Abstract
High-resolution magnetic resonance microscopy (MRM) was used to determine regional brain volumetric changes in a mouse model of Alzheimer's disease. These transgenic (Tg) mice overexpress human mutant amyloid precursor protein (APP) V717F under control of platelet-derived growth factor promoter (PDAPP mice), and cortical and hippocampal β-amyloid (Aβ) deposits accumulate in heterozygotes after 8–10 mos. We used MRM to obtain 3D volumetric data on mouse brains imaged in their skulls to define genotype- and age-related changes. Hippocampal, cerebellar, and brain volumes and corpus callosum length were quantified in 40-, 100-, 365-, and 630-day-old mice. Measurements taken at age 100 days, before Aβ deposition, revealed a 12.3% reduction of hippocampus volume in Tg mice compared with WT controls. This reduction persisted without progression to age 21 mos. A significant 18% increase in hippocampal volume occurred between 40 and 630 days in WT mice, and no corresponding significant increase occurred in Tg mice. Cavalieri volume estimates of hippocampal subfields from 100-day-old Tg mice further localized a 28% volume deficit in the dentate gyrus. In addition, corpus callosum length was reduced by ≈25% in Tg mice at all ages analyzed. In summary, reduced hippocampal volume and corpus callosum length can be detected by MRM before Aβ deposition. We conclude that overexpression of APP and amyloid may initiate pathologic changes before the appearance of plaques, suggesting novel targets for the treatment of Alzheimer's disease and further reinforcing the need for early diagnosis and treatment.
The essential neuropathologic features of Alzheimer's disease (AD) include the progressive deposition of amyloid plaques and neurofibrillary tangles in neocortical and hippocampal structures and a parallel global decrease in cortical volume (1, 2). Extensive data from analysis of postmortem human brain and mouse models of AD associate the neuropathology of AD with alterations in the expression, distribution, and deposition of β amyloid protein (Aβ). In AD brains, Aβ levels are increased, and the protein can be found in fibrillar chains within compact plaques, aggregated in diffuse plaques, or as oligomers and monomers in regions outside of plaques (3–6). Some of the known human mutations associated with AD affect the processing or cleavage of amyloid precursor protein (APP) and can cause increased Aβ levels or increase the relative amount of the primary plaque component Aβ1–42 compared with Aβ1–40 (3, 5–10). Other human mutations have been identified within the coding region of Aβ1–42, which can increase Aβ neurotoxicity (11). Transgenic (Tg) mouse models have shown that high levels of Aβ can cause AD-like amyloid plaque pathology. Tg mice that overexpress APP have some but not all deficits observed in AD (12, 13). Other Tg mouse models that have mutations associated with AD resulting in up-regulation of Aβ production have many, but not all, of the observed deficits seen in AD, including reduced hippocampal volume, reduced synapse densities, plaque formation, and cognitive deficits (14–21).
In one mouse model of familial early-onset AD, mice express the human V717F mutation (22) in the APP gene under the control of the platelet-derived growth factor-β promoter (PDAPP mice) (15). In heterozygous mice, Aβ-containing plaques begin to accumulate after 8–10 mos. Hippocampal synaptophysin and mitogen-activated protein 2 immunoreactivity are reduced at this time point and beyond (15, 16). Aβ-containing plaques accumulate substantially in the hippocampus and neocortex of aged 18- to 20-mo-old mice, corresponding to behavioral deficits associated with the memory loss seen in such aged mice (23). Passive and active vaccination with anti-Aβ antibodies has been reported to reduce tissue Aβ levels (24), demonstrating the potential for testing therapeutic strategies for AD with this model. However, there is currently insufficient rigorous quantitative data from PDAPP ± (Tg) mice to demonstrate whether additional therapeutic strategies may be advantageous. Specifically, we sought to define the earliest time point at which volumetric alterations could be detected, and whether such changes were progressive.
We initially hypothesized that a hippocampal volume deficit would be detected by magnetic resonance microscopy (MRM) in aged 21-mo-old Tg mice, corresponding to a time when plaque accumulation and behavioral deficits are maximally evident. We anticipated that such hippocampal volume changes would not be evident in younger 40- or 100-day-old Tg mice, corresponding to time points before significant plaque deposition.
To obtain accurate estimates of volumes of intracerebral structures, we used MRM to acquire high-resolution 3D digital datasets with which to identify and quantify the volume of defined brain regions without the distortion associated with brain removal and sectioning. The MRM technique has not been previously used to quantify volume changes in mouse models of AD. Accordingly, we applied MRM to determine the volumes of the hippocampus, brain, and cerebellum of Tg mice before removal of the brain from the skull at four developmental ages: 40 days, 100 days, 12 mos (365 days), and 21 mos (630 days), in comparison to similar measurements in WT mice of the same strain.
Materials and Methods
Mice.
Heterozygous male PDAPP and WT littermate mice (Taconic Farms) were used for these studies. Twelve 40-day-old (six Tg, six WT), 13 100-day-old (six Tg, seven WT), 12 12-mo-old (five Tg, seven WT), and 12 21-mo-old (six Tg, six WT) mice were used for MRM analysis; a subset of the identical 100-day-old mice was used for stereologic volume estimation of the hippocampus (five Tg, six WT). Mouse body weights were monitored at all ages, and no significant difference was found between groups (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org).
Tissue Preparation.
