Abstract
In a screen to identify novel cellulose deficient mutants, three lines were shown to be allelic and define a novel complementation group, irregular xylem5 (irx5). IRX5 was cloned and encodes a member of the CesA family of cellulose synthase catalytic subunits (AtCesA4). irx5 plants have an identical phenotype to previously described mutations in two other members of this gene family (IRX1 and IRX3). IRX5, IRX3, and IRX1 are coexpressed in exactly the same cells, and all three proteins interact in detergent solubilized extracts, suggesting that three members of this gene family are required for cellulose synthesis in secondary cell walls. The association of IRX1 and IRX3 was reduced to undetectable levels in the absence of IRX5. Consequently, these data suggest that IRX5, IRX3, and IRX1 are all essential components of the cellulose synthesizing complex and the presence of all three subunits is required for the correct assembly of this complex.
Cellulose is a polymer of β-(1,4)-linked glucose with each glucose residue oriented 180° to its neighbor. This orientation allows the chain to adopt a flat, ribbon like structure and these chains crystallize to form microfibrils. These microfibrils are synthesized by large plasma membrane bound complexes, which have been observed in electron micrographs as hexameric structures known as rosettes. These rosettes appear to be associated with the ends of microfibrils (1).
A link between cellulose synthesis and rosettes has come from the identification of genes required for cellulose synthesis in higher plants. At the restrictive temperature, Arabidopsis plants carrying the temperature-sensitive rsw1 mutation have decreased crystalline cellulose. The rsw1 mutation is caused by an alteration in the AtCesA1 gene believed to be a catalytic subunit of the cellulose synthase complex (2). rsw1 plants also exhibit reduced numbers of rosettes in the plasma membrane. Furthermore, by using an antibody recognizing the central, catalytic region of a cotton CesA protein, it has been demonstrated that CesA proteins localize to these plasma membrane rosettes (3). These experiments demonstrate that rosettes are the site of cellulose synthesis and that the CesA protein is central to the organization of rosette structure.
Arabidopsis contains 10 CesA genes (named AtCesA1-10) considered the “true” cellulose synthases that form a subfamily of the cellulose synthase like genes (http://cellwall.stanford.edu/cesa; ref. 4). It is currently unclear why plants contain so many CesA genes. In addition to rsw1, mutations in the CesA6 gene results in a deficiency in cellulose in primary cell walls, suggesting that both AtCesA1 and AtCesA6 are required for cellulose synthesis in the primary cell wall (5). An isoxaben-resistant mutant (ixr2) is also caused by a mutation in AtCesA6 (6). Similarly the isoxaben-resistant mutant ixr1 is caused by a mutation in another member of the CesA gene family (AtCesA3) suggesting that this gene may also be required for primary cell wall cellulose synthesis (7). The irregular xylem (irx) mutants (irx1-3) of Arabidopsis exhibit a collapsed xylem phenotype that is caused by a decrease in cellulose content in secondary cell walls (8). irx1 and irx3 affect exactly the same cell types and are caused by mutations in the AtCes8 and AtCesA7 genes, respectively. Furthermore, coprecipitation experiments with an epitope-tagged version of IRX3 demonstrate that IRX3 and IRX1 function within the same enzyme complex (9, 10). These data are consistent with the idea that at least two CesA gene products are required for cellulose synthesis in both the primary and secondary cell wall of higher plants.
A better understanding of why multiple CesA genes are required to make cellulose in higher plants and how these gene products are organized into rosette structures is essential for both a proper understanding of how single β-(1,4) chains of glucose are synthesized and how these chains become organized into crystalline microfibrils. This is emphasized by the recent report that CesA proteins are required for both the formation of short cellodextrin primers as well as long cellulose chains, suggesting that different CesA genes may have different functions (11).
In the present study, we describe the identification of a novel mutant, irx5, which has severely reduced secondary cell wall cellulose. We show that this phenotype is caused by a mutation in another member of the AtCesA gene family, AtCesA4. AtCesA4 is expressed in cells undergoing secondary cell wall deposition. We demonstrate that IRX5, IRX1 and IRX3 are distinct cellulose synthase catalytic subunits all essential for secondary cell wall cellulose synthesis in the same cells. Furthermore, all three subunits are required for correct assembly of the protein complex. These findings provide further insight into the synthesis of cellulose and the organization and assembly of rosette structures in plants.
