Abstract
Background
Landmark based geometric morphometrics (GM) allows the quantitative comparison of organismal shapes. When applied to systematics, it is able to score shape changes which often are undetectable by traditional morphological studies and even by classical morphometric approaches. It has thus become a fast and low cost candidate to identify cryptic species. Due to inherent mathematical properties, shape variables derived from one set of coordinates cannot be compared with shape variables derived from another set. Raw coordinates which produce these shape variables could be used for data exchange, however they contain measurement error. The latter may represent a significant obstacle when the objective is to distinguish very similar species.
Results
We show here that a single user derived dataset produces much less classification error than a multiple one. The question then becomes how to circumvent the lack of exchangeability of shape variables while preserving a single user dataset. A solution to this question could lead to the creation of a relatively fast and inexpensive systematic tool adapted for the recognition of cryptic species.
Conclusions
To preserve both exchangeability of shape and a single user derived dataset, our suggestion is to create a free access bank of reference images from which one can produce raw coordinates and use them for comparison with external specimens. Thus, we propose an alternative geometric descriptive system that separates 2-D data gathering and analyzes.
Findings
Morphometric techniques measure size, shape and the relation between size and shape (allometry). In practice, size and shape refer to a measurable part of the organism under study. A few anatomical landmarks (LM) available on a wing (or any measurable part of the body) do not completely describe the shape. However, provided there is operational homology [1] among individual LM, only a partial capture of shape is needed to allow valid comparisons among species
Anatomical landmarks (LM)
Shape is described by new variables derived from raw coordinates of LM after Procrustes superimposition. These variables describing the shape of each specimen depend on the composition of the group under study. If other specimens (i.e. coordinates) are added to the analysis, shape variables must be recomputed accordingly [2,3].
Size
To avoid the problem of multidimensionality, traditional systematists often select one single dimension to represent body size. For an insect, the length of the wing along its largest axis is frequently used as an estimator of body size [4-6]. Such relationship is often assumed rather than demonstrated [5].
Size variable: the centroid size
The centroid size (CS) is the square root of the sum of the squared distances from the centroid to each LM (see Gower, 1971 in [7]). It is a global size estimator informing about size changes in various directions. It is expressed in pixels, i.e. units relative to the resolution of the viewing device (most often a computer display). As a scalar it is less sensible to small digitization errors, and can be shared among systematists provided the pixels can be converted into absolute length units (inches, centimeters, millimeters, etc.).
Shape
Not only in entomology, but also in many fields where morphometrics is applied, shape has been traditionally described as the ratio of one dimension to another. Although intuitively the ratio may appear capable of scaling for size, it often does not [8-11]. Moreover, the ratios introduce some well-known statistical drawbacks [9]. Angles also do not improve the situation since they are another kind of ratio [10].
Shape variables: the Procrustes residuals, the partial warps, the relative warps
In geometric morphometrics (GM), the shape of a configuration of LM is represented by their relative positions as contained in their coordinates after correction for size, position and orientation [7]. The statistical procedure is called Generalized Procrustes Analysis (GPA) [12]. Residual coordinates produced by GPA lie in a curved space [13,14], they must be further modified by a rigid rotation so that they can be studied using classical statistical techniques [15]. Resulting shape variables are called "partial warps" scores (PW). The PW, or their principal components, namely the "relative warps" (RW), may be used in classical statistical analyzes (a complete glossary of the many technical terms related to GM can be found at http://life.bio.sunysb.edu/morph). These transformations are computed relative to the consensus configuration derived from a specific group of samples, this thwarts mixing the final variables with other such variables computed from other individuals.
Allometry
Geometric shape variables (PW) are not allometry-free variables (they are isometry-free variables). The tentative removal of the allometric effect on shape can be justified for intraspecific studies [8,16,17] and less so for interspecific comparisons, where allometric variation is likely to be part of the evolutionary differences relevant to systematics.