Mice were anesthetized with avertin (0.5 mg/g body weight) and transcardially perfused at 15 ml/min, first with 20 ml of 1% paraformaldehyde in 0.1 M NaPO4 buffer, pH 7.4, at room temperature, followed by 100 ml of 4% paraformaldehyde in the same buffer at 4°C. The mouse head was removed and postfixed in 4% paraformaldehyde for 5–8 h, then transferred to buffer and stored at 4°C. For stereologic volume estimation, the brains were dissected, cryoprotected with 30% sucrose, frozen, and stored at −20°C. Fifty-micrometer sections were cut with a sliding microtome, mounted on gelatin-coated slides, and stained with 0.1% thionin followed by 0.24% cresyl violet.
MRI.
T2-weighted spin-echo 3D datasets were acquired from the mouse heads (brain intact within the skull) by using a vertical bore 11.7 T Bruker Avance imaging spectrometer with a microimaging gradient insert and 25 mm birdcage RF coil (Bruker, Billerica, MA). Samples were immersed in fomblin (Ausimont, Thorofare, NJ) and maintained at 4°C. In each case, low-resolution (110-μm isotropic resolution) and high-resolution (55-μm isotropic resolution) data sets were acquired. Imaging parameters: 3D spin echo Rapid Acquisition with Relaxation Enhancement (RARE) (25) acquisition protocol, matrix dimensions = 256 × 128 × 128 and 512 × 256 × 256 (high-resolution set); field of view = 2.8 × 1.4 × 1.4 cm; repetition time (TR) = 3.0 s and 1.5 s (high-resolution set); effective echo time (TE) = 18.3 and 12.4 ms (high-resolution set); RARE factor = 4, number averages = 2 and 1 (high-resolution set). Total imaging time per head was 6 h 53 min (low-resolution set) and 13 h 45 min (high-resolution set).
Volume Determination.
3D MRM files with matrix dimensions = 256 × 128 × 128 were analyzed by using amira software (TGS, Richmond, TX) with the observer blind to genotypes. The acquisition scheme described above provided clear white/gray matter borders that were confirmed by histological analyses. The borders of the brain, cerebellum, and hippocampus were drawn and checked in three orthogonal planes to ensure accuracy. The rostral border of the cerebellum included the flocculus, middle cerebellar peduncle, and central lobule two, and the ventral border was drawn dorsal to the fourth ventricle. The border of the hippocampus was drawn at the gray/white matter border with the fimbria/corpus callosum. The border of the brain was drawn at the pial surface (including olfactory bulbs), and the medulla/spinal cord border was drawn down from the most caudal point of the cerebellum. Brain volumes reported exclude the volume of hippocampus and cerebellum. For each brain, the hippocampus was segmented three times, and the mean of three trials was used as the value for hippocampal volume. After image segmentation, borders were smoothed (amira smooth setting of “3”), and a generalized marching cubes (GMC) calculation was performed to generate a 3D surface reconstruction to obtain volumes (all means and coefficients of variance are presented in Table 1, which is published as supporting information on the PNAS web site). A voxel count was also performed, and volumes derived from voxel counts and GMC calculation were similar (data not shown). In addition, the mean hippocampal volume obtained from the 256 × 128 × 128 matrix file from 18 brains was compared with hippocampal volume obtained from one segmentation trial from 512 × 256 × 256 matrix file of the same brains, and the volumes were similar and were correlated (Fig. 7, which is published as supporting information on the PNAS web site).
Cavalieri Volume Estimation.
Volume estimation using the Cavalieri principle on Tg (n = 5) and WT (n = 6) mice was performed with stereo investigator software (MicroBrightField, Williston, VT) driving a motorized stage (Ludl, Hawthorne, NY) on a Zeiss Axioplan 2ie with a ×5 objective (numerical aperture = 0.25). The final magnification used for grid point counting was ×190. Before Cavalieri analysis, a Nissl-stained atlas was generated with contours drawn for dentate gyrus, CA1/CA2, CA3, and subiculum subfields in the hippocampus, which was used as a guide for all analyses. Cavalieri analysis was performed with the investigator blind to genotype. Volumes of each subfield were estimated by using a one-in-four systematic random series of 50-μm Nissl-stained sections using a 200-μm2 point-counting grid. Between 350 and 1,051 grid points from 6 and 16 sections were counted for each subfield per hippocampus, which provided coefficient of error (CE) estimates of <0.1. Means and Gunderson CE estimates are provided (Table 2, which is published as supporting information on the PNAS web site).
Corpus Callosum Length Measurement.
By using 512 × 256 × 256 MRM files, a point was placed at the most rostral position of the genu of the corpus callosum where white matter appeared continuous across the midline. A second point was placed at the most caudal position of the splenium of the corpus callosum, where white matter last appears as continuous. The length of the line between these two points was used to determine the corpus callosum length.
Statistical Analysis.
All volume data along with corpus callosum length were simultaneously analyzed with multiple ANOVA (MANOVA) followed by Bonferroni post hoc test. Tukey's honestly significant difference post hoc test was also used and provided statistical significance of pairwise comparisons that matched results obtained with Bonferroni's (data not shown). Cavalieri volume estimations of Tg hippocampal subfield volumes were compared with WT subfield volumes by using an unpaired Student's t test.
Results
MRM Analysis.