Methods
Plant Material, Mutant Isolation, and Genetic Analysis.
Arabidopsis thaliana plants were grown in soil under continuous illumination as described (8). Plants from ethylmethylsulphonate (EMS)-mutated seeds were screened by snapping the stem by hand. Plants exhibiting weak stems were then sectioned and stained with toluidine blue to determine the structure of the vascular bundles (8). Mutant plants were assigned an unambiguous name which identifies the batch of mutagenized seed from which the mutants were isolated (e.g., NGT20-28).
Ac and Ds parent lines (Nottingham Arabidopsis Stock Centre stock nos. CS8043 and CS8047, respectively) were crossed and plants showing transposition in the F2 generation selfed. The F3 generation was screened for phenotypic mutants. pbl3-41 was identified as being dwarfed with dark green leaves.
Cellulose Analysis.
The lower half of mature stems from mutant and Landsberg erecta plants were used for cellulose measurements as described (8).
Thermal Asymmetric Interlaced (TAIL)-PCR.
DNA was extracted from plants by using the method of ref. 12; 1 μl was used for TAIL-PCR essentially according to protocols described in ref. 13. Primers used were as follows: 3′ and 5′ specific primers for primary, secondary, and tertiary reactions, respectively: T3-1 (5′-ATTTCGACTTTAACCCGACCGGAT-3′); T3-2 (5′-TCGTATCGGTTTTCGATTACCGTA-3′); T3-3 (5′-TTCCGTCCCGCAAGTTAAATATGA-3′); T5-1 (5′-ACGGTCGGGAAACTAGCTCTA-3′); T5-2 (5′-CGTTTTGTATATCCCGTTTCCGTT-3′); T5-3 (5′-AAATCGGTTATACGATAACGGTCG-3′). Nonspecific primers used were: AD2 (5′-NGTCGASWGANAWGAA-3′) and AD3 (5′-WGTGNAGWANCANAGA-3′). PCR was carried out with HotStarTaq (Qiagen) according to the manufacturer's instructions in an Eppendorf Mastercycler. After removal of excess primer and dNTPs by incubation with 10 units of ExoI and 2 units of shrimp alkaline phosphatase (Amersham Pharmacia) for 30 min at 37°C followed by 15 min at 80°C tertiary PCR products were sequenced directly, as described below, using either the T5-3 or T3-3 primers.
PCR and RT-PCR.
For RT-PCR, total RNA was isolated from mature stems by using an RNeasy Plant Mini kit (Qiagen). Five hundred nanograms of this RNA was subjected to RT-PCR using Reverse-iT One Step (Abgene, Epsom, Surrey, U.K.). For the 5′ portion of the gene the primers used were IRX55′For (5′-GCTCAGTGTACCTCGCCAT-3′) and IRX55′Rev (5′-CCTCCGCCGCAACAACAGCA-3′). For the 3′ portion of the gene, primers IRX53′For (5′-GCCATGTGATTGTTGGCCGT-3′) and IRX53′Rev (5′-GCGCCAAGCAAAATGGCTCAAA-3′) were used, with the reverse primer acting as the gene-specific primer for the reverse transcription reaction in each case. After incubation for 60 min at 47°C and inactivation of the reverse transcription for 5 min at 94°C, the reactions were subjected to 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, followed by incubation at 72°C for 5 min. RT-PCR products were gel purified before cloning into the vector pGEM-T Easy (Promega) for sequencing.
For PCR amplification from plant genomic DNA, DNA was extracted from leaf tissue according to the method of ref. 14. PCR primers IRX5-2600+ (5′-CCGGTGGAGTGGTGTAAGCAT-3′) and IRX53′Rev, and IRX5-450+ (5′-TAAATGGAAAGCGAGGCAAGA-3′) and IRX5-950− (5′-CGTCATCAGACACATAGCAGC-3′) were used to amplify the mutated region from irx5-2 and irx5-3 genomic DNA respectively under the following conditions: 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. PCR was performed with TaqDNA polymerase (Immunogen International, Sunderland, U.K.) according to the manufacturer's instructions in a PTC100 thermal cycler (MJ Research, Cambridge, MA). Again, the PCR products were gel-purified and cloned into pGEM-T Easy for sequencing.