Measurement error
Measurement error (ME) can be introduced at various steps of morphometric analysis [18]. The mounting technique of specimens or organs, the photographing conditions, and the user's skill to collect LM coordinates may produce artefactual variation. Generally, similar techniques are used to process similar organisms, and digital techniques of modern photography provide adequate resolution for correct recognition of LM under different conditions.
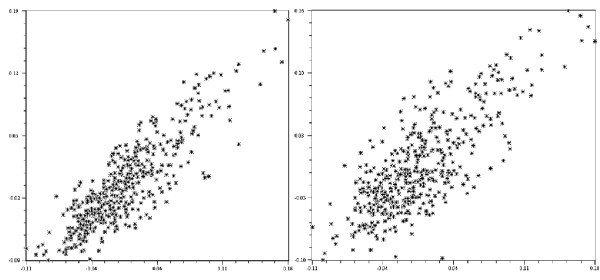
The "user effect" When a single user repeats the measurements on the same specimens, the ME is generally not important. The "user effect" refers to the divergence between two users digitizing the same LM. Between two different users, the error is generally due to small but persistent differences in pointing to the exact location of some LM. We show the results of a repeatability [18] study on three different insect species (Table 1). The repeatability (R) could vary according to the user's skill and the quality of the anatomical LM [19], but it systematically decreased when two users were compared (Table 1). The effect was visibly amplified when looking at the final computation of Procrustes distances (Figure 1).
Table 1.
Mean repeatability of landmark collection from the wings of three insect species
| One user | Two users | |
|---|---|---|
| mean ± stdev | mean ± stdev | |
| Glossina palpalis palpalis | 0.8053 ± 0.1101 | 0.6744 ±0.1623 |
| Glossina fuscipes fuscipes | 0.8099 ± 0.1315 | 0.6153 ± 0.2054 |
| Aedes aegypti [43] | 0.9206 ± 0.0303 | 0.8811 ±0.0632 |
Mean repeatability between the first and second measurements by the same user (first column "One user"), and mean repeatability between the measurements of a user versus the measurements of another user (second column "Two users"). The repeatability was measured as a Model II oneway ANOVA on repeated measures, where R is provided by the ratio of the between-individual variance and the total variance (module VAR of the CLIC package). stdev, standard deviation. The test was performed on the RW, and the average R over the first five RW are presented. The relatively high between-users repeatability in Aedes aegypti did not prevent a visible "user effect" on the Procrustes distances estimations (See Figure 1)
Figure 1.
Distances evaluated two times, by either the same user (left) or two different users (right). Dots (stars) are pair-wise Procrustes distances between female Aedes aegypti processed at 11 landmarks of the right wing. The distances were evaluated two times on the same set of specimens, by either the same user (left) or two different users (right). The repeatability of the measurements were estimated and shown Table 1. The user effect can be visualized going from left to right as a larger scattering of the distances around their expected values (identity). Procrustes distances were evaluated using the module COV of the CLIC software. The graph was obtained from the NTSYSpc-2.02™software.
Reducing ME generally requires averaging repeated collections of the data [18]. However, such a laborious task might not be satisfactory when comparing very close specimens or groups, and ME may become a significant obstacle for different users [20,21].
The taxonomic power of GM
The most important objection to the morphological concept of species is the existence of sibling (or isomorphic) species [22]. Sibling (or also cryptic) species are morphologically identical or nearly identical entities recognized as different species according to other, modern concept(s) of species. However, this objection to the typological concept (i.e. to "morphospecies") is weakened by the possibilities of modern quantitative shape comparisons [23-25]. Shape comparisons detect minimal morphological variations, which often are undetectable by traditional morphological studies and even by classical morphometric approaches. Cryptic species of insects showed distinct shapes in Triatominae [26-28], sandflies [29], parasitoid hymenoptera [23,30,31], fruit flies [32] and screwworm flies [33]. Morphometric discrimination is not confined to species determination, it has also been used to question species boundaries [34], or to synonymize controversial taxa [35].
