Abstract
In this study, a putative esterase, designated EstMY, was isolated from an activated sludge metagenomic library. The lipolytic gene was subcloned and expressed in Escherichia coli BL21 using the pET expression system. The gene estMY contained a 1,083 bp open reading frame (ORF) encoding a polypeptide of 360 amino acids with a molecular mass of 38 kDa. Sequence analysis indicated that it showed 71% and 52% amino acid identity to esterase/lipase from marine metagenome (ACL67845) and Burkholderia ubonensis Bu (ZP_02382719), respectively; and several conserved regions were identified, including the putative active site, GDSAG, a catalytic triad (Ser203, Asp301, and His327) and a HGGG conserved motif (starting from His133). The EstMY was determined to hydrolyse p-nitrophenyl (NP) esters of fatty acids with short chain lengths (≤C8). This EstMY exhibited the highest activity at 35°C and pH 8.5 respectively, by hydrolysis of p-NP caprylate. It also exhibited the same level of activity over wide temperature and pH spectra and in the presence of metal ions or detergents. The high level of stability of esterase EstMY with unique substrate specificities makes it highly valuable for downstream biotechnological applications.
Introduction
Lipolytic enzymes are ubiquitous α/β hydrolyzing enzymes existing in animals, plants, and microbes. The enzymes contain esterases (EC3.1.1.1) and lipases (EC3.1.1.3) which catalyze the hydrolysis and synthesis of fatty acid esters including acylglycerides [1]. Due to some useful features such as broad substrate specificity, stability in organic solvents and regio-/enantioselectivity, lipolytic enzymes of microbial origin are widely used in industrial biotechnology, such as production of fine chemicals, pharmaceuticals, and fine chemicals synthesis [2-4].
Modern biotechnology has a steadily increasing demand for novel biocatalysts, thereby prompting the development of new experimental approaches to find and identify novel biocatalyst-encoding genes. Based on the direct cloning of the metagenome [5] for the construction of large clone libraries, metagenomics allows access to new sequences, genes, complete pathways and their products by multiple screening possibilities. With the advent of the metagenome approach, the so far uncultured microorganisms (estimated to more than 99%) [6-10] are now more readily accessible, resulting in an exponential increase in the number of potential biocatalysts. Indeed, the metagenomic approach was useful in mining novel lipolytic enzymes from environmental samples, and also, several genes encoding esterases have been isolated in metagenomic libraries prepared from highly diverse bacterial communities, including marine sediment [11-13], soils [8,10,14,15], drinking water biofilm [10], pond and lake water [16,17], and tidal flat sediment [18]. Some of these enzymes display enhanced characteristics, therefore, searching for novel lipolytic enzymes still attracts considerable attention.
Pre-studies based on 16S rDNA library have extensively expanded our knowledge of microbial diversity in activated sludge from sewage treat plant, including members of varied un-culturable groups (unpublished data). Here, we report the cloning, sequence analysis, and biochemical enzymatic characterization of a novel esterase, EstMY, from an activated sludge derived metagenomic library. Our report demonstrates that metagenomics is a powerful approach in mining new industrial enzymes. The esterase EstMY constituted a new member of family IV of bacterial lipolytic enzymes.
Materials and methods
Sampling
Activated sludge was collected from a sewage treatment plant treating nitrogen-containing aromatic wastewater on September 2008 in Mianyang City, SiChuan Province.
Bacterial strains, plasmids, and culture
The starting strains and plasmids used in this study are listed in Table 1. E. coli was grown at 37°C in Luria-Bertani (LB) medium supplemented with appropriate antibiotics [19]. When required, ampicillin was added at a final concentration of 100 μg/ml, kanamycin at 25 μg/ml, and chloramphenicol, at 12.5 μg/ml.
Table 1.
Starting bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | lacх74 recA1 deoR F - mcrA ∆ (mrr-hsdRMS-mcrBC) ϕ80 lacZ∆M15∆ araD139∆ (ara-leu)7697 galU galK | Transgen |
| E. coli EPI300™-T1R | [F- e14-(McrA-) D(mcrC-mrr) (TetR) hsdR514 supE44 supF58 lacY1 or D(lacIZY)6 galK2 galT22 metB1 trpR55 l-] | Epicentre |
| E. coli BL21(DE3) | F-, ompT, hsdSB (rB-, mB-), dcm, gal, λ(DE3), pLysS, Cmr | Novogen |
| E. coli EPI300-FosD11L2 | Positive clone from Fosmid genomic library, which carries the lipolytic gene | This study |
| E. coli TOP10-EstMY | Positive clone from sublibrary, which carries the EstMY gene fragment | This study |
| E. coli BL21(DE3)-EstMY | Positive clone, which carries the pEstMY-His expression vector | This study |
| Plasmids | ||
| pCC1FOS | Cloning vector; Chlr | Epicentre |
| pUC18 | Cloning vector; Apr | Takara |
| pET28a | Expression vector; Kmr | Novagen |
| FosD11L2 | pCC1FOS, which carries the estMY gene cluster (31 kb) | This study |
| pUC18-EstMY | pUC18, which carries the complete lipolytic gene (estMY) | This study |
| pEstMY-His | pET28a carrying amplified HindIII -NdeI fragment containing lipolytic gene (estMY) | This study |
Apr, ampicillin resistant; Chlr, chloramphenicol resistant; Kmr, kanamycin resistant.