DNA Sequencing.
Plasmid templates purified by Qiagen QIAprep spin miniprep kits were primed with either universal or gene-specific primers of high-purity salt-free grade (MWG Biotech, Milton Keynes, Buckinghamshire, U.K.) and were sequenced automatically by using ABI Prism Big Dye Terminators (Applied Biosystems). DNA sequences were analyzed by using programs available for use on the internet.
Production of IRX5-Specific Polyclonal Antibodies.
The region encoding the first variable region of IRX5 (amino acids 96–175) was amplified by PCR with primers IRX5VR1For (5′-CGCATATGAATATCAAATATCGCCAGGA-3′) and IRX5VR1Rev (5′-GCCTCGAGAGTCACTAAACCTCTCTTCTCT-3′) and cloned into the NdeI and XhoI sites of pET24a (Novagen). IRX5VR1 was overexpressed, purified, and used to immunize sheep as described (10), and the antiserum was affinity purified as described.
Immunological Techniques.
Whole stem extracts were prepared by grinding under liquid nitrogen and denaturing in an equal volume of 2× loading buffer for 30 min at 37°C followed by clarification by centrifugation. Protein extracts were then electrophoresed through SDS/7.5% polyacrylamide gels (15). After transfer to Immuno-blot poly(vinylidine difluoride) membrane (Bio-Rad), protein gel blots were performed according to standard protocols (16). IRX5 and IRX1 antisera were used at 1 in 1,000 dilutions, and IRX3 antisera at 1 in 5,000 (10). Alkaline-phosphatase conjugated donkey anti-sheep antibody (Sigma) was used, followed by colorimetric detection of alkaline phosphatase activity to detect antibody binding.
Serial tissue prints were performed by repeatedly pressing the freshly cut section of a stem onto damp, prewetted poly(vinylidine difluoride) membrane for 4 s. After allowing the membranes to stand for 5 min, they were treated the same as protein gel blots.
Immunoprecipitations were carried out by using stem extracts ground in an equal volume of IP buffer (50 mM Tris⋅HCl, pH 8.0/150 mM NaCl) solubilized in a final concentration of 2% Triton X-100, and clarified by centrifugation. Polyclonal antisera was added to a final concentration of ≈5 μg/ml and incubated at 4°C for 1 h with end over end rotation. After the addition of 40 μl/ml of protein G plus/protein A agarose (Calbiochem) and a further 1 h incubation at 4°C with end over end rotation, samples were centrifuged briefly to pellet the agarose, which was washed with three changes of IP buffer. All buffers contained protease inhibitors (protease inhibitor mixture for mammalian cell extracts; Sigma). After centrifugation and aspiration, immunoprecipitated proteins were boiled in 2× SDS/PAGE loading buffer for 5 min before chilling on ice. Samples were then clarified by centrifugation and analyzed by SDS/PAGE and protein gel blotting.
Interaction Between IRX5 and IRX3.
The interactions between IRX5 and IRX3 were performed exactly as described (10).
Results
Mutant Isolation.
A population of ≈1,000 Arabidopsis lines containing randomly transposed Ds elements was generated by using an enhancer/gene trapping system (17). A number of lines were identified with visible phenotypes, including pbl3-41, which was dwarfed and dark green in appearance, a phenotype previously observed in irx mutants (8). Stem sections exhibited xylem elements with an irregular and collapsed appearance characteristic of the irx phenotype.