The MRM acquisition protocol provided an in-plane resolution of ≈100 μm (Fig. 1). From these 3D image files, the hippocampus, cerebellum, and remaining brain were easily detected and segmented. Once complete, manual segmentation of each structure was checked for accuracy in three orthogonal planes, and surface reconstructions and corresponding volumes of hippocampus, cerebellum, and “rest-of-brain” (excluding hippocampus and cerebellum) were obtained (Fig. 2). MRM-derived volume and coefficients of variance values are provided (Table 1), and comparison of volumes obtained with analysis of low- vs. high-resolution files is also provided (Fig. 7).
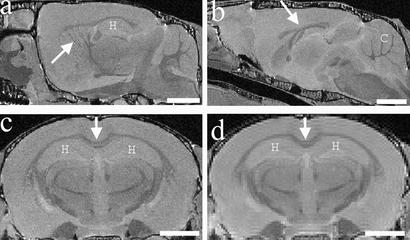
Figure 1.
Anatomical structures visible with MRM. (a–c) Examples of structures visible with high-resolution files (512 × 256 × 256 matrix). (a) Pencil fibers in the striatum (arrow) are visible, as well as the hippocampus (H) in a sagittal section. (b) Corpus callosum (arrow) and cerebellum (C) are visible in this midsagittal plane. (c) Corpus callosum (arrow) and hippocampus (H) are visible in a coronal plane. (d) A coronal plane of a lower-resolution file (256 × 128 × 128 matrix), where corpus callosum (arrow) and hippocampus (H) are clearly visible, compared with a matching coronal plane of a high-resolution file of the same brain seen in c. (Bar = 2 mm.)
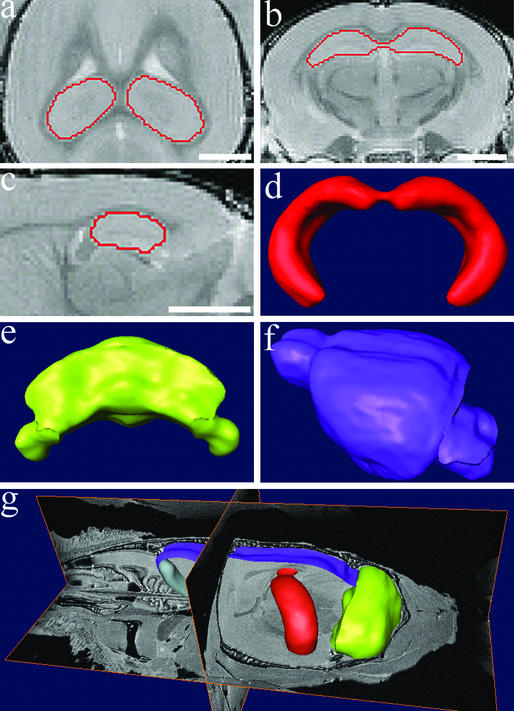
Figure 2.
Image segmentation and surface reconstruction were used to obtain volumes of brain structures. (a–c) Examples of image segmentation of hippocampus on a 3D volumetric MRM file. Accurate borders were drawn and verified in three dimensions simultaneously as shown in the horizontal (a), coronal (b), and sagittal (c) planes. 3D surface reconstructions were generated from the image segmentation of the hippocampus (d), as well as the cerebellum (e) and brain (f). The brain volumes reported excluded the volumes of the hippocampus and cerebellum. (g) 3D surface reconstructions within a 3D volumetric MRM file. Note the brain is undissected and still lies within the mouse head. (Bar = 2 mm.)
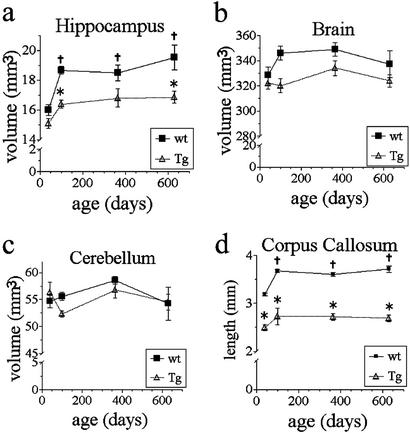
A MANOVA showed a main effect of genotype on hippocampal volume [F (1, 49) = 31, P < 0.001]. A Bonferroni post hoc test showed there was no volume difference detectable when comparing Tg and WT mice at 40 days. However, hippocampal volume was statistically significantly smaller by 12.3% in Tg vs. WT mice at 100 days (Fig. 3). This early time point precedes any accumulation of Aβ and precedes the formation of Aβ-containing plaques. This loss of hippocampal volume begins between 40 and 100 days of age, but as shown for the two older sampling periods, the volume loss does not progress between ages 100 and 630 days. At 630 days, there was a significant 13.7% volume deficit in Tg hippocampal volume that was not statistically different from volumes obtained at 100 or 365 days.
Figure 3.
Quantitative analysis of MRM images of Tg and WT mice at 40, 100, 365, and 630 days of age. (a) Bonferroni's post hoc test showed hippocampal volumes of Tg and WT mice were not different at 40 days, but Tg were significantly smaller at 100 and 630 days (21 mos) (*, P < 0.05). WT hippocampal volumes at 100, 365, and 630 days were significantly larger than 40-day WT volumes (†, P < 0.05). No statistically significant volume changes occurred in the Tg mice over time. (b) Although there was a main effect of genotype on brain volume, a post hoc test showed that no specific time point was significantly reduced in Tg compared with WT mice. (c) Cerebellum volumes did not differ between groups and did not change over time. (d) Corpus callosum length was significantly reduced in Tg mice at all time points (*, P < 0.0001). Corpus callosum lengths in WT mice at 100, 365, and 630 days were significantly longer compared with 40 days WT (†, P < 0.05). Corpus callosum length in Tg mice did not significantly change over time. (Error bars, SEM.)