A geometric characterization system
Traditionally, morphometric traits have been introduced in dichotomous keys in the form of ratios, e.g. "the second antennal segment larger than the first one". GM does not use ratios, it is a powerful multi-characters approach able to derive quantitative information about morphological similarities. However, the consensus-dependent construction of shape variables prevents GM to be converted into a straightforward taxonomic tool [25,36,37].
Zelditch et al. [36] suggested identifying anatomical parts showing differences on D'Arcy Thompson visualization grids, then introducing ratios to taxonomic key. This proposition could be acceptable as long the GPA accurately identifies each LM displacement. However, the GPA considers the whole configuration and not individual LM. Moreover, extracting localized difference would mean some loss of information about shape variation, an unwanted effect when comparing conspecific populations or morphologically "indistinguishable" species.
Admittedly, the simplicity of classical taxonomic keys cannot be achieved with modern morphometric methods, and if one wants to use the full metric properties of the organisms, an analytical step cannot be avoided. Our suggestion for a geometric characterization tool is to separate the analytical step from the constitution of the data, in line with "partial disarticulation" of Bowker [38].
Circumventing the "user effect"
A drastic solution to eliminate the user's source of ME is to eliminate the human user. The task of collecting LM is then automatized by dedicated software [39-41]. Nonetheless, it might be expected that various algorithms of image recognition could differ and show unequal performances. In the same way we describe a "user effect", a possible "software effect" could exist too. Since this effect (Table 1) is amplified in the final distances computation (Figure 1), and because the classification is based on distances, more errors are expected when data are derived from two users.
Our results (Table 2) show the assignation errors using either Procrustes or Mahalanobis distances. As expected, the error rate increased when coordinates were collected by two different users. In total, this "user effect" produced a two times increase in total error rate after Procrustes classification, and a more than ten times increase using Mahalanobis classification (Table 2).
Table 2.
Assignation errors using landmark collection from the wings of two Glossina species
| Species | Distances | One user | Two users |
|---|---|---|---|
| Glossina p. palpalis | Procrustes | 2/44 (5%) | 2/44 (5%) |
| Mahalanobis | 1/44 (2%) | 8/44 (18%) | |
| Glossina f. fuscipes | Procrustes | 5/44 (11%) | 10/44 (23%) |
| Mahalanobis | 1/44 (2%) | 14/44 (32%) | |
| Total errors | Procrustes | 7/88 (8%) | 12/88 (14%) |
| Mahalanobis | 2/88 (2%) | 22/88 (25%) | |
Assignation errors according to the "one user" and "two users" procedures, and according to Procrustes and Mahalanobis distances. "One user" means that the same user digitized both the reference and the external specimens. "Two users" means that reference and external individuals were digitized by two different persons. Absolute values are the number of wrong species attributions out of the total of "unknown" specimens examined. For instance, "2/44" ( first row) means that 2 of the 44 external individuals known to be G. p. palpalis were wrongly assigned to G. f. fuscipes, according to the Procrustes distances.
The solution to the multiple users problem which is immediately applicable is limiting image digitization to a single user, either a human or a software (Table 3, steps 3 and 4), while still allowing images to be shared (Table 3, step 1).
Table 3.