DNA preparation and manipulation
E. coli cells were transformed by the calcium chloride procedure [19]. Recombinant plasmid DNA was isolated by the method of Birnboim and Doly [20]. For sequencing, this DNA was further purified by polyethylene glycol precipitation [19]. Restriction enzymes, T4 DNA ligase and calf intestinal alkaline phosphatases were purchased from New England Biolabs (Ipswich, USA) or Takara (Tokyo, Japan) and used according to the manufacturers' instructions. BugBuster Ni-NTA His. Bind Purification Kit was purchased from Novagen (Code No. NV70751-3, Novagen).
Construction of metagenomic DNA library and related sublibrary
Activated sludge DNA extraction was carried out as previously described using SDS and proteinase K treatment [21], and removing humic acids (HAs) prior to DNA extraction was conducted by removing HAs buffer, 100 mmol/L Tris-HCl pH 10.0, 100 mmol/L Na4P2O7 100 mM, Na2EDTA, 1.0% PVP, 100 mM NaCl, 0.05% Triton X-100 [22]. Approximately 150 μg of metagenomic DNA was run on a preparative pulsed-field gel (Bio-Rad CHEF DR®III; 0.1-40 s switch time, 6 V/cm, 0.5× TBE buffer, 120° included angle, 16 h) and the appropriate size of DNA ranging from 30-45 kb was isolated, electroeluted and dialyzed against 0.5× TE buffer for further Fosmid library construction. The purified DNA fragments were end-repaired by End-repaired enzyme mix. After drop dialysis and concentration, the blunt-ended, 5'-phosphorylated DNA was ligated into the cloning-ready Copycontrol pCC1FOS vector, and the recombinant molecules were packaged into ྔ phage followed by phage transfection to E. coli EPI300 by using protocols described in MaxPlax™ Lambda packaging kit (Epicentre Biotechnologies, Madison, Wisconsin, USA). A fosmid clone showing strong lipolytic enzyme activity on a tributyrin agar plate was selected for further characterization and designated FosD11L2. The DNA was purified from the selected clone, partially digested with Sau3AI in order to obtain 3-5 kb DNA fragments, ligated to the pUC18 vector and transformed into E. coli TOP10 cells (Transgen). Transformants were selected on LB (ampicillin, 100 μg/ml) plates containing 1% (v/v) tributyrin as the indicator substrate [23].
Genetic characterization and sequence analysis
The lipolytic DNA fragment obtained from positive clone E.coli TOP10-EstMY was sequenced with primer walking method by SinoGenoMax Co. Ltd (Chinese National Human Genome Center, Beijing). The ORFs were analyzed using DNASTAR (Lynnon Biosoft) software and ORF finder online analysis http://www.ncbi.nlm.nih.gov/projects/gorf/, Database searches for protein sequences was performed using BLAST and FASTA programs [24,25]. Peptide sequences of various enzymes or subunits were extracted from National Center for Biotechnology Information (Washington, D.C).
Phylogenetic analysis
Deduced amino acid sequences of 12 lipolytic enzymes were subjected to protein phylogenetic analysis. A phylogenetic tree was generated using the neighbor joining method of Saitou and Nei [26] with MEGA 4.0 software [27]. A total of 6 sequences were aligned with the CLUSTAL_W program [28] and visually examined with BoxShade Server program. The length of each branch pair represents the evolutionary distance between the sequences.
Heterologous expression of gene estMY and purification of recombinant EstMY
To express EstMY, the full length of the estMY gene was amplified by PCR with a pair of primers estMY-f and estMY-r (Table 2), in which the high fidelity PrimeSTAR™HS DNA Polymerase (code: DR010SA, Takara) was used. The integrity of the nucleotide sequence of all newly constructed plasmids was confirmed by DNA sequencing. The primer pairs with restriction enzyme sites (underlined) for HindIII and NdeI were designed to generate an N-terminal His-tag of the recombinant esterase. The estMY gene was cloned into an expression vector, pET28a (+) and the recombinant plasmid pestMY-His was transformed into E. coli BL21 (DE3) cells. When the cell density at 600 nm reached around 0.6, expression of recombinant EstMY protein was initiated by addition of 0.6 mM isopropylthio-β-D-galactoside and continued cultivation for additional 4 h. Cells were harvested by centrifugation at 5,000 ×g for 5 min, washed twice with ice-cold 50 mM sodium phosphate buffer (pH 8.0) and resuspended in the same buffer containing 10 mM imidazole, disrupted by sonification in an ice-water bath (60 times, 5s). Recombinant EstMY esterase was applied to metal-chelating chromatography using Ni-NTA affinity chromatography (Novagen) according to the manufacturer's instructions.
Table 2.