We also unexpectedly found continued growth in hippocampal volume in WT mice between 40 and 100 days. MANOVA showed a main effect of age on hippocampal volume [F (3, 49) = 11.5, P < 0.001]. There was an 18% increase in hippocampal volume in WT mice between 40 and 630 days of age. The post hoc analyses showed that the hippocampus of 40-day-old WT mice was smaller than that of 100-, 365-, or 630-day-old WT mice. However, the Tg mice showed no significant growth during this same time frame (Fig. 3).
MANOVA also showed a main effect of genotype on brain volume [F (3, 49) = 10.8, P < 0.01]; however, post hoc tests did not detect a significant deficit in Tg mice at any age. Finally, there was no change in cerebellar volume detected between genotypes or across time within a genotype (Fig. 3).
Cavalieri Volume Estimation.
In addition to MRM analysis, five Tg and six WT 100-day-old animals that had been analyzed with MRM were also histologically prepared for stereologic volume estimation of hippocampal subfields from Nissl-stained sections. This additional analysis allowed, (i) comparison of MRM-derived volumes versus Cavalieri volume estimations; and (ii) analysis of hippocampal subfields to localize further possible regions of volume loss within the hippocampus. Total hippocampal volumes obtained from MRM analysis were highly correlated with the total hippocampal volumes obtained from Cavalieri volume estimates of the same brains (r2 = 0.57, P = 0.007, n = 11) (Fig. 4). Cavalieri volume estimation also detected a significant decrease in total hippocampal volume of 12.6% at 100 days in Tg mice compared with WT by an unpaired Student's t test (P = 0.04), which corresponds closely with the 12.3% volume deficit obtained with MRM (P < 0.001 by unpaired Student's t test to compare Tg vs. WT) (Fig. 4).
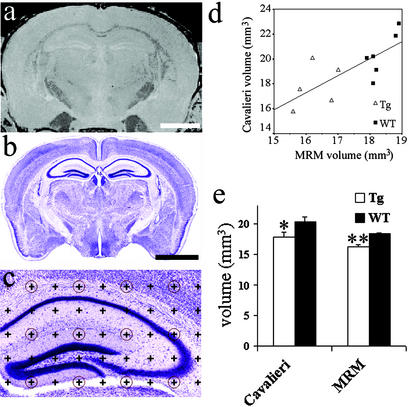
Figure 4.
Comparison of 3-mo MRM-derived hippocampal volumes with Cavalieri volume estimation of hippocampus from the same brains. (a) A coronal section through the hippocampus from a WT brain with a 512 × 256 × 256 matrix MRM file. (b) A Nissl-stained section through the hippocampus of the same WT brain. (c) An example of a Nissl-stained hippocampus with a 200-μm grid overlay used for Cavalieri volume estimation. (d) The correlation between hippocampal volumes of 11 brains derived with MRM analysis and Cavalieri volume estimation (r2 = 0.57, P < 0.01). Six WT brains (■) and five Tg brains (▵) were used for this analysis. (e) Hippocampal volumes obtained with Cavalieri volume estimation of 100-day Tg mice were significantly smaller compared with WT mice (12.6%, *, P = 0.04 with unpaired Student's t test), compared with the previously shown 12.3% smaller Tg hippocampal volume obtained with MRM analysis (**, P < 0.001 with unpaired Student's t test). Although a significant difference was found with both methods, note the larger error bars and the higher P value using the Cavalieri method. (Bar, 2 mm; error bars, SEM.)
Hippocampal volumes obtained from MRM analysis were slightly smaller and had a much tighter interindividual range (i.e., less variability) compared with the Cavalieri-obtained volumes on the sectioned brains. The SD of hippocampal volumes within groups was smaller from MRM analysis (Tg SD ± 0.61, WT SD ± 0.41) compared with Cavalieri (Tg SD ± 1.76, WT SD ± 1.75).
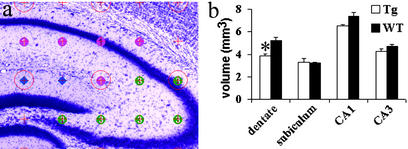
The Cavalieri analysis also provided valuable hippocampal subfield volumes based on anatomical borders detectable by microscopic analysis with Nissl stain, which included CA1/CA2, CA3, subiculum, and dentate gyrus. Analysis of the dentate gyrus as measured included both the granule cell and molecular layers but did not include the hilus. Cavalieri analysis demonstrated a significant and selective volume reduction of 28% in Tg dentate gyrus compared with WT (Fig. 5). It is important to emphasize that the volume reduction observed was specific to Tg dentate gyrus. No significant volume reduction was observed in the CA1, CA3, and subiculum subfields of the Tg hippocampus in the 100-day animal analyses (subfield volume fractions given in Fig. 8, which is published as supporting information on the PNAS web site).
Figure 5.
One hundred-day-old hippocampal subfield volume estimation. (a) An example of a Nissl-stained 100-day WT mouse hippocampus with a 200-μm grid overlay used for Cavalieri volume estimations. Subfields are marked as CA1 (pink circles), CA3 (green circles), and dentate gyrus (blue diamonds). Dentate gyrus volumes were obtained excluding the hilus. (b) There was a significantly smaller dentate gyrus (28%) in Tg (n = 5) compared with WT mice (n = 6) (*, P = 0.003 with unpaired Student's t test). No other subfield was found to be significantly different. (Error bars, SEM.)