A "one user" procedure of metric identification
| Images (provided by multiple users) | |
|---|---|
| Step 1 | Obtaining reference images from a web data base |
| Step 2 | Obtaining images of unknown specimens |
| Digitization (performed by a single user) | |
| Step 3 | Digitizing the images of reference (reference coordinates) |
| Step 4 | Digitizing the images of the unknown specimens (unknown coordinates) |
| Classification | |
| Procrustes | |
| Step 5 | Pairwise Procrustes distances between each unknown and each reference image |
| Mahalanobis | |
| Step 6* | Computing shape variables on the combined sets of coordinates obtained from step 3 and one unknown specimen obtained from step 4 |
| Step 7 | Computing a discriminant model using the reference shape variables, exclusively (a partition of data from step 6) |
| Step 8 | Entering to the discriminant model the shape variables of the unknown specimen (a partition of data from step 6) |
| Go to Step 6 for the next unknown specimen. | |
General steps implemented in the CLIC package, relevant to the geometric approach: digitization (Steps 3 and 4, module COO of the CLIC package), the Procrustes classification (Step 5, module MOG of CLIC) and the discriminant analysis (Steps 6 and 7, module MOG) to identify organisms using mean reference pictures (Step 1) and own pictures (Step 2). The step 2 refers to the field and/or laboratory activities of the biologist: for entomologists, it generally requires the traditional tasks of collecting, dissecting and mounting insects. (*) To reduce multidimensionality, the number of shape variables (Step 6) can be reduced by selecting a set of few first relative warps (RW, i.e. principal components of partial warps). The MOG module automatically selects a number of RW lower than the smallest sample group.
2-D pictures database
Instead of coordinates which are affected by the measurement error, a reference database would contain the digital pictures from which coordinates can be collected. Then a single user having access to these reference pictures could include them with her/his own images and analyze the images together. This procedure eludes the production of coordinates by different users, though it does not address the errors due to different mounting and photographing techniques.
Thus, to identify morphologically close species and characterize populations, we suggest for GM a procedure separating data gathering from analyzes, i.e. a system consisting of a 2-D pictures database (Table 3, step 1), the extraction of relevant data (Table 3, step 3) and a related model of individual classification (Table 3, step 2 and steps 4 to 8; see next paragraph).
Conditions to provide useful images, such as a size scale (reticule), separation of sexes or the need for published references, are described at the CLIC web page http://www.mpl.ird.fr/morphometrics/clic/index.html. Since the CLIC bank is dedicated to cryptic species, only images which have been the material of a published work would be accepted. Furthermore, to take into account the environment, reference images should be labeled with not only the species but, ideally, the geographic origin, the date of capture, and other parameters defining their habitats.
Classification
Where specific canalization of shape is efficient, we expect any specimen to be more similar to other specimens of the same species than to specimens belonging to different species [42,43]. The species classification as implemented in the CLIC package would then rely on the estimation of metric distances and related attribution algorithm. Classification techniques making use of artificial intelligence [44-46] are not considered here. When adding supplementary data to an analysis performed on reference data, the supplementary data are assigned to the reference group with which they have the shortest distance. The shortest distance might be however an important one and actually outside the mean distance among the members of that group. Thus, assignment to a given class, i.e. "discrimination" in a statistical sense [47], does not necessarily mean belonging to that class ("identification" in the biological sense).
Procrustes distances
The Procrustes distances are based on a minimum criterion (GPA is based on a least-squares algorithm). They are computed in a curved space, so that they are not Euclidean distances. An Euclidean distance is simply a line drawn between two points on a plane, and can be computed from the coordinates of these points by the well-known Pythagorean theorem [36].
The MOG module of the CLIC package allows the introduction of unknown specimens, and then performs a first classification named "Procrustes classification". It is based on pair-wise Procrustes distances of each unknown with the average image of each reference species, as well as with each reference image separately. The direct shape comparison between individual configurations could appear as a relevant technique for classification of unknown specimens. In our example, the total error rate ranged from 8% to 14% according to the "one user" or "two users" modes, respectively (Table 2). However, this classification does not take into account the dispersion ellipses of the reference groups. In our approach, we want to assign unknown individuals to reference groups. Their dispersion ellipses may differ for artefactual (sampling process) or biological reasons (different correlations among variables), and produce undue overlapping or similarities with other specimens.
Mahalanobis distances
This influence of intragroup variation is taken into account with Mahalanobis distances by standardizing the within group variance [47]. Mahalanobis distances may be presented as Euclidean distances computed using the discriminant factors derived from either PW or RW as input variables. In the CLIC procedure (Table 3), the discriminant model is computed between the reference images only (Table 3, step 7), the unknown specimens are then added as supplementary data one by one (Table 3, step 8), and the shape variables used in this classification technique are computed relative to the consensus including the single unknown specimen (Table 3, step 6). The "one by one" procedure is mandatory. Should a large number of unknown, external individuals be entered at once and shape computed from the grand total, the external individuals would modify the total consensus, which could reduce the discrimination between references and alter the classification power.