Primers used in the study
| Primer | Sequence 5'-3' | Description |
|---|---|---|
| HTFP061 | GTACAACGACACCTAGAC | Sequencing primer for pCC1FOS™ |
| HTRP062 | CAGGAAACAGCCTAGGAA | Sequencing primer for pCC1FOS™ |
| M13 primer RV | CAGGAAACAGCTATGAC | Sequencing primer for pUC18 |
| M13 primer M2 | AGCTGTTCACCGAAGTGCTG | Sequencing primer for pUC18 |
| EstMY -W1F | CGCCCCTTTCGACCAGCAACG | Genomic walking primer for estMY gene |
| EstMY -W2F | CTACGCCGACCTCACCGGCCT | Genomic walking primer for estMY gene |
| EstMY -W1R | GAGGGGTGTGCGGGGATGCG | Genomic walking primer for estMY gene |
| EstMY -W2R | GACGTAGCCGCCGCCGTGAAG | Genomic walking primer for estMY gene |
| EstMY -F | GGCATATGGCCGCGCCCGTTCCGCCCATCAG NdeI | Forward primer for estMY gene |
| EstMY -R | GGAAGCTTCTACGCTGCCGCCCTAGCGCCGATGHindIII | Reverse primer for estMY gene |
The NdeI and HindIII sites are underlined. The start codon is in bold.
Polyacrylamide gel electrophoresis of enzyme in the presence of sodium dodecyl sulfate (SDS) was carried out by the method of Sambrook and Russell [19].
Characterization of recombinant EstMY and biochemical properties
The purified EstMY was subjected to a series of biochemical analysis, including determing the pH optimum, temperature optimum, substrate specificity, and effects of various detergents and metal ions. All measurements were carried out in triplicate. The values were the mean of the data. The substrate specificity of the purified EstMY protein was performed using the following substrates of p-NP-fatty acyl esters [23,29]: acetate (C2), butyrate (C4), hexanoate (C6), caprylate (C8), decanonate (C10), laurate (C12), myristate (C14) and palmitate (C16). The enzyme was incubated with the ester derivatives (0.5 mM) in 5 ml Tris-HCl buffer (50 mM, pH 8.0) at 30°C for 10 min. The reaction was quenched by adding 5 ml trichloroacetic acid (0.5 mM) and then recovered the original pH value with 5.15 ml NaOH (0.5 mM). The enzymatic activity was measured by monitoring the p-nitrophenoxide production by absorbance at 405 nm against an enzyme-free blank, which was measured using a Ultraspec 3000 UV/vis spectrometer (Amersham Biosciences, Sweden) [30,31]. One unit of enzyme activity was defined as the amount of activity required to release 1 μmol p-NP per minute under the above condition. The highest activities of enzyme assay using the substrate (i. e. p-NP-caprylate) was defined as the 100%. To determine the presence of esterase activity, the triglyceride derivative 1,2-di-O-lauryl-rac-glycero-3-glutaric acid 6'-methylresorufin ester (DGGR) (Sigma Aldrich) was used as a chromogenic substrate, and the formation of methylresorufin was analyzed spectrophotometrically at 580 nm [32-34]. Candida rugosa lipase (Sigma Aldrich) was used as a positive control.
The optimum temperature of purified EstMY was determined by assaying lipolytic enzyme activities in a 50 mM Tris-HCl buffer (pH 8.0) for a temperature range of 20-65°C, in which p-NP-caprylate (0.5 mM) acted as substrate. Optimal pH was determined by examining the activity of the enzyme after incubation at 35°C for 10 min using p-NP-caprylate (0.5 mM) as substrate. The buffers used were: 50 mM phosphate buffer (pH 5.0-7.5), 50 mM Tris-HCl (pH 8.0-10.5).
Various metal ions (CoCl2, CaCl2, ZnCl2, MgCl2, K2SO4, FeSO4, CuCl2, Ni(NO3)2, and FeCl3), and chelating agent EDTA at final concentrations of 5 mM were added to the enzyme in 50 mM Tris-HCl (pH 8.0), whereafter it was assayed for esterase activity following preincubation at 35°C. Effect of detergents or reductors on esterase activity was determined by incubating the enzyme for 30 min at 35°C in 50 mM Tris-HCl (pH 8.0), containing Triton X-100, Tween 20, Tween 80, β-mercaptoethanol, 1,4-dithiothreitol (DTT), sodium dodecyl sulfate (SDS), cetyltrimethyl ammonium bromide (CTAB), phenylmethanesulfonyl fluoride (PMSF), diethylpyrocarbonate (DEPC). The concentrations of metal ions, EDTA, detergents, and surfactants used were 5 mM, 3 mM, and 0.5% (v/v), respectively. The activity of the enzyme preparation in the absence of metal ions and detergents before incubation was defined as the 100% level.
Nucleotide sequence accession number
The DNA sequence of EstMY from activated sludge was deposited in GenBank under accession number of HM366454.