Corpus Callosum Length.
In addition to the regional volume deficits observed, we also detected a white-matter deficit by using MRM indicated by a shorter corpus callosum in Tg mice. MANOVA showed a main effect of genotype on corpus callosum length [F (1, 49) = 252, P < 0.0001; Fig. 3]. The corpus callosum length was reduced at all time points examined in Tg mice. MANOVA also showed a main effect of age on corpus callosum length [F (3, 49) = 9.7, P < 0.0001; Fig. 3]. The main effect of age was due to an increase in length over time in the WT group. There was no progression of corpus callosum length deficit over time observed in Tg mice. Other white matter tracts such as anterior commissure and fornix displayed no apparent qualitative differences across Tg and WT mice (not shown).
Discussion
Contrary to our initial hypothesis, we have shown that Tg mice exhibit a significant hippocampal volume deficit at 100 days of age compared with WT mice, yet no such difference was apparent at 40 days. The hippocampal volume reduction emerges between 40 and 100 days of age and is predominantly localized to the dentate gyrus. This volume deficit occurs before any histologically evident accumulation of Aβ or formation of Aβ-containing diffuse or compact plaques (at 6 mos, plaques occupy <0.1% of hippocampal volume; J.F.R., unpublished data). The hippocampal volume deficit in 100- and 630-day-old Tg mice is not progressive and is due to a continued postnatal increase in hippocampal volume in WT mice that does not occur in Tg mice.
Validation/Advantages of the MRM Method.
This is the first example, to our knowledge, of applying quantitative 3D MRM to detect brain volume changes over time in a mouse model of AD. Volume determination from a complete 3D hippocampal image produced precise and reproducible data with low variation compared with Cavalieri volume estimation. Also, to our knowledge, this is the first example of a direct comparison between MRM- and Cavalieri-derived volumes of sectioned brain. These data confirm the value of MRM as a tool for quantifying brain pathology in the rodent and support the notion of a dependable relationship between MRM volume analysis and Cavalieri volume analysis for the mouse hippocampus. Cavalieri hippocampal volumes were ≈10% larger than MRM volumes, perhaps due to changes during tissue handling or storage. The quantitative histological analysis was required to determine how pervasive within the hippocampal formation was this loss of volume. The Cavalieri analysis pinpointed that one subfield was selectively affected, namely the dentate gyrus. The MRM analysis also exhibited such a consistent and accurate set of measurements that the volume loss was apparent at the level of whole hippocampus, reinforcing the utility of the MRM approach for revealing small but meaningful pathological changes. The 15% growth of WT mice hippocampus after 40 days has not been quantified previously with 3D volume determination in mice but confirms an earlier report that the wet weight of dissected mouse hippocampus increases by 15% between 30 and 300 days of age (26).
Neurobiologic Basis for the Anatomic Specificity of the Neuropathologic Findings.
This combined MRM/stereologic analysis not only established a highly specific and surprisingly early temporal window for the detection of an inferred onset of pathology but also targeted the region of earliest hippocampal pathology with great precision, i.e., the molecular layer of dentate gyrus. This decrement in volume could reflect pathology in afferents, in dentate granule cell dendrites, or both. That the molecular layer of the dentate gyrus exhibits such an extensive and specific loss is particularly important, given that it is the site of termination of the perforant path from the entorhinal cortex, a circuit that is highly vulnerable in aging and AD (27–29). The early dentate gyrus pathology in this mouse model is consistent with the early pathology of the entorhinal cortex in AD (30), in that the entorhinal cortex projects heavily to the molecular layer of the dentate gyrus through the perforant path. This circuit will need to be analyzed with higher-resolution techniques to define cellular pathology associated with volume loss, but such consistency with the selective vulnerability apparent in humans further validates this mouse model of AD.
The age of onset of potentially subtle pathology and related neuronal dysfunction in mouse models of AD is not well characterized. These studies pinpoint hippocampal pathology in Tg mice that is not present at 40 days but is clearly detectable in the dentate gyrus of the hippocampus by 100 days of age. Volume changes in PDAPP mice have not been previously detected at this early time point with 3D volume analysis examining the entire hippocampus. These data complement other studies that report hippocampal electrophysiological, behavioral, or metabolic changes in PDAPP mice at young ages preceding significant plaque deposition but at ages beyond 40 days old (19, 31–35). Early localized pathology may also occur in the human brain in normal aging (28) as well as in AD (36–44). Future studies designed to assess therapeutic interventions should therefore consider targeting very early time points in both animal models and humans.
Mechanisms Underlying PDAPP-Induced Neuropathology.
Comparisons of MRM-derived hippocampal volumes demonstrate that between 40 and 100 days of age, WT mice showed continued growth, whereas Tg mice showed no significant growth during the same time. Continued growth within the normal mouse hippocampus at this stage has not been well characterized and may indicate that maturation is still ongoing well after 40 days of age. The negligible growth observed between 40 and 100 days in the Tg mice may reflect the influence of increased soluble Aβ, before the appearance of plaques, on the normal maturation of neurons and synapses. Aβ can bind to neuronal receptors (45–47), and Aβ, whether in oligomeric or fibrillar form, can block long-term potentiation in hippocampal neurons or cause neuronal damage in vitro (48, 49). However, whether this interaction occurs in vivo in a way that blocks or degrades normal synaptic connections remains to be tested. Aβ can also bind extracellular molecules that normally promote dendritic branching or extension, such as the heparan sulfate proteoglycan agrin (50–52). Agrin is especially noteworthy because it promotes dendritic branching, dendritic elongation, microtubule assembly, and synaptic differentiation (53–55). If elevated levels of Aβ interfere with the normal function of extracellular matrix molecules such as agrin, then synaptic formation as well as maintenance may be impaired.