The Mahalanobis classification is a powerful technique, but very sensible to possible artifacts and/or outliers: it produced the best result in the "one user" procedure (2%, see Table 2), the worst one elsewhere (25%, see Table 2).
Software
The modules of the optional CLIC package http://www.mpl.ird.fr/morphometrics/clic/index.html have been shortly described previously [11,25]. Similar, complementary or additional analyzes can be performed using other freely available software, most of them listed in the main GM web page: http://life.bio.sunysb.edu/morph.
Conclusion
Information systematists could expect from GM is determining whether populations are drawn from multiple species and how they can be discriminated [36]. Here we suggest the use of GM to classify unknown specimens according to known reference species, and we show that to reduce artefactual classification errors, both unknown and reference specimens should be digitized by a single user. This is possible if the reference material is made available to the user from a free access online bank of images. Instead of transporting specimens from one user to another, their images can be made available thanks to web based technologies. The solution proposed here is mostly applicable to 2-D data that can be collected from photographs.
We suggest that the pictures deposited in the bank of reference images be labeled with geographic locality and data of collection time, this will provide the possibility for investigating intraspecific variation. Population structure studies are then also possible.
Availability and requirements
1. Project name: CLIC (Collection of Landmarks for Identification and Characterization)
2. Project home page: http://www.mpl.ird.fr/morphometrics/clic/index.html
3. Operating system(s): Uploading images is platform independent. The CLIC package is currently available for Windows and Linux platforms only.
4. Programming language: HTML, TclTk (CLIC package)
5. Other requirements: The uploading process is currently performed by the author of the CLIC initiative. Images can be sent using for instance an online file sharing software connected to the dujardinbe@gmail.com email address.
6. License: The CLIC package is under GPL license.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JPD designed the HTML CLIC page at http://www.mpl.ird.fr/morphometrics/clic/index.html, and wrote the source code of the CLIC package modules. JPD and AH redacted the paper. DK made the repeatability study on tsetse flies, and AH on Aedes mosquitoes. The authors read and approved the final manuscript.
Contributor Information
Jean-Pierre AL Dujardin, Email: dujardinbe@gmail.com.
Dramane Kaba, Email: kaba_dramane@yahoo.fr.
Amy B Henry, Email: amybethhenry@gmail.com.
Acknowledgements
To Abdoulaye Diarrassouba and Panpim Thongsripong for operational help. This study has been supported by IRD grants number HC3165-3R165-GABI-ENT2 and HC3165-3R165-NV00-THA1, as well as by the EU INCO-DEV/TFCASS (0318490) grant.