Results and discussion
Construction and screening of a metagenomic library
One hundred micrograms of prokaryotic DNA was extracted per gram of wet-weight activated sludge, and 1.5 μg of size-selected, pulsed field gel-purified high-molecular-weight (HMW) DNA suitable for fosmid library construction was obtained. Three hundred nanograms of 30-45 kb purified metagenomic DNA was ligated into the copy control pCC1FOS vector and then tranfected into E. coli EPI300-T1R, producing a metagenome library of more than 7,0000 fosmids with insert size ranging from 27 kb to 38 kb, with an average size of 32 kb, covering approximately 2.1 Gbp of the total metagenomic DNA. Given an average prokaryotic genome of approximately 5 Mbp, the metagenome library theoretically reached the size of over 400 prokaryotic genomes. The prokaryotic origin of the library was confirmed by end-sequencing of randomly selected fosmids and comparison with known ORFs in NCBI. Expression screening of the fosmid library for hydrolytic activity based on the hydrolysis of emulsified tributyrin (1%) resulted in the finding of a recombinant clone, FosD11L2, forming a clear zone on the indicator plate. In order to identify the hydrolytic gene within a fragment of 31 kb, the insert was subject to further subcloning.
Subcloning and identification of the esterase gene
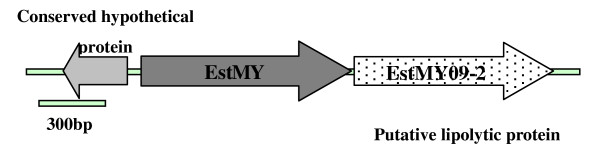
The DNA insert (31 kb) of fosmid D11L2 was partial digested by Sau3AI and subcloned into prepared pUC18 vector, producing a subclone library of more than 3,000 clones with an average insert size of 3.5 kb. One hundred and fifty subclones were screened for lipolytic activity. Among the 9 positive sub-clones forming a clear zone on the indicator plates, one sub-clone that expressed extracellular lipase/esterase activity was sequenced from both ends and the sequences were assembled into a contig of 2,680 bp. An ORF of 1,083 bp encoding a putative lipase/esterase (named EstMY) of 360 amino acids was identified. A second ORF encoding a putative lipolytic enzyme, designated EstMY-092, was identified as well as an additional putative ORF encoding a conserved hypothetical protein (Figure 1).
Figure 1.
Sequencing of subclones (FosD11L2) expressing lipolytic activity resulted in the assembly of a 2,609 bp contig. Three major ORFs with conserved domains were identified: estMY, encoding a novel esterase EstMY; a putative conserved hypothetical protein, with homology to a cytidylate kinase; and an ORF (EstMY09-2) encoding a putative lipolytic protein.
Amino acid sequence alignment indicated that this EstMY exhibited low identity with other esterase/lipases. EstMY shared the highest (71%) sequence identity with the ACL67845 esterase/lipase isolated from a marine metagenome library, 65% sequence identity to Est25 screened from a soil metagenomic library [35], followed by the putative lipase/esterase from other environmental samples (50-65% identity), the putative alpha/beta hydrolase from Burkholderia ubonensis Bu and Parvibaculum lavamentivorans DS-1 (ZP_02382719, 52% identity; and YP_001412150, 49% identity, respectively), members of the family IV hydrolases.
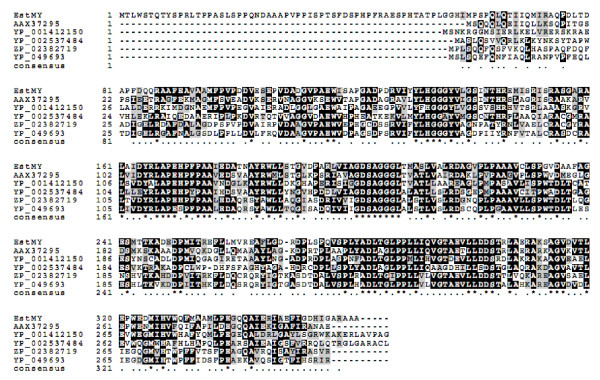
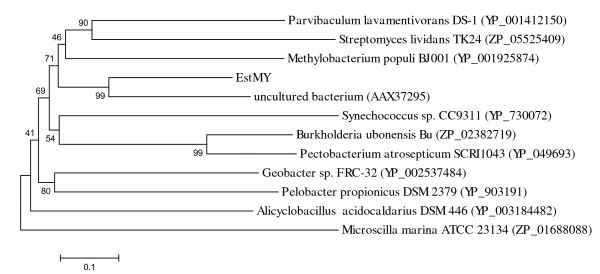
Various lipases and esterases contain the conserved active site motif of the pentapeptide GXSXG with a serine acting as the catalytic nucleophile, a conserved aspartate or glutamate and a histidine, together constituting a catalytic triad [2], organized in the α/β hydrolase fold [36]. The amino acid sequence alignment to bacterial lipolytic enzymes retrieved from GenBank http://www.ncbi.nlm.nih.gov, identified the conserved motifs, including the putative active site GDSAG (Figure 2). Thus, EstMY probably uses a catalytic triad consisting of the serine (Ser203) in the GDSAG active site, the aspartate (Asp301) and the highly conserved histidine (His327) for catalysis. Moreover, EstMY contains a HGGG conserved blocks (starting from His133), which corresponds to a family IV characteristic motif (HGG), which is in close proximity to the active site contributing to the formation of the oxyanion hole that is likely to participate directly in the catalytic process [2,11,37]. Furthermore, to clarify the phylogenetic relationship of the EstMY with other esterases or lipases, a neighbour joining phylogenetic tree was constructed using the amino acid sequence of the lipolytic enzymes. As shown in Figure 3. In this tree, EstMY formed a distinct group with the uncultured bacterium protein (AAX37295), which is located closest to the branch of putative acetyl-hydrolase (accession number ZP_02382719) of strain Burkholderia ubonensis Bu, esterase (accession number ZP_05525409) from Streptomyces lividans TK24, and also, alpha/beta hydrolase domain-containing protein (accession number YP_001412150 and YP_001925874 respectively) from Parvibaculum lavamentivorans DS-1 and Methylobacterium populi BJ001 respectively, which constitute family IV lipases. These results suggest that the EstMY is a new member of family IV lipases.