Simultaneously, ≈5,000 plants from several independently generated EMS-mutagenized pools were prescreened for stem strength by using subjective tests to determine the effort required to snap stems by hand. Plants exhibiting an irx phenotype were confirmed by using sections of the inflorescence stem vascular tissue. In addition to further alleles of existing complementation groups (irx1 and irx3), two novel mutants (lines NGT13-4 and NGT20-28) were identified, which formed a new complementation group, irx5. These lines were isolated from different pools of EMS-mutagenized Landsberg erecta plants, indicating that they were independent mutations. Reciprocal crosses among pbl3-41, NGT13-4, and NGT20-28 yielded only mutant plants confirming that these lines fell into the same complementation group. The Ds insertion line, pbl3-41, has been named irx5-1 and the EMS-derived alleles NGT13-4 and NGT20-28 were named irx5-2 and irx5-3, respectively. Crosses between these lines and wild-type yielded only wild-type plants in the F1 population, indicating that these mutations are fully recessive.
Mutant Phenotype.
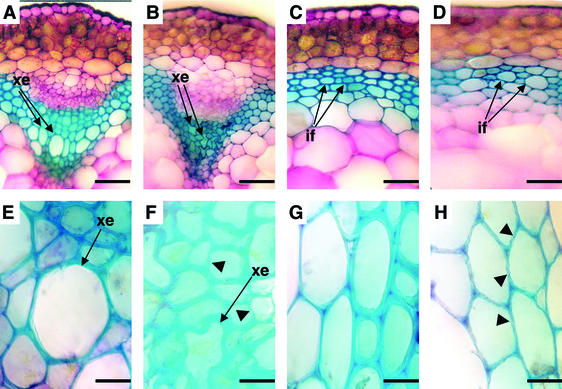
Fig. 1 A and B shows that irx5 plants have an irregular xylem phenotype that has been described for mutations affecting secondary cell wall cellulose synthesis (9, 10) and lignin synthesis (18). Fig. 1 C and D shows that cells in the interfascicular region also have thinner walls, similar to those seen in irx3 plants (8).
Figure 1.
Cross sections of wild-type and irx5-1 stems. Sections of wild type (A, C, E, and G) and irx5-1 (B, D, F, and H) were stained with Toluidine blue. xe, xylem elements; if, interfascicular cells. Arrowheads indicate irregular cell walls. (Bars in A–D represent 0.05 mm. Bars in E–H represent 0.01 mm.)
A closer examination of the cell walls in the xylem and interfascicular region reveals that irx5 affects the same cells as irx3. Fig. 1E shows that wild-type xylem cells have a characteristic well defined, smooth edge. In contrast, irx5 xylem cell walls are thinner, and have an irregular, unevenly stained appearance (Fig. 1F). This difference is also seen in cells from the interfascicular region (Fig. 1 G and H). These phenotypes are identical to those described for irx3 (8).
Cellulose measurements of cell wall extracts from irx5 stems demonstrate that the cellulose content is reduced to ≈30% of wild type (Table 1). This figure is comparable with irx3 that had previously been reported to have a severe cellulose-deficient phenotype (8). Previous studies have suggested that irx1-1 mutants only reduce the cellulose content to ≈50% of wild type. We have recently identified an allele of this locus (irx1-2) (10) in which the cellulose content is reduced to levels very similar to irx3 and irx5 (Table 1). Because irx1-2 is caused by a change of S679 to L, it still may not completely abolish activity. Consequently it is likely that IRX1, IRX3, and IRX5 are all equally important for cellulose synthesis in the secondary cell wall.
Table 1.
Percentage of cellulose in an ethanol-insoluble cell wall fraction from mature stems
| Line | % Cellulose* |
|---|---|
| Landsberg erecta | 29.1 ± 3.6 |
| irx1-1 | 15.1 ± 3.0 |
| irx1-2 | 10.4 ± 2.8 |
| irx3-1 | 7.5 ± 0.78 |
| irx5-1 | 8.0 ± 1.1 |
Values are the mean of measurements from six samples ± SD.
Identification of the irx5 Gene.
We used thermal asymmetric interlaced PCR to amplify a fragment from the 5′ end of the Ds insertion from irx5-1. Comparison of the DNA sequence of this fragment with the Arabidopsis genome showed the insertion to be between nucleotides 49098 and 49099 of P1 clone MRH10 (GenBank accession no. AB006703). The gene, designated MRH10.14, in which this insertion was found corresponds to the cellulose synthase gene AtCesA4 (http://cellwall.stanford.edu/). This places the 