References
- Smith GR. In: Proceedings of the Michigan Morphometrics Workshop. Special Publiation Number 2. Rohlf FJ, Bookstein FL, editor. The University of Michigan Museum of Zoology. Ann Arbor MI; 1990. Homology in morphometrics and phylogenetics; p. 380. pp. 325-338. [Google Scholar]
- Rohlf FJ, Marcus LF. A revolution in morphometrics. TREE. 1993;8(4):129–132. doi: 10.1016/0169-5347(93)90024-J. [DOI] [PubMed] [Google Scholar]
- Adams DC, Rohlf FJ, Slice DE. Geometric morphometrics: Ten years of progress following the "revolution". Ital J Zool. 2004;71:5–16. doi: 10.1080/11250000409356545. [DOI] [Google Scholar]
- Nasci RS. Relationship of wing length to adult dry weight in several mosquito species (Diptera: Culicidae) Journal of Medical Entomology. 1990;27:716–719. doi: 10.1093/jmedent/27.4.716. [DOI] [PubMed] [Google Scholar]
- Siegel JP, Novak RJ, Lampman RL, Steinly BA. Statistical appraisal of the weight-wing length relationship of mosquitoes. J Med Entomol. 1992;29(4):711–714. doi: 10.1093/jmedent/29.4.711. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Dalton R, Kim E, Dahl E, Diabate A, Dabire R, Dujardin J. Genetic contribution to variation in larval development time, adult size, and longevity of starved adults of Anopheles gambiae. Infection, Genetics and Evolution. 2006;6(5):410–416. doi: 10.1016/j.meegid.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. In: Proceedings of the Michigan Morphometrics Workshop. Special Publiation Number 2. The University of Michigan Museum of Zoology. Ann Arbor, MI, pp380. Rohlf F, Bookstein F, editor. University of Michigan Museums, Ann Arbor; 1990. Rotational fit (Procrustes) methods; pp. 227–236. [Google Scholar]
- Klingenberg CP. In: Advances in Morphometrics Proceedings of the 1993 NATO-ASI on Morphometrics. Marcus LF, Corti M, Loy A, Naylor GJP, Slice D, editor. New York: Plenum Publ NATO ASI, seR. A, Life Sciences; 1996. Multivariate allometry; pp. 23–49. [Google Scholar]
- Albrecht GH, Gelvin BR, Hartman SE. Ratios as a size adjustment in morphometrics. American Journal of Physical Anthropology. 1993;4(91):441–468. doi: 10.1002/ajpa.1330910404. [DOI] [PubMed] [Google Scholar]
- Burnaby TP. Growth-invariant discriminant functions and generalized distances. Biometrics. 1966;22:96–110. doi: 10.2307/2528217. [DOI] [Google Scholar]
- Dujardin JP, Slice D. In: Encyclopedia of Infectious Diseases. Modern Methodologies. Tibayrenc M, editor. Chapter 25. Wiley & Sons; 2007. Geometric morphometrics. Contributions to medical entomology; pp. 435–447. full_text. [Google Scholar]
- Rohlf FJ. In: Advances in Morphometrics. Proceedings of the 1993 NATO-ASI on Morphometrics. Marcus LF, Corti M, Loy A, Naylor G, Slice D, editor. New York: Plenum Publ. NATO ASI, ser. A, Life Sciences; 1996. Morphometric spaces, shape components and the effects of linear transformations; pp. 117–129. [Google Scholar]
- Kendall DG. Shape-manifolds, Procrustean metrics, and complex projective spaces. Bulletin of the London Mathematical Society. 1984;16:81–121. doi: 10.1112/blms/16.2.81. [DOI] [Google Scholar]
- Slice DE. Landmark coordinates aligned by Procrustes analysis do not lie in Kendall's shape space. Systematic Biology. 2001;50(1):141–149. doi: 10.1080/10635150119110. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ, Bookstein FL. Computing the uniform component of shape variation. Systematic Biology. 2003;52(1):66–69. doi: 10.1080/10635150390132759. [DOI] [PubMed] [Google Scholar]
- Morales Vargas ER, Ya-umphan P, Phumala-Morales N, Komalamisra N, Dujardin JP. Climate associated size and shape changes in Aedes aegypti (Diptera: Culicidae) populations from Thailand. Infections, Genetics and Evolution. 2010;10(4):580–585. doi: 10.1016/j.meegid.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Caro-Riaño H, Jaramillo N, Dujardin JP. Growth changes in Rhodnius pallescens under simulated domestic and sylvatic conditions. Infection, Genetics and Evolution. 2009;9(2):162–8. doi: 10.1016/j.meegid.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Arnqvist G, Martensson T. Measurement error in geometric morphometrics: empirical strategies to assess and reduce its impact on measure of shape. Acta Zoologica Academiae Scientarum Hungaricae. 1998;44(1-2):73–96. [Google Scholar]
- Bookstein FL. Morphometric tools for landmark data: Geometry and Biology. Cambridge University Press, Cambridge; 1991. p. 435. [Google Scholar]
- Jordaens K, Van Dongen S, Van Riel P, Geenen S, Verhagen R, Backeljau T. Multivariate morphometrics of soft body parts in terrestrial slugs: comparison between two datasets, error assessment and taxonomic implications. Biological Journal of the Linnean Society. 2002;75(4):533–542. doi: 10.1046/j.1095-8312.2002.00040.x. [DOI] [Google Scholar]
- Rasmussen P, Wheeler W, Moser T, Vine L, Sullivan B, Rusch D. Measurements of Canada goose morphology - Sources of error and effects on classification of subspecies. Journal of Wildlife Management. 2001;65(4):716–725. doi: 10.2307/3803022. [DOI] [Google Scholar]
- Mayr E. In: Species concepts and phylogenetic theory. A debate. Quentin D Wheeler, Rudolf Meier, editor. New York: Columbia University Press; 2000. The Biological Species Concept; pp. 17–29. [Google Scholar]
- Baylac M, Villemant C, Simbolotti G. Combining geometric morphometrics with pattern recognition for the investigation of species complexes. Biological Journal of the Linnean Society. 2003;80(1):89–98. doi: 10.1046/j.1095-8312.2003.00221.x. [DOI] [Google Scholar]
- Becerra JM, Valdecasas AG. Landmark superimposition for taxonomic identification. Biological Journal of the Linnean Society. 2004;81:267–274. doi: 10.1111/j.1095-8312.2003.00286.x. [DOI] [Google Scholar]
- Dujardin JP. Morphometrics applied to Medical Entomology. Infection, Genetics and Evolution. 2008;8:875–890. doi: 10.1016/j.meegid.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Villegas J, Feliciangeli MD, Dujardin JP. Wing shape divergence between Rhodnius prolixus from Cojedes (Venezuela) and R. robustus from Mérida (Venezuela) Infection, Genetics and Evolution. 2002;2:121–128. doi: 10.1016/S1567-1348(02)00095-3. [DOI] [PubMed] [Google Scholar]
- Matias A, De la Riva JX, Torrez M, Dujardin JP. Rhodnius robustus in Bolivia identified by its wings. Memorias do Instituto Oswaldo Cruz. 2001;96(7):947–950. doi: 10.1590/s0074-02762001000700010. [DOI] [PubMed] [Google Scholar]
- Dujardin JP, Costa J, Bustamante D, Jaramillo N, Catalá S. Deciphering morphology in Triatominae: The evolutionary signals. Acta Tropica. 2009;110:101–111. doi: 10.1016/j.actatropica.2008.09.026. [DOI] [PubMed] [Google Scholar]
- De la Riva J, Le Pont F, Ali V, Matias R, Mollinedo S, Dujardin JP. Wing geometry as a tool for studying the Lutzomyia longipalpis (Diptera: Psychodidae) complex. Memórias do Instituto O. Cruz. 2001;96(8):1089–1094. doi: 10.1590/s0074-02762001000800011. [DOI] [PubMed] [Google Scholar]
- Villemant C, Simbolotti G, Kenis M. Discrimination of Eubazus (Hymenoptera, Braconidae) sibling species using geometric morphometrics analysis of wing venation. Systematic Entomology. 2007;32(4):625–634. doi: 10.1111/j.1365-3113.2007.00389.x. [DOI] [Google Scholar]
- Kitthawee S, Dujardin JP. Diachasmimorpha longicaudata: reproductive isolation and geometric morphometrics of the wings. Bilogical Control. 2009;51(1):191–197. doi: 10.1016/j.biocontrol.2009.06.011. [DOI] [Google Scholar]
- Kitthawee S, Dujardin JP. The geometric approach to explore the Bactrocera tau complex (Diptera: Tephritidae) in Thailand. Zoology. 2010. in press . [DOI] [PubMed]
- Lyra ML, Hatadani LM, de Azeredo-Espin AM, Klaczko LB. Wing morphometry as a tool for correct identification of primary and secondary New World screwworm fly. Bulletin of Entomological Research. 2009;23:1–8. doi: 10.1017/S0007485309006762. [DOI] [PubMed] [Google Scholar]
- Aytekin AM, Alten B, Caglar S, Ozbel Y, Kaynas S, Simsek FM, Kasap OE, Belen A. Phenotypic variation among local populations of phlebotomine sand flies (Diptera: Psychodidae) in southern Turkey. Journal of Vector Ecology. 2007;32(2):226–234. doi: 10.3376/1081-1710(2007)32[226:PVALPO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Gumiel M, Catalá S, Noireau F, de Arias AR, Garcia A, Dujardin JP. Wing geometry in Triatoma infestans (Klug) and T. melanosoma Martinez, Olmedo and Carcavallo (Hemiptera: Reduviidae) Systematic Entomology. 2003;28(2):173–179. doi: 10.1046/j.1365-3113.2003.00206.x. [DOI] [Google Scholar]
- Zelditch ML, Swiderski DL, Sheets HD, Fink WL. Geometric morphometrics for biologists: a primer. Elsevier, Academic Press. New-York; 2004. [Google Scholar]
- Gurgel-Goncalves R, Abad-Franch F, Ferreira JBC, Santana DB, Cuba CAC. Is Rhodnius prolixus (Triatominae) invading houses in central Brazil? Acta Tropica. 2008;107:90–98. doi: 10.1016/j.actatropica.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Bowker GC. Biodiversity Datadiversity. Social Studies of Sciences. 2000;30:643–683. doi: 10.1177/030631200030005001. [DOI] [Google Scholar]
- Weeks PJD. Species-identification of wasps using principal component associative memories. Image and Vision Computing. 1999;17:861–866. doi: 10.1016/S0262-8856(98)00161-9. [DOI] [Google Scholar]
- Houle D, Mezey J, Galpern P, Carter A. Automated measurement of Drosophila wings. BMC Evolutionary Biology. 2003;3:25. doi: 10.1186/1471-2148-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniswamy S, Thacker NA, Klingenberg CP. Automatic Identification of Morphometric Landmarks in Digital Images. 2008. http://www.tina-vision.net/docs/memos/2007-007.pdf
- Dujardin JP, Le Pont F, Baylac M. Geographic versus interspecific differentiation of sand flies: a landmark data analysis. Bulletin of Entomological Research. 2003;93:87–90. doi: 10.1079/BER2002206. [DOI] [PubMed] [Google Scholar]
- Henry A, Thongsripong P, Fonseca-Gonzalez I, Jaramillo-Ocampo N, Dujardin JP. Wing shape of dengue vectors from around the world. Infection, Genetics and Evolution. 2010. [DOI] [PubMed]
- Marcondes CB, Borges PSS. Distinction of Males of the Lutzomyia intermedia (Lutz & Neiva, 1912) Species Complex by Ratios between Dimensions and by an Artificial Neural Network (Diptera: Psychodidae, Phlebotominae) Memorias of Oswaldo Cruz. 2000;95(5):685–688. doi: 10.1590/s0074-02762000000500012. [DOI] [PubMed] [Google Scholar]
- Baylac M, Villemant C, Simbolotti G. Combining geometric morphometrics with pattern recognition for the investigation of species complexes. Biological Journal of the Linnean Society. 2003;80(1):89–98. doi: 10.1046/j.1095-8312.2003.00221.x. [DOI] [Google Scholar]
- MacLeod N, O'Neill M, Walsh SA. A comparison between morphometric and artificial neural network approaches to the automated species recognition problems in systematics. Biodiversity Databases: Techniques, Politics, and Applications (Systematics Association Special Volume) -US - ISBN:9780415332903 (Curry, Gordon B. and Humphries, Chris J., eds) 2007;Chapter V:37–62. [Google Scholar]
- Albrecht GH. Multivariate analysis of the study of form with special reference to canonical variate analysis. American Zoologist. 1980;20:679–693. [Google Scholar]