Figure 2.
Conserved sequence blocks from multiple sequence alignment of EstMY from activated sludge metagenomic library and other related proteins. Sequences alignment was carried out with CLUSTALW [28] and BoxShade Server http://www.ch.embnet.org/software/BOX_form.html. AAX37295, lipase/esterase from marine metagenome; YP_001412150, alpha/beta hydrolase domain-containing protein from Parvibaculum lavamentivorans DS-1; YP_002537484, alpha/beta hydrolase domain-containing protein from Methylobacterium populi BJ001; ZP_02382719, putative acetyl-hydrolase from Burkholderia ubonensis Bu; YP_049693, putative acetyl-hydrolase from Pectobacterium atrosepticum SCRI1043.
Figure 3.
Phylogenetic analysis of EstMY and closely related proteins. Phylogenetic analysis was performed using the program MEGA4.0. Except for EstMY, the protein sequences for bacterial lipolytic enzymes were retrieved from GenBank http://www.ncbi.nlm.nih.gov. The numbers at node indicate the bootstrap percentages of 1000 resamples.
Expression and purification of recombinant EstMY
To investigate the property of this EstMY, estMY gene was expressed as an N-terminal His-tag fusion protein using pET-28a(+) expression system in E. coli BL21(DE3). The recombinant protein was analyzed by SDS-PAGE and Coomassie brilliant blue staining (Figure 4). These results indicate that recombinant EstMY protein is expressed (Mw, about 38 kDa), as which correlated well to the predicted full length of EstMY. The purity of the purified protein was more than 98% according to SDS-PAGE analysis.
Figure 4.
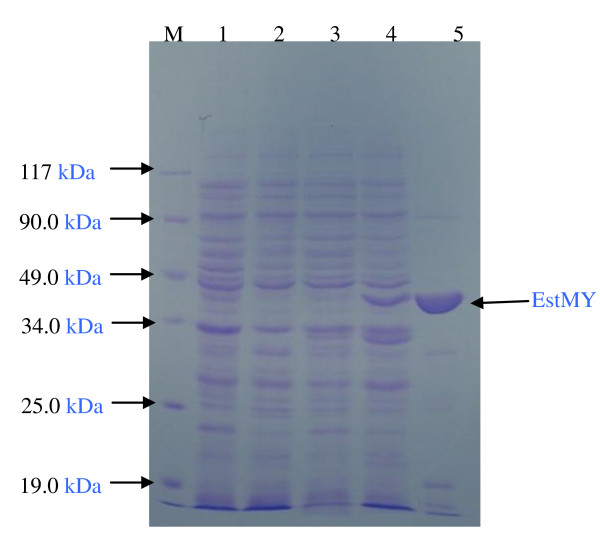
SDS-polyacrylamide gel of overexpressed esterase EstMY in E. coli. Lane 1: molecular weight protein marker (Tiangen, Cat. No: MP203); lane 2, E. coli/pET28a: total protein extract, as negative control; lane 3: induced culture of E. coli/pET28a, as negative control; lane 4: total protein extract E. coli/pEstMY-His; lane 5, total protein extract, induced culture of E. coli/pEstMY-His; lane 6: purified EstMY (38 kDa).
Substrate specificity of EstMY
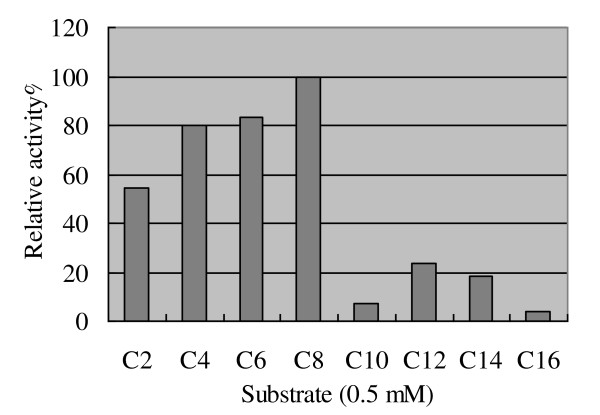
We expressed EstMY as a hexahistidine-tagged (His-tagged) protein and investigated its chain length substrate specificity using p-nitrophenyl esters (Sigma). Results showed EstMY was able to hydrolyse p-nitrophenyl esters with acyl chains up to 14 carbons (p-nitrophenyl myristate), with the highest activity towards short-chain fatty acids (C2, C4, C6 and C8), while much lower towards long-chain fatty acids (>C8) (Figure 5). Moreover, the EstMY was not able to hydrolyse the triglyceride derivative 1, 2-di-O-lauryl-rac-glycero-3-glutaric acid 6'-methylresorufin ester (DGGR) (data not shown), while DGGR was able to form chromogenic product methylresorufin by the true lipase from Candida rugosa (positive control). Furthermore, EstMY showed no fluorescence on olive oil plates with rhodamine B, which indicated that EstMY is a true esterase [29,32,38,39].