There is also evidence that Aβ (or APP) facilitates neurite outgrowth and synapse formation (56–59). Although only culture experiments but not in vivo models (60) support this possibility to date, it is interesting to note that APP-deficient or knockout mice have a corpus callosum deficit (61), as is seen in PDAPP mice overexpressing APP, and aged APP knockout mice have a 12% volume deficit in the dentate gyrus of the hippocampus (60). This apparently dichotomous effect on neuronal connections may be evidence that one normal function of Aβ may be to modulate synaptic connections.
In AD, corpus callosum deficits have been observed and localized to specific regions and have been correlated with deficits in cognitive function and metabolic activity in corresponding cortical regions (62, 63). Although the corpus callosum deficit in AD may be progressive (64–66), the etiology and the cell populations contributing to this deficit are unknown in this mouse model. The deficit could be a result of reduced collateral crossing axons, a comigrating factor unrelated to APP expression resulting from the triple strain background (61), or even a result of Aβ affecting glial cell function.
If the time course in this mouse model is similar to the time course in the corresponding familial AD, more longitudinal prospective studies are warranted, and early diagnosis strategies/criteria will have to be developed. Ultimately, AD prevalence may be reduced by combined strategies of early diagnosis and preventive drug therapies.
Supplementary Material
Acknowledgments
We thank Trevor McCardle for excellent technical assistance with tissue preparation and Patrick R. Hof for helpful suggestions on the manuscript.
Abbreviations
- AD
Alzheimer's disease
- APP
amyloid precursor protein
- Aβ
β-amyloid protein
- MRM
magnetic resonance microscopy
- MANOVA
multiple ANOVA
- Tg
transgenic
References
- 1.Hardy J, Selkoe D. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Taylor J, Hardy J, Fischbeck K. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 3.Kuo Y M, Emmerling M R, Vigo-Pelfrey C, Kasunic T C, Kirkpatrick J B, Murdoch G H, Ball M J, Roher A E. J Biol Chem. 1996;271:4077–4081. doi: 10.1074/jbc.271.8.4077. [DOI] [PubMed] [Google Scholar]
- 4.Lue L F, Kuo Y M, Roher A E, Brachova L, Shen Y, Sue L, Beach T, Kurth J H, Rydel R E, Rogers J. Am J Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borchelt D R, Thinakaran G, Eckman C B, Lee M K, Davenport F, Ratovitsky T, Prada C M, Kim G, Seekins S, Yager D, et al. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 6.Lemere C A, Lopera F, Kosik K S, Lendon C L, Ossa J, Saido T C, Yamaguchi H, Ruiz A, Martinez A, Madrigal L, et al. Nat Med. 1996;2:1146–1150. doi: 10.1038/nm1096-1146. [DOI] [PubMed] [Google Scholar]
- 7.Citron M, Oltersdorf T, Haass C, McConlogue L, Hung A Y, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe D J. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- 8.Cai X D, Golde T E, Younkin S G. Science. 1993;259:514–516. doi: 10.1126/science.8424174. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki N, Cheung T T, Cai X D, Odaka A, Otvos L, Eckman C, Golde T E, Younkin S G. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 10.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird T D, Hardy J, Hutton M, Kukull W, et al. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 11.Murakami K, Irie K, Morimoto A, Ohigashi H, Shindo M, Nagao M, Shimizu T, Shirasawa T. Biochem Biophys Res Commun. 2002;294:5–10. doi: 10.1016/S0006-291X(02)00430-8. [DOI] [PubMed] [Google Scholar]
- 12.Higgins L S, Rodems J M, Catalano R, Quon D, Cordell B. Proc Natl Acad Sci USA. 1995;92:4402–4406. doi: 10.1073/pnas.92.10.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mucke L, Masliah E, Yu G Q, Mallory M, Rockenstein E M, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold K H, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti P A, et al. Proc Natl Acad Sci USA. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C, Carr T, Clemens J, Donaldson T, Gillespie F, et al. Nature. 1995;373:523–527. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- 16.Dodart J C, Mathis C, Saura J, Bales K R, Paul S M, Ungerer A. Neurobiol Dis. 2000;7:71–85. doi: 10.1006/nbdi.1999.0278. [DOI] [PubMed] [Google Scholar]
