24]. More evidence is needed to confirm whether three isoforms actually exist in lepidopteron insects.
Figure 8. A phylogenic tree of EcR-A and EcR-B genes in insects and other organisms.
A phylogenic tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA) software version 4.1 with the sequences obtained from GenBank. The robustness of each cluster was verified in 50,000 replicates. The scale on the x-axis represents the estimated branch lengths and numbers indicate the bootstrap values.
The regulation of EcR on insect development has been well demonstrated by mutational analysis and by using inducible expression of double stranded RNA (dsRNA) of EcR isoforms in D. melanogaster, which resulted in a block in neuronal remodeling and ecdysone responses during larval molting and metamorphosis [25], [26], [27], developmental arrest during embryogenesis and failure of pupation [28]. The results of delayed development duration and reduced food-intake, which were presented herein by using RNAi in vivo during the last larvae instar of S. exigua, indicated that SeEcR is necessary for appropriate developmental behavior of the insect. The effect of EcR deficiency on survival rate varied in different insects. D.melanogaster carrying mutations that inactivate all three isoforms of EcR are embryonically lethal [29], and EcR RNAi studies in Tribolium castaneum showed nearly 100% lethality rate in injected larvae [30]. In our study, as high as a 95% mortality prior to eclosion was observed (Fig. 3A). In addition, the SeEcR knockdown larvae show defective phenotypes, including the typical “half pupation” phenotype (Fig. 4E), which resembles misshaped structures that were observed after the ingestion of dsRNA for SeCHSA [31].
The effect of RNAi on the target gene SeEcR in this study was effective (Fig. 3). To confirm that the observed RNAi effect is not due to an off-target effect, a second different SeEcR dsRNA fragment (362 bp), designed from the DBD (DNA binding domain) domain and the D (hinge) domain of two EcR isoforms, was used for injection. Similar results were observed in survival rates (such as sharp decline during the larvae to prepupae period) and in the typical phenotypes (e.g. half pupation and abnormal eclosion) (Data not shown). This proved that the RNAi of SeEcR is gene-specific.
The five genes (except SeTre-2) in the chitin biosynthesis pathway are regulated by SeEcR
EcR acts as an ecdysteroid-inducible transcription factor by directly interacting with USP via the DNA-response elements in the promoters of target genes [32], [33]. Apparently, the EcR plays a vital role in the transcription of multifarious genes [34], [35]. However, the role of the EcR in controlling the transcription of the genes responsible for the key enzymes involved in the chitin biosynthesis processes remains enigmatic. Measurement of the transcript levels following dsEcRcom injection allowed us to identify distinct gene-specific responses of SeTre-1, SeTre-2, SeG6PI, SeUAP, SeCHSA and SeCHSB.
Most likely, SeEcR continuously regulated the transcription of SeTre-1, SeCHSA and SeCHSB (Fig. 5A, E and F). In the bamboo borer Omphisa fuscidentalis, expression of EcR-A and EcR-B1 was followed by a decrease in trehalose concentration and an increase in trehalase activity [36]. The CHSA and CHSB mRNA levels could be up-regulated in a stage-specific manner during the onset of metamorphosis in D. melanogaster [18]. Whereas, the change of SeG6PI and SeUAP transcripts in S. exigua were more complicated (Fig. 5C and D). The undisturbed mRNA levels of SeG6PI at 24 hr and an unexpectedly increased expression of SeUAP at 108 hr after dsEcRcom injection led to the hypothesis that catalyzed products of G6PI or UAP might be potential feedback inhibitory substances in the pathway. It is not suprising that the role of UAP and G6PI might be far more complex than we expected [37]. Both our studies (Figs. 5B and 6) and those of Tatun et al. revealed 20E and EcR hardly had effect on a transmembrane enzyme gene (Tre-2) at early detection point, which substantially dued to the uninvolvement of Tre-2 in the dynamic changes in the hemolymph trehalose concentration that occurred during the larval–pupal transformation [38]. In short, the loss of SeEcR results in not only reduced abundance of the five genes transcripts responsible for the enzymatic reactions in chitin biosynthesis pathway, but also obviously suppressed chitin metabolism (Table 1) in a stage- and tissue-specific response to insect hormone during insect metamorphosis.
The four genes in the pathway are ecdysone-regulated late genes
Ecdysone has been shown to negatively regulate transcription of pupal cuticle protein genes in imaginal disks and is required for apical extracellular matrix formation and epithelial morphogenesis in D. melanogaster [39]. In an in vitro system of lepidopteron imaginal wing discs, ecdysone had been reported to stimulate the incorporation of N-acetyl-D-glucosamine into glycoproteins, and upregulate the expression of adult cuticular genes in cell lines derived from Tenebrio molitor [40]. This data provides important clues in regards to an intriguing extension of the ecdysone-mediated genetic regulatory network, and the question as to whether the five genes in the chitin biosynthesis pathway are among the ecdysone-induced genes arises.
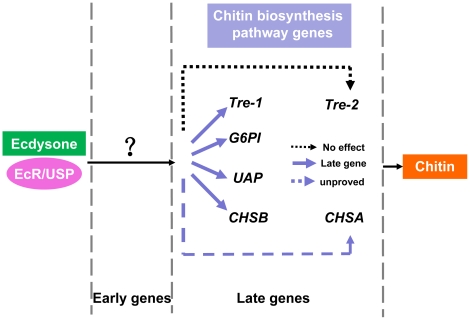
Here in our study, significant up-regulation of SeCHSA and SeCHSB was detectable at 4 hr and durative to 36 hr after 20E injection into the larvae of S. exigua, and high levels of SeTre-1, SeG6PI and SeUAP expression were detected at 12 hr post-injection of 20E. This implies that the expression of the five genes are induced by 20E (Fig. 7). In agreement with our present results, previous in vivo studies had demonstrated that hemolymph trehalose concentration was sensitive to 20E level in Omphisa fuscidentalis [41]. In vitro up-regulation of CHSA and CHSB was due to an indirect interaction between the ecdysone receptor complex and ecdysone-responsive elements within the CHSA and CHSB promoters in D. melanogaster and these two genes act as members of the late genes [18]. The expressiong of the mmy gene encoding UDP-Nacetylglucosamine pyrophosphorylase (UAP) in Drosophila could be altered by the moulting hormone 20-Hydroxyecdysone [42]. Discriminatively, SeTre-2 seems insensitive to 20E, which was more similar to the ecdysone-independent genes. Taken together, we speculate that the five genes SeTre-1, SeTre-2, SeG6PI, SeUAP, SeCHSA and SeCHSB excluding SeTre-2 are among the ecdysone-induced genes hierarchies (Fig. 9).
Figure 9. Summary of the 20-hydroxyecdysone late-response genes in the chitin biosynthesis pathway in S.exigua.
See text for details.
Different 20E-responsive genes seem to be differentially regulated by ecdysone. This has been experimentally demonstrated by the in vitro culture experiments in larval salivary glands of D.melanogaster [2], [8], [9]. When there is a continuouse high level of 20E, early genes are activated with a quick response, reach their maximum expression and then regress within 4 hours. Due to the inhibition by cycloheximide (Chx) on their induced expression and a delayed reaction when incubating with ecdysone, the 20E late-response genes could be classified at a lower hierarchy position [1], [5], [43], [44]. In the in vitro cultured midguts, the four genes (SeTre-1, SeG6PI, SeUAP and SeCHSB) showed a slow peak in activation by 20E at least 4 hr after incubation, but no continuous increase in their mRNA expression was detected during the 12 hours when cultured in the presence of both Chx and 20E (Fig. 7). This response of gene transcription to 20E and Chx in this manner is characteristic of the “late” genes analogous to genes of the ecdysone regulatory hierarchy as described by Ashburner and his colleagues in D. melanogaster (Fig. 9).
Further studies to directly identify the relationship between the expression of the chitin biosynthesis interrelated genes and the hormone governing the stages of insect metamorphosis is currently in progress in our laboratory, and the regulation of other 20E early-response genes is required to identify in order to be verify for this connection.
Materials and Methods
Insects
S.exigua were reared in the laboratory at 25°C±2°C and 75%±5% r.h. (relative humidity) on a 14L:10D photoperiod using an artificial diet [45]. All dissections and tissue sampling of specimens were carried out in ice-cold saline and stored at −80°C for later use.
RNA isolation and cDNA synthesis
Total RNA was extracted from individual specimens of S. exigua using Trizol reagent (Invitrogen, USA). Total RNA (5 µg) was used for synthesizing cDNA with the AMV reverse transcriptase XL (Takara, Japan) with the following reaction conditions: 37°C for 10 min, 42°C for 1 h, 99°C for 5 min, and 5°C for 5 min [46].
Cloning and analysis of SeEcR cDNAs
The ABD SMART RACE cDNA amplification kit (BD Bioscience Clontech, CA, USA) was used to obtain the full-length SeEcR cDNA. To clone the S. exigua EcR gene, degenerate primers based on the conserved amino acids sequences of “PRQQEE” (forward primer, SeEcR-F) and “QPSXE” (reverse primer, SeEcR-R) were used to obtain a common fragment of EcR (Table 2). The touch-down PCR amplification reactions were carried out as follows: 94°C for 5 min, 5 cycles of 94°C for 40 s, 48°C for 40 s and 72°C for 40 s, 25 cycles of 94°C for 40 s, 54°C for 40 s and 72°C for 40 s, and with an extension step at 72°C for 10 min at the end. The amplified product was then separated on an agarose gel and purified using the Gel Extraction Kit (OMEGA, USA). The amplified fragment was subcloned into the pMD-18T vector (Takara, Japan) and sequenced. To get a longer cDNA sequence, we performed 3′-RACE and 5′-RACE using nested gene-specific primers for SeEcR (Table 2) and anchor primers supplied in a SMART RACE cDNA Amplification Kit (Clontech).
Table 2. Primers used in this study.
| Information about the primers | |||
| For dsRNA | |||
| Name of primers | Nucleotide sequence (5′–3′) | ||
| dsEcR-F* | GGATCCTAATACGACTCACTATAGG GTGTCGGTTGAAGAAATGTCT | ||
| dsEcR-R* | GGATCCTAATACGACTCACTATAGG CGCAACATCATCACCTCACT | ||
| dsEcR-SF | GTGTCGGTTGAAGAAATGTCT | ||
| dsEcR-SR | CGCAACATCATCACCTCACT | ||
| dsGFP-F* | GGATCCTAATACGACTCACTATAGG AAGGGCGAGGAGCTGTTCACCG | ||
| dsGFP-R* | GGATCCTAATACGACTCACTATAGG CAGCAGGACCATGTGATCGCGC | ||
| dsGFP-SF | AAGGGCGAGGAGCTGTTCACCG | ||
| dsGFP-SR | CAGCAGGACCATGTGATCGCGC | ||
| For SeEcR cloning | |||
| Name of primers | Degenerate primers (5′–3′) | ||
| SeEcR-F | CCNCGGCARCARGAGGAGC | ||
| SeEcR-R | TCCTCBTCNGANGGCTG | ||
| Nested PCR primers (5′–3′) | |||
| 5-SeEcR-1 | GAATCGTGGCACCACCTCGTGAAT | ||
| 5-SeEcR-2 | GAGGCGGTGGATCACACTGCAT | ||
| 3-SeEcR-1 | GTGTCGGTTGAAGAAATGTCT | ||
| 3-SeEcR-2 | ATGCAGTGTGATCCACCGCCTC | ||
| For quantitative real-time RT-PCR | |||
| Gene | Forward (5′–3′) | Reverse (5′–3′) | |
| SeEcRcom | GTGAGGTGATGATGTTGCGAGTA | AGAAAATGACGATGGCAGTGAG | |
| SeTre-1 | GGTTGGGACTTCTCAACGCGC | GACAGCATCCCGGTGATTCCC | |
| SeTre-2 | GGCAGGCGTCGGGATTACTTC | GGCCAGGCATTCGGATAGTCC | |
| SeG6PI | ATCAGTGGACAGTGGAAAGGGTA | TTGAGGTGGTTGGCGTAAGG | |
| SeUAP | CCTGATGGCAACGGAGGACTG | CAACTTTCGCCGCACAGTCAG | |
| SeCHSA | GGCTCGGGTCCTCATAACGTC | GTACCTGGTCCATCGTTGGCG | |
| SeCHSB | GCTCGGTTCTGCGGTTGTGTC | CCATAGCCACGTCACACGCTC | |
* The sequence in bond at the 5′ end of the primer is T7 promoter sequence.
On the National Center for Biotechnology Information (NCBI) website, the sequence of the two EcR cDNAs were compared with the other EcR sequences deposited in the GenBank using the “BLAST-X” tool. Amino acid sequences were deduced from the corresponding cDNA sequences using the translation tool at the ExPASy Proteomics website (http://expasy.org/tools/dna.html). A protein sequence analysis tool from the ExPASy Proteomics website (http://expasy.org/) was used to predict molecular weight. Multiple sequence alignments of the deduced amino acid sequences were made using Multiple Alignment software (http://www.ebi.ac.uk/clustalw/index.html) [47]. Phylogenic and evolutionary analyses were conducted using Molecular Evolutionary Genetics Analysis (MEGA) software version 4.1.
dsRNA synthesis
A 546 bp fragment from the EcR common region (Fig. 2A) and a 714 bp fragment from GFP (ACY56286) were PCR amplified. dsRNA synthesis was performed using the T7 RiboMAX™ Express RNAi System Kit (Promega, USA) according to the manufacturer's instructions. Primers for EcR and GFP were designed by adding the T7 polymerase promoter sequence to their 5′ends (Table 2). The dsRNA was resuspended in an appropriate amount of nuclease-free water (DEPC water). The purified dsRNAs were quantified by spectroscopy and examined by agarose gel electrophoresis to ensure their integrity.
RNA interference bioassays and sampling
In vivo RNAi in S. exigua larvae was performed as previously described [48]. The 2nd day of fifth-instar (the last instar) larvae were anaesthetized on ice for 2–3 min and used for the injection experiment. Five micrograms of dsRNA dissolved in 5 µl of DEPC water were injected into larvae between the second and the third abdominal segment using 10 µl microliter syringes (Hamilton), and the injection point was sealed immediately with wax. Both the dsRNA of GFP and an equivalent volume of buffer were used as controls.
For phenotype observation, feeding and developmental duration experiments, each group contained 30 individual larvae in each set of repetition and three replicates were used. Observation was performed every 12 hours after injection. The following three controls were included: the dsGFP group (an equivalent amount of GFP dsRNA), the buffer group (the same volume of DEPC water) and the CK group (control check with no treatment). The artificial diet pellets were cut into appropriate analogous sizes. All diets were replaced daily and each piece of food was weighed on an accurate electronic scale to measure the food intake of each individual insect. The results are expressed as the mean±standard errors (SE) of three independent replicates (n = 3), each representing 30 fifth-instar larvae. The statistical significance of data from the survival rates, food-intake and developmental duration were determined by one-way analysis of variance (ANOVA) and analyzed by Duncan multiple comparison test. Percentage data were arcsine square root transformed prior to ANOVA. For chitin content assay, each group included 50 individuals and 8 abnormal larvae were chosen for analysis (n = 8). The cuticle or midgut of individual misshapen specimens was disesscted as the chitin analysis sample. For the target gene detection experiment, each group included 60 individuals with three replicates, and the SeEcR mRNA levels of randomly chosen individual samples were measured at 12 hr, 24 hr, 36 hr, 72 hr and 108 hr after the injection of SeEcR dsRNA. For the pathway gene detection assay, each treatment included 60 individuals with three replicates and three misshapen samples were selected at 12 hr, 24 hr, 36 hr, 72 hr and 108 hr post-injection for the independent detection.
Western blot analysis
Protein samples were made by homogenizing 1 larva per 150 µl of PBS (20 mM sodium phosphate and 130 mM NaCl, pH 7.4) plus protease inhibitor cocktail Phenylmethyl sulfonylfluoride (PMSF). After freeze thawing for three times, centrifuged at 12,000 g, 4°C for 30 min. The protein content in supernatant was determined by BCA kit, and the supernatant containing equivalent protein was mixed with 5×loading buffer for SDS polyacrylamide gel electrophoresis and boiled for 10 minutes, After centrifugation at 12,000 g for 5 min, protein samples were cooled to room temperature before loading on a 10% polyacrylamide SDS gel. The process of western blotting were modified from the method previously described [49]. A purified primary antibody (anti-SeEcRcom antibody, 1∶750 dilution) and an lgG goat anti-rabbit antibody conjugated with HRP was used for secondary antibody (BOSTER, 1∶5000 dilution).
In vivo ecdysteroid injection
For the in vivo ecdysteroid injection experiment, 20E (Sigma, St. Louis, MO, USA) was dissolved in 5 µl of DEPC water at doses of 2 µg per insect, and each group included 30 individuals with six replicates. Three individual larvae were randomly chosen for the experiment 4 hr, 12 hr and 36 hr after injection.
In vitro midgut culture experiments
The 2nd day of fifth-instar larvae were dissected in saline. The midguts together with the peritrophic membranes were ligated tightly using cotton thread at the anterior and posterior ends, and then cut to the ligatures. After the isolated midguts were rinsed four times with medium, they were independently cultured in Grace's Insect Tissue Culture Medium at 25°C±2°C for 12 hr.
Four groups were included in this culture experiments: the CK control group (control check group with no addictive in culture medium); the 20E group (added 20E hormone to final concentration of 10−6 M); the Chx group (added cycloheximide to a final concentration of 10−3 M); the 20E+Chx group (added 20E to final concentration 10−6 M and cycloheximide to 10−3 M). Each group included 10 individual tissues with three replicates. Samples for mRNA expression analysis were selected randomly at 0 hr, 4 hr, 8 hr and 12 hr after incubation.
Quantitative real-time RT-PCR
The relative mRNA expression of genes was assessed by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) by using the SYBR Green based detection system (SYBR Premix Ex Taq, Takara, Japan) with LightCycler480 system (Roche, Germany). This assay allows for precise quantification of weakly expressed genes. The oligonucleotides used in qRT-PCR are indicated in Table 2. Specificity of the reaction was checked by analysis of the melting curve of the final amplified product. The housekeeping gene and target gene from each sample were run in parallel on the same PCR plate. All of the qRT-PCR reactions were performed using the following common program: pre-incubation at 95°C for 10 s, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 20 s, and elongation at 72°C for 15 s. At the end of each cycle, a fluorescence reading were used to determined the extent of amplification. Standard curves were obtained using a 10-fold serial dilution of pooled cDNA from all stages.
As described above, cDNA was synthesized from specimens of S. exigua from experimental group and the GFP dsRNA group at every detection point using the AMV reverse transcriptase XL (Takara, Japan). The qRT-PCR measurement was performed in triplicate and normalized to an internal control for each sample (the S. exigua β-actin gene AY507963, Table 2). The relative expression level of the target gene was calculated by the mRNA level in the treated group divided by the level in the control group.
Chitin analysis
A modified analysis procedure was used in the chitin content measurement experiment as previously described [50]. For chitinase digestion, each pellet was resuspended in 250 µl of McIlvaine's buffer, and Streptomyces plicatus chitinase (Sigma, 5 mg/ml in PBS, 50 µg for cuticle and 25 µg for midgut) was added and incubated for 72 h at 37°C to hydolyse chitin to GlcNAc. Standard curves were generated for data processing.
Acknowledgments
We are grateful to Dr. Sheng Li (Institute of Plant Physiology and Ecology, Shanghai, China), Dr. Jian Wang (University of Maryland at Colleage Park, USA) and Dr. Xiaoqiang Yu (University of Missouri-Kansas City, USA) for their suggestions and reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Natural Science Foundation of China (30871634), National Basic Research Program of China (2006CB102001) and Science Foundation of the State Key Laboratory of Biocontrol (SKLBC 10A 01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev Biol. 1974;39:141–157. doi: 10.1016/s0012-1606(74)80016-3. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M, Richards G. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster, III. Consequences of ecdysone withdrawal. Developmental Biology. 1976;54:241–255. doi: 10.1016/0012-1606(76)90302-x. [DOI] [PubMed] [Google Scholar]
- 3.Dubrovsky EB. Hormonal cross talk in insect development. Trends Endocrinol Metab. 2005;16:6–11. doi: 10.1016/j.tem.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Kapitskaya MZ, Li C, Miura K, Segraves W, Raikhel AS. Expression of the early-late gene encoding the nuclear receptor HR3 suggests its involvement in regulating the vitellogenic response to ecdysone in the adult mosquito. Mol Cell Endocrinol. 2000;160:25–37. doi: 10.1016/s0303-7207(99)00253-1. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner M. Puffs, genes, and hormones revisited. Cell. 1990;61:1–3. doi: 10.1016/0092-8674(90)90205-s. [DOI] [PubMed] [Google Scholar]
- 6.Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- 7.Huet F, Ruiz C, Richards G. Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development. 1995;121:1195–1204. doi: 10.1242/dev.121.4.1195. [DOI] [PubMed] [Google Scholar]
- 8.Gariou-Papalexiou A, Mintzas AC, Zacharopoulou A. Ecdysone-regulated chromosome puffing in the salivary glands of the Mediterranean fruit fly, Ceratitis capitata. Genome. 2001;44:752–762. doi: 10.1139/g01-068. [DOI] [PubMed] [Google Scholar]
- 9.Thummel CS. Ecdysone-regulated puff genes 2000. Insect Biochemistry and Molecular Biology. 2002;32:113–120. doi: 10.1016/s0965-1748(01)00112-6. [DOI] [PubMed] [Google Scholar]
- 10.Andres AJ, Thummel CS. The Drosophila 63F early puff contains E63-1, an ecdysone-inducible gene that encodes a novel Ca2+-binding protein. Development. 1995;121:2667–2679. doi: 10.1242/dev.121.8.2667. [DOI] [PubMed] [Google Scholar]
- 11.Andres AJ, Thummel CS. Hormones, puffs and flies: the molecular control of metamorphosis by ecdysone. Trends Genet. 1992;8:132–138. doi: 10.1016/0168-9525(92)90371-A. [DOI] [PubMed] [Google Scholar]
- 12.Huet F, Ruiz C, Richards G. Puffs and PCR: the in vivo dynamics of early gene expression during ecdysone responses in Drosophila. Development. 1993;118:613–627. doi: 10.1242/dev.118.2.613. [DOI] [PubMed] [Google Scholar]
- 13.Stowers RS, Garza D, Rascle A, Hogness DS. The L63 Gene Is Necessary for the Ecdysone-Induced 63E Late Puff and Encodes CDK Proteins Required for Drosophila Development. Developmental Biology. 2000;221:23–40. doi: 10.1006/dbio.2000.9685. [DOI] [PubMed] [Google Scholar]
- 14.Wright LG, Chen T, Thummel CS, Guild GM. Molecular characterization of the 71E late puff in Drosophila melanogaster reveals a family of novel genes. J Mol Biol. 1996;255:387–400. doi: 10.1006/jmbi.1996.0032. [DOI] [PubMed] [Google Scholar]
- 15.Stowers RS, Russell S, Garza D. The 82F late puff contains the L82 gene, an essential member of a novel gene family. Dev Biol. 1999;213:116–130. doi: 10.1006/dbio.1999.9358. [DOI] [PubMed] [Google Scholar]
- 16.Kato N, Mueller CR, Fuchs JF, Wessely V, Lan Q, et al. Regulatory mechanisms of chitin biosynthesis and roles of chitin in peritrophic matrix formation in the midgut of adult Aedes aegypti. Insect Biochemistry and Molecular Biology. 2006;36:1–9. doi: 10.1016/j.ibmb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Binnington KC. Ultrastructural changes in the cuticle of the sheep blowfly, Lucilia, induced by certain insecticides and biological inhibitors. Tissue Cell. 1985;17:131–140. doi: 10.1016/0040-8166(85)90021-7. [DOI] [PubMed] [Google Scholar]
- 18.Gagou ME, Kapsetaki M, Turberg A, Kafetzopoulos D. Stage-specific expression of the chitin synthase DmeChSA and DmeChSB genes during the onset of Drosophila metamorphosis. Insect Biochemistry and Molecular Biology. 2002;32:141–146. doi: 10.1016/s0965-1748(01)00101-1. [DOI] [PubMed] [Google Scholar]
- 19.Braquart C, Bouhin H, Quennedey A, Delachambre J. Up-regulation of an adult cuticular gene by 20-hydroxyecdysone in insect metamorphosing epidermis cultured in vitro. Eur J Biochem. 1996;240:336–341. doi: 10.1111/j.1432-1033.1996.0336h.x. [DOI] [PubMed] [Google Scholar]
- 20.Zimoch L, Hogenkamp DG, Kramer KJ, Muthukrishnan S, Merzendorfer H. Regulation of chitin synthesis in the larval midgut of Manduca sexta. Insect Biochemistry and Molecular Biology. 2005;35:515–527. doi: 10.1016/j.ibmb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Koelle M, Talbot W, Segraves W, Bender M, Cherbas P, et al. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 22.Minakuchi C, Nakagawa Y, Kiuchi M, Tomita S, Kamimura M. Molecular cloning, expression analysis and functional confirmation of two ecdysone receptor isoforms from the rice stem borer Chilo suppressalis. Insect Biochem Mol Biol. 2002;32:999–1008. doi: 10.1016/s0965-1748(02)00036-x. [DOI] [PubMed] [Google Scholar]
- 23.Perera SC, Ladd TR, Dhadialla TS, Krell PJ, Sohi SS, et al. Studies on two ecdysone receptor isoforms of the spruce budworm, Choristoneura fumiferana. Mol Cell Endocrinol. 1999;152:73–84. doi: 10.1016/s0303-7207(99)00058-1. [DOI] [PubMed] [Google Scholar]
- 24.Kamimura M, Tomita S, Kiuchi M, Fujiwara H. Tissue-specific and stage-specific expression of two silkworm ecdysone receptor isoforms — ecdysteroid-dependent transcription in cultured anterior silk glands. Eur J Biochem. 1997;248:786–793. doi: 10.1111/j.1432-1033.1997.t01-1-00786.x. [DOI] [PubMed] [Google Scholar]
- 25.Lam G, Thummel CS. Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr Biol. 2000;10:957–963. doi: 10.1016/s0960-9822(00)00631-x. [DOI] [PubMed] [Google Scholar]
- 26.Davis MB, Carney GE, Robertson AE, Bender M. Phenotypic analysis of EcR-A mutants suggests that EcR isoforms have unique functions during Drosophila development. Developmental Biology. 2005;282:385–396. doi: 10.1016/j.ydbio.2005.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schubiger M, Wade AA, Carney GE, Truman JW, Bender M. Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development. 1998;125:2053–2062. doi: 10.1242/dev.125.11.2053. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Bender M. A conditional rescue system reveals essential functions for the ecdysone receptor (EcR) gene during molting and metamorphosis in Drosophila. Development. 2000;127:2897–2905. doi: 10.1242/dev.127.13.2897. [DOI] [PubMed] [Google Scholar]
- 29.Bender M, Imam FB, Talbot WS, Ganetzky B, Hogness DS. Drosophila Ecdysone Receptor Mutations Reveal Functional Differences among Receptor Isoforms. Cell. 1997;91:777–788. doi: 10.1016/s0092-8674(00)80466-3. [DOI] [PubMed] [Google Scholar]
- 30.Tan A, Palli SR. Ecdysone [corrected] receptor isoforms play distinct roles in controlling molting and metamorphosis in the red flour beetle, Tribolium castaneum. Mol Cell Endocrinol. 2008;291:42–49. doi: 10.1016/j.mce.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian H, Peng H, Yao Q, Chen H, Xie Q, et al. Developmental Control of a Lepidopteran Pest Spodoptera exigua by Ingestion of Bacteria Expressing dsRNA of a Non-Midgut Gene. PLoS ONE. 2009;4:e6225. doi: 10.1371/journal.pone.0006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao T, Segraves W, Oro A, McKeown M, Evans R. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 33.Yao T, Forman B, Jiang Z, Cherbas L, Chen J, et al. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 34.Martin D, Wang SF, Raikhel AS. The vitellogenin gene of the mosquito Aedes aegypti is a direct target of ecdysteroid receptor. Mol Cell Endocrinol. 2001;173:75–86. doi: 10.1016/s0303-7207(00)00413-5. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto KR, Alberts BM. Steroid Receptors: Elements for Modulation of Eukaryotic Transcription. Annual Review of Biochemistry. 1976;45:721–746. doi: 10.1146/annurev.bi.45.070176.003445. [DOI] [PubMed] [Google Scholar]
- 36.Tatun N, Singtripop T, Sakurai S. Dual control of midgut trehalase activity by 20-hydroxyecdysone and an inhibitory factor in the bamboo borer Omphisa fuscidentalis Hampson. Journal of Insect Physiology. 2008;54:351–357. doi: 10.1016/j.jinsphys.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Zimoch L, Hogenkamp DG, Kramer KJ, Muthukrishnan S, Merzendorfer H. Regulation of chitin synthesis in the larval midgut of Manduca sexta. Insect Biochem Mol Biol. 2005;35:515–527. doi: 10.1016/j.ibmb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 38.Tatun N, Singtripop T, Tungjitwitayakul J, Sakurai S. Regulation of soluble and membrane-bound trehalase activity and expression of the enzyme in the larval midgut of the bamboo borer Omphisa fuscidentalis. Insect Biochem Mol Biol. 2008;38:788–795. doi: 10.1016/j.ibmb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Apple RT, Fristrom JW. 20-Hydroxyecdysone is required for, and negatively regulates, transcription of Drosophila pupal cuticle protein genes. Developmental Biology. 1991;146:569–582. doi: 10.1016/0012-1606(91)90257-4. [DOI] [PubMed] [Google Scholar]
- 40.Porcheron P, Moriniere M, Coudouel N, Oberlander H. Ecdysteroid-stimulated synthesis and secretion of an N-acetyl-D-glucosamine-rich glycopeptide in a lepidopteran cell line derived from imaginal discs. Arch Insect Biochem Physiol. 1991;16:257–271. doi: 10.1002/arch.940160405. [DOI] [PubMed] [Google Scholar]
- 41.Singtripop T, Oda Y, Wanichacheewa S, Sakurai S. Sensitivities to juvenile hormone and ecdysteroid in the diapause larvae of Omphisa fuscidentalis based on the hemolymph trehalose dynamics index. Journal of Insect Physiology. 2002;48:817–824. doi: 10.1016/s0022-1910(02)00104-x. [DOI] [PubMed] [Google Scholar]
- 42.Tonning A, Helms S, Schwarz H, Uv AE, Moussian B. Hormonal regulation of mummy is needed for apical extracellular matrix formation and epithelial morphogenesis in Drosophila. Development. 2006;133:331–341. doi: 10.1242/dev.02206. [DOI] [PubMed] [Google Scholar]
- 43.Basso LR, Jr, Vasconcelos C, Fontes AM, Hartfelder K, Silva JA, Jr, et al. The induction of DNA puff BhC4-1 gene is a late response to the increase in 20-hydroxyecdysone titers in last instar dipteran larvae. Mech Dev. 2002;110:15–26. doi: 10.1016/s0925-4773(01)00589-5. [DOI] [PubMed] [Google Scholar]
- 44.Basso LR, Jr, Neves MdC, Monesi N, Paçó-Larson ML. Broad-Complex, E74, and E75 early genes control DNA puff BhC4-1 expression in prepupal salivary glands. genesis. 2006;44:505–514. doi: 10.1002/dvg.20239. [DOI] [PubMed] [Google Scholar]
- 45.Chen X, Yang X, Senthil Kumar N, Tang B, Sun X, et al. The class A chitin synthase gene of Spodoptera exigua: molecular cloning and expression patterns. Insect Biochem Mol Biol. 2007;37:409–417. doi: 10.1016/j.ibmb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Kumar NS, Tang B, Chen X, Tian H, Zhang W. Molecular cloning, expression pattern and comparative analysis of chitin synthase gene B in Spodoptera exigua. Comp Biochem Physiol B Biochem Mol Biol. 2008;149:447–453. doi: 10.1016/j.cbpb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Tang B, Chen X, Liu Y, Tian H, Liu J, et al. Characterization and expression patterns of a membrane-bound trehalase from Spodoptera exigua. BMC Molecular Biology. 2008;9:51. doi: 10.1186/1471-2199-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Tian H, Zou L, Tang B, Hu J, et al. Disruption of Spodoptera exigua larval development by silencing chitin synthase gene A with RNA interference. Bull Entomol Res. 2008;98:613–619. doi: 10.1017/S0007485308005932. [DOI] [PubMed] [Google Scholar]
- 49.Song Q, Alnemri ES, Litwack G, Gilbert LI. An Immunophilin is a Component of the Insect Ecdysone Receptor (EcR) Complex. Insect Biochemistry and Molecular Biology. 1998;27:973–982. doi: 10.1016/s0965-1748(97)00080-5. [DOI] [PubMed] [Google Scholar]
- 50.Arakane Y, Muthukrishnan S, Kramer KJ, Specht CA, Tomoyasu Y, et al. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol Biol. 2005;14:453–463. doi: 10.1111/j.1365-2583.2005.00576.x. [DOI] [PubMed] [Google Scholar]