Figure 5.
Substrate specificity of overexpressed and purified esterase EstMY. Specific activity of hydrolysis of different p-nitrophenyl esters. p-NP acetate (C2), p-NP butyrate (C4), p-NP hexanoate (C6), p-NP caprylate (C8), p-NP decanoate (C10), p-NP laurate (C12), p-NP myristate (C14), and p-NP palmitate (C16). Relative activity was shown as the percentage of the activity of the activity towards 4-nitrophenyl caprylate. All measurements were performed in triplicate.
Effect of temperature and pH on EstMY
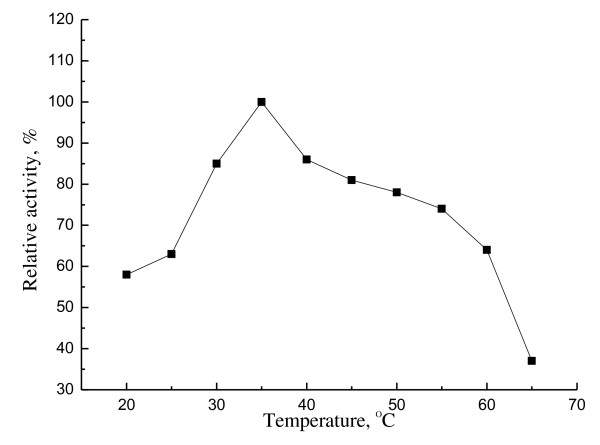
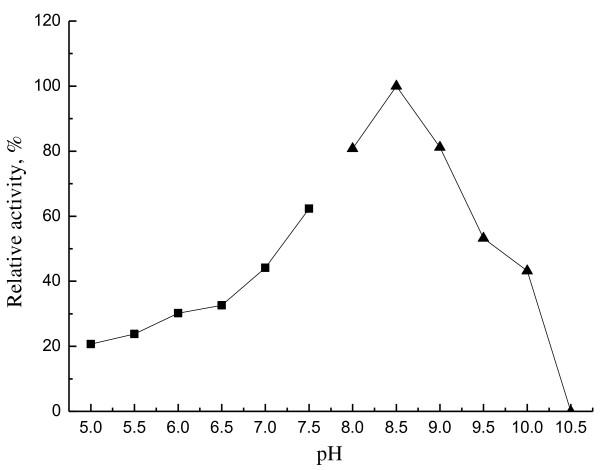
Esterase activity of EstMY was determined from 20°C to 65°C. The purified EstMY showed highest activity at 35°C. It showed a broader temperature spectrum and retained over 37% activity at 65°C (Figure 6). However, h1Lip1 from marine sediment metagenome showed a bad thermostability because there was no activity left after incubation at 40°C for 30 minutes [29]. And also, the esterase showed activity in a rather broader pH range of 7.0-10.0. Maximal activity was observed at pH 8.5 and lost activity at pH 10.5 (Figure 7).
Figure 6.
Apparent temperature optimum of esterase EstMY. Relative activity of p-NP-caprylate hydrolysis at different temperatures by purified EstMY. The activity was determined at different temperatures at pH 8.0 in 50 mM Tris-HCl buffer. The activity at 35°C was set as 100% (3,936 U/ml). All measurements were performed in triplicate.
Figure 7.
Effect of pH on the purified esterae EstMY. Relative activity of p-NP-caprylate hydrolysis was performed in various pH buffers at 35°C (pH 5.0-7.5, 50 mM phosphate buffer; pH 8.0-10.5, 50 mM Tris-HCl buffer). The activity at pH 8.5 was set as 100% (4,615 U/ml). All measurements were performed in triplicate.
Effect of metal ions on esterase
The effects of metal ions and ethylenediamine tetraacetic acid (EDTA) on the EstMY esterase activity were investigated by measuring the residual enzyme activity in their presence and depicted in Table 3. Among metal ions tested, the esterase activity was slightly increased by Co2+ (126%), Ca2+ (104%) and K+ (103%). Furthermore, the esterase activity was inhibited by Ni2+, Zn2+, and Mg2+, moreover, almost totally inhibited by Cu2+ , and Fe3+ (7% and 10% residual acitivity respectively), while the chelating agent EDTA had no effect, suggesting this esterase is not a metalloenzyme.
Table 3.
Effect of metal ions on esterase activity
| Compounds | Concentration (mM) | Relative activity (%) |
|---|---|---|
| Control | 0 | 100.0 ± 2.9 |
| CoCl2 | 5 | 126.4 ± 2.1 |
| K2SO4 | 5 | 103.2 ± 3.6 |
| FeSO4 | 5 | 100.9 ± 2.6 |
| CuCl2 | 5 | 7.8 ± 2.7 |
| Ni(NO3)2 | 5 | 36.2 ± 4.3 |
| EDTA | 5 | 102.7 ± 3.2 |
| FeCl3 | 5 | 10.9 ± 3.4 |
| CaCl2 | 5 | 104.1 ± 3.7 |
| ZnCl2 | 5 | 23.7 ± 1.8 |
| MgCl2 | 5 | 79.7 ± 2.6 |
Activity without metal ions was set as 100% (4,897 U/ml). All measurements were repeated three times.
Effect of detergents and reductors on esterase
The effects of detergents and reductors on esterase activity are shown in Table 4. A significant increase in lipolytic activity was observed with addition of 3 mM CTAB (130%), 0.5% Triton X-100 (129%), Tween 80 (138%), and Tween 20 (156%), after 0.5 h preincubation with detergents at 35°C. Moreover, 3 mM β-mercaptoethanol and DTT did not affect the lipolytic activity (101% and 106%, respectively), whereas DEPC and SDS had a strong inhibitory effect on esterase activity. In accordance to our results, Nawani et al. [40] also found a total inactivation of activity in the presence of SDS but an enhanced activity in the presence of Triton X-100, Tween 80, and Tween 20. Interestingly, the esterase EstMY activity was not impacted by 3 mM PMSF, suggesting EstMY may possess a lid structure, which could eliminate the inhibition effect of PMSF. This is a special characteristic of carboxylesterases [11,41,42] and site-directed mutagenesis of amino acid Ser203 will be carried out to confirm the function of Ser203.
Table 4.
Effect of detergents and enzyme inhibitors on esterase activity
| Compounds | Concentration | Relative activity (%) |
|---|---|---|
| Control | 0 | 100.0 ± 2.1 |
| β-mercaptoethanol | 3 mM | 101.7 ± 2.6 |
| DTT | 3 mM | 106.9 ± 4.9 |
| CTAB | 3 mM | 129.7 ± 2.2 |
| DEPC | 3 mM | 38.6 ± 2.7 |
| PMSF | 3 mM | 101.3 ± 4.1 |
| SDS | 3 mM | 12.3 ± 2.9 |
| Triton X-100 | 0.5% | 129.6 ± 4.6 |
| Tween 80 | 0.5% | 138.4 ± 2.1 |
| Tween 20 | 0.5% | 156.7 ± 3.3 |
Activity without detergents and enzyme inhibitors was set as 100% (4,970 U/ml). All measurements were repeated three times.
In conclusion, we identified a new esterase EstMY belonging to family IV lipases, whose encoding gene was isolated from activated sludge of a sewage treatment plant treating nitrogen-containing aromatic wastewater. EstMY is expected to show high potential for downstream biotechnological applications including synthetic organic chemistry. This was confirmed by its extensive biochemical characterization, which revealed the enzymes substrate specificity, wide pH and temperature spectra, and also, stability towards addictives including metal ions and detergents. Future work will establish the structure of this enzyme to gain more information about its catalytic mechanism. Our research also demonstrated the potential of metagenome strategy in bioprospecting novel genes and biocatalysts and expanded our knowledge of biocatalyst diversity, especially for bacterial esterases. Enlargement of the lipases/esterases pool can be an immediate source of genetic modification, or yield enzymes that can be further specialized by directed evolution, and also, this would optimize their industrial applications.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JGL participated in the design of experiments, and carried out the study and drafted the manuscript. KGZ carried out the SDS-PAGE experiment, sequence alignment and enzyme characteristics analysis. WJH conceived the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Li JunGang, Email: lijungang999@sina.cn.
Zhang KeGui, Email: keguizh@yahoo.com.cn.
Han WenJun, Email: hanwenjun_79@hotmail.com.
Acknowledgements
This work was supported by the grant No. 2005B049 from the Scientific Reserch Fund of Sichuan Provincial Education Department.
References
- Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–183. doi: 10.1042/0264-6021:3430177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15(1):29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409(6817):258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- Straathof AJ, Panke S, Schmid A. The production of fine chemicals by biotransformations. Curr Opin Biotechnol. 2002;13(6):548–556. doi: 10.1016/S0958-1669(02)00360-9. [DOI] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68(4):669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Lee SW, Won K, Lim HK, Kim JC, Choi GJ, Cho KY. Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl Microbiol Biotechnol. 2004;65(6):720–726. doi: 10.1007/s00253-004-1722-3. [DOI] [PubMed] [Google Scholar]
- Elend C, Schmeisser C, Hoebenreich H, Steele HL, Streit WR. Isolation and characterization of a metagenome-derived and cold-active lipase with high stereospecificity for (R)-ibuprofen esters. J Biotechnol. 2007;130(4):370–377. doi: 10.1016/j.jbiotec.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD, Steele HL, Reymond JL, Jaeger KE, Streit WR. Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl Environ Microbiol. 2006;72(5):3637–3645. doi: 10.1128/AEM.72.5.3637-3645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, He H, Guo C, Sun B. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl Microbiol Biotechnol. 2008;80(4):615–625. doi: 10.1007/s00253-008-1566-3. [DOI] [PubMed] [Google Scholar]
- Park HJ, Jeon JH, Kang SG, Lee JH, Lee SA, Kim HK. Functional expression and refolding of new alkaline esterase, EM2L8 from deep-sea sediment metagenome. Prot Expr Purif. 2007;52(2):340–347. doi: 10.1016/j.pep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Jeon JH, Kim JT, Kim YJ, Kim HK, Lee HS, Kang SG, Kim SJ, Lee JH. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl Microbiol Biotechnol. 2009;81(5):865–874. doi: 10.1007/s00253-008-1656-2. [DOI] [PubMed] [Google Scholar]
- Henne A, Schmitz RA, Bomeke M, Gottschalk G, Daniel R. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol. 2000;66(7):3113–3116. doi: 10.1128/AEM.66.7.3113-3116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang K, Liu YH. Molecular cloning and characterization of a novel pyrethroid-hydrolyzing esterase originating from the Metagenome. Microb Cell Fact. 2008;7:38. doi: 10.1186/1475-2859-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HC, Grant S, Jones B, Grant WD, Heaphy S. Detecting cellulase and esterase enzyme activities encoded by novel genes present in environmental DNA libraries. Extremophiles. 2003;7(5):415–421. doi: 10.1007/s00792-003-0339-2. [DOI] [PubMed] [Google Scholar]
- Ranjan R, Grover A, Kapardar RK, Sharma R. Isolation of novel lipolytic genes from uncultured bacteria of pond water. Biochem Biophys Res Commun. 2005;335(1):57–65. doi: 10.1016/j.bbrc.2005.07.046. [DOI] [PubMed] [Google Scholar]
- Wu C, Sun BL. Identification of novel esterase from metagenomic library of Yangtze river. J Microbiol Biotechnol. 2009;19(2):187–193. doi: 10.4014/jmb.0804.292. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62(2):316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi F, Fu LY, Wang GZ, Zheng TL. A simple method for removing humic acids from marine sediment samples prior to DNA extraction. Chin High Technol Lett. 2006;16(5):539–544. [Google Scholar]
- Roh C, Villatte F. Isolation of a low-temperature adapted lipolytic enzyme from uncultivated microorganism. J Appl Microbiol. 2008;105(1):116–123. doi: 10.1111/j.1365-2672.2007.03717.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Rapid and sensitive sequence comparison with Fastp and Fasta. Method Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. full_text. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor-Joining Method - a new method for reconstructing phylogenetic Trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeman F, Sjoling S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol. 2007;59(2):524–534. doi: 10.1111/j.1574-6941.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- FJiang YQW H, Liu CG. Comparison and improvement of three determination methods for lipase activity. Chem Eng. 2007;24(8):72–75. [Google Scholar]
- Pignede G, Wang HJ, Fudalej F, Gaillardin C, Seman M, Nicaud JM. Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteriol. 2000;182(10):2802–2810. doi: 10.1128/JB.182.10.2802-2810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- Panteghini M, Bonora R, Pagani F. Measurement of pancreatic lipase activity in serum by a kinetic colorimetric assay using a new chromogenic substrate. Ann Clin Biochem. 2001;38:365–370. doi: 10.1258/0004563011900876. [DOI] [PubMed] [Google Scholar]
- Zandonella G, Haalck L, Spener F, Faber K, Paltauf F, Hermetter A. Enantiomeric perylene-glycerolipids as fluorogenic substrates for a dual wavelength assay of lipase activity and stereoselectivity. Chirality. 1996;8(7):481–489. doi: 10.1002/(SICI)1520-636X(1996)8:7<481::AID-CHIR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Choi GS, Kim SB, Yoon GS, Kim YS, Ryu YW. Screening and characterization of a novel esterase from a metagenomic library. Protein Expres Purif. 2006;45(2):315–323. doi: 10.1016/j.pep.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J. et al. The alpha/beta-hydrolase fold. Protein Eng. 1992;5(3):197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Laurell H, Contreras JA, Castan I, Langin D, Holm C. Analysis of the psychrotolerant property of hormone-sensitive lipase through site-directed mutagenesis. Protein Eng. 2000;13(10):711–717. doi: 10.1093/protein/13.10.711. [DOI] [PubMed] [Google Scholar]
- Verger R. 'Interfacial activation' of lipases: Facts and artifacts. Trends Biotechnol. 1997;15(1):32–38. doi: 10.1016/S0167-7799(96)10064-0. [DOI] [Google Scholar]
- Jaeger KE, Eggert T. Lipases for biotechnology. Curr Opin Biotechnol. 2002;13(4):390–397. doi: 10.1016/S0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- Nawani N, Dosanjh NS, Kaur J. A novel thermostable lipase from a thermophilic Bacillus sp.: Characterization and esterification studies. Biotechnol Lett. 1998;20(10):997–1000. doi: 10.1023/A:1005430215849. [DOI] [Google Scholar]
- De Simone G, Menchise V, Manco G, Mandrich L, Sorrentino N, Lang D, Rossi M, Pedone C. The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus. J Mol Biol. 2001;314(3):507–518. doi: 10.1006/jmbi.2001.5152. [DOI] [PubMed] [Google Scholar]
- De Simone G, Galdiero S, Manco G, Lang D, Rossi M, Pedone C. A snapshot of a transition state analogue of a novel thermophilic esterase belonging to the subfamily of mammalian hormone-sensitive lipase. J Mol Biol. 2000;303(5):761–771. doi: 10.1006/jmbi.2000.4195. [DOI] [PubMed] [Google Scholar]