- 17.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Haute C V, Checler F, et al. J Biol Chem. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- 18.Chui D H, Tanahashi H, Ozawa K, Ikeda S, Checler F, Ueda O, Suzuki H, Araki W, Inoue H, Shirotani K, et al. Nat Med. 1999;5:560–564. doi: 10.1038/8438. [DOI] [PubMed] [Google Scholar]
- 19.Hsia A Y, Masliah E, McConlogue L, Yu G Q, Tatsuno G, Hu K, Kholodenko D, Malenka R C, Nicoll R A, Mucke L. Proc Natl Acad Sci USA. 1999;96:3228–3233. doi: 10.1073/pnas.96.6.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar-Singh S, Dewachter I, Moechars D, Lubke U, De Jonghe C, Ceuterick C, Checler F, Naidu A, Cordell B, Cras P, et al. Neurobiol Dis. 2000;7:9–22. doi: 10.1006/nbdi.1999.0272. [DOI] [PubMed] [Google Scholar]
- 21.Fitzjohn S M, Morton R A, Kuenzi F, Rosahl T W, Shearman M, Lewis H, Smith D, Reynolds D S, Davies C H, Collingridge G L, et al. J Neurosci. 2001;21:4691–4698. doi: 10.1523/JNEUROSCI.21-13-04691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murrell J, Farlow M, Ghetti B, Benson M D. Science. 1991;254:97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Chen K, Knox J, Inglis J, Bernard A, Martin S, Justice A, McConlogue L, Games D, Freedman S, et al. Nature. 2000;408:975–979. doi: 10.1038/35050103. [DOI] [PubMed] [Google Scholar]
- 24.Bard F, Cannon C, Barbour R, Burke R, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, et al. Nat Med. 2000;6:916–919. doi: 10.1038/78682. [DOI] [PubMed] [Google Scholar]
- 25.Hennig J, Nauerth A, Friedburg H. Magn Reson Med. 1986;3:823–833. doi: 10.1002/mrm.1910030602. [DOI] [PubMed] [Google Scholar]
- 26.Lu L, Airey D C, Williams R W. J Neurosci. 2001;21:3503–3514. doi: 10.1523/JNEUROSCI.21-10-03503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyman B T, Van Horsen G W, Damasio A R, Barnes C L. Science. 1984;225:1168–1170. doi: 10.1126/science.6474172. [DOI] [PubMed] [Google Scholar]
- 28.Morrison J H, Hof P R. Science. 1997;278:412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- 29.Bussière T, Friend P D, Sadeghi N, Wicinski B, Lin G I, Bouras C, Giannakopoulos P, Robakis N K, Morrison J H, Perl D P, Hof P R. Neuroscience. 2002;112:75–91. doi: 10.1016/s0306-4522(02)00056-8. [DOI] [PubMed] [Google Scholar]
- 30.Killiany R J, Hyman B T, Gomez-Isla T, Moss M B, Kikinis R, Jolesz F, Tanzi R, Jones K, Albert M S. Neurology. 2002;58:1188–1196. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- 31.Giacchino J, Criado J R, Games D, Henriksen S. Brain Res. 2000;876:185–190. doi: 10.1016/s0006-8993(00)02615-9. [DOI] [PubMed] [Google Scholar]
- 32.Huitron-Resendiz S, Sanchez-Alavez M, Gallegos R, Berg G, Crawford E, Giacchino J L, Games D, Henriksen S J, Criado J R. Brain Res. 2002;928:126–137. doi: 10.1016/s0006-8993(01)03373-x. [DOI] [PubMed] [Google Scholar]
- 33.Larson J, Lynch G, Games D, Seubert P. Brain Res. 1999;840:23–35. doi: 10.1016/s0006-8993(99)01698-4. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Alavez M, Gallegos R A, Kalafut M A, Games D, Henriksen S J, Criado J R. Neurosci Lett. 2002;330:45–48. doi: 10.1016/s0304-3940(02)00732-2. [DOI] [PubMed] [Google Scholar]
- 35.Dodart J C, Mathis C, Bales K R, Paul S M, Ungerer A. Neurosci Lett. 1999;277:49–52. doi: 10.1016/s0304-3940(99)00847-2. [DOI] [PubMed] [Google Scholar]
- 36.Jack C R, Petersen R C, Xu Y C, Waring S C, O'Brien P C, Tangalos E G, Smith G E, Ivnik R J, Kokmen E. Neurology. 1997;49:786–794. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jack C, Petersen R, Xu Y, O'Brien P, Smith G, Ivnik R, Boeve B, Waring S, Tangalos E, Kokmen E. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Killiany R J, Gomez-Isla T, Moss M, Kikinis R, Sandor T, Jolesz F, Tanzi R, Jones K, Hyman B T, Albert M S. Ann Neurol. 2000;47:430–439. [PubMed] [Google Scholar]
- 39.Fox N, Crum W, Scahill R, Stevens J, Janssen J, Rossor M. Lancet. 2001;358:201–205. doi: 10.1016/S0140-6736(01)05408-3. [DOI] [PubMed] [Google Scholar]
- 40.Gomez-Isla T, Price J L, McKeel D W, Morris J C, Growdon J H, Hyman B T. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Convit A, De L M, Tarshish C, De S S, Tsui W, Rusinek H, George A. Neurobiol Aging. 1997;18:131–138. doi: 10.1016/s0197-4580(97)00001-8. [DOI] [PubMed] [Google Scholar]
- 42.Price J L, Ko A I, Wade M J, Tsou S K, McKeel D W, Morris J C. Arch Neurol. 2001;58:1395–1402. doi: 10.1001/archneur.58.9.1395. [DOI] [PubMed] [Google Scholar]
- 43.Masliah E, Mallory M, Alford M, De Teresa R, Hansen L A, McKeel D W, Morris J C. Neurology. 2001;56:127–129. doi: 10.1212/wnl.56.1.127. [DOI] [PubMed] [Google Scholar]
- 44. Hof, P. R., Bussiere, T., Gold, G., Kovari, E., Giannakopoulos, P., Bouras, C., Perl, D. P. & Morrison, J. H. (2003) J. Neuropathol. Exp. Neurol. 62, in press. [DOI] [PubMed]
- 45.Wang H Y, Lee D H, D'Andrea M R, Peterson P A, Shank R P, Reitz A B. J Biol Chem. 2000;275:5626–5632. doi: 10.1074/jbc.275.8.5626. [DOI] [PubMed] [Google Scholar]
- 46.Wang H Y, Lee D H, Davis C B, Shank R P. J Neurochem. 2000;75:1155–1161. doi: 10.1046/j.1471-4159.2000.0751155.x. [DOI] [PubMed] [Google Scholar]
- 47.Yaar M, Zhai S, Fine R E, Eisenhauer P B, Arble B L, Stewart K B, Gilchrest B A. J Biol Chem. 2002;277:7720–7725. doi: 10.1074/jbc.M110929200. [DOI] [PubMed] [Google Scholar]
- 48.Walsh D M, Klyubin I, Fadeeva J V, Cullen W K, Anwyl R, Wolfe M S, Rowan M J, Selkoe D J. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 49.Wang H W, Pasternak J F, Kuo H, Ristic H, Lambert M P, Chromy B, Viola K L, Klein W L, Stine W B, Krafft G A, et al. Brain Res. 2002;924:133–140. doi: 10.1016/s0006-8993(01)03058-x. [DOI] [PubMed] [Google Scholar]
- 50.Berzin T M, Zipser B D, Rafii M S, Kuo-Leblanc V, Yancopoulos G D, Glass D J, Fallon J R, Stopa E G. Neurobiol Aging. 2000;21:349–355. doi: 10.1016/s0197-4580(00)00121-4. [DOI] [PubMed] [Google Scholar]
- 51.Cotman S L, Halfter W, Cole G J. Mol Cell Neurosci. 2000;15:183–198. doi: 10.1006/mcne.1999.0816. [DOI] [PubMed] [Google Scholar]
- 52.Verbeek M M, Otte-Holler I, van den Born J, van den Heuvel L P, David G, Wesseling P, de Waal R M. Am J Pathol. 1999;155:2115–2125. doi: 10.1016/S0002-9440(10)65529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bose C M, Qiu D, Bergamaschi A, Gravante B, Bossi M, Villa A, Rupp F, Malgaroli A. J Neurosci. 2000;20:9086–9095. doi: 10.1523/JNEUROSCI.20-24-09086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferreira A. J Cell Sci. 1999;112:4729–4738. doi: 10.1242/jcs.112.24.4729. [DOI] [PubMed] [Google Scholar]
- 55.Mantych K B, Ferreira A. J Neurosci. 2001;21:6802–6809. doi: 10.1523/JNEUROSCI.21-17-06802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allinquant B, Hantraye P, Mailleux P, Moya K, Bouillot C, Prochiantz A. J Cell Biol. 1995;128:919–927. doi: 10.1083/jcb.128.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mattson M P. J Neurobiol. 1994;25:439–450. doi: 10.1002/neu.480250409. [DOI] [PubMed] [Google Scholar]
- 58.Mucke L, Abraham C R, Masliah E. Ann NY Acad Sci. 1996;777:82–88. doi: 10.1111/j.1749-6632.1996.tb34405.x. [DOI] [PubMed] [Google Scholar]
- 59.Perez R G, Zheng H, Van der Ploeg L H, Koo E H. J Neurosci. 1997;17:9407–9414. doi: 10.1523/JNEUROSCI.17-24-09407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phinney A L, Calhoun M E, Wolfer D P, Lipp H P, Zheng H, Jucker M. Neuroscience. 1999;90:1207–1216. doi: 10.1016/s0306-4522(98)00645-9. [DOI] [PubMed] [Google Scholar]
- 61.Magara F, Muller U, Li Z W, Lipp H P, Weissmann C, Stagljar M, Wolfer D P. Proc Natl Acad Sci USA. 1999;96:4656–4661. doi: 10.1073/pnas.96.8.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hampel H, Teipel S J, Alexander G E, Horwitz B, Teichberg D, Schapiro M B, Rapoport S I. Arch Neurol. 1998;55:193–198. doi: 10.1001/archneur.55.2.193. [DOI] [PubMed] [Google Scholar]
- 63.Hampel H, Teipel S J, Alexander G E, Pogarell O, Rapoport S I, Moller H J. J Neural Transm. 2002;109:837–855. doi: 10.1007/s007020200069. [DOI] [PubMed] [Google Scholar]
- 64.Pantel J, Schroder J, Jauss M, Essig M, Minakaran R, Schonknecht P, Schneider G, Schad L R, Knopp M V. Psychiatry Res. 1999;90:181–192. doi: 10.1016/s0925-4927(99)00018-9. [DOI] [PubMed] [Google Scholar]
- 65.Teipel S J, Hampel H, Pietrini P, Alexander G E, Horwitz B, Daley E, Moller H J, Schapiro M B, Rapoport S I. Arch Neurol. 1999;56:467–473. doi: 10.1001/archneur.56.4.467. [DOI] [PubMed] [Google Scholar]
- 66.Teipel S J, Bayer W, Alexander G E, Zebuhr Y, Teichberg D, Kulic L, Schapiro M B, Moller H J, Rapoport S I, Hampel H. Arch Neurol. 2002;59:243–248. doi: 10.1001/archneur.59.2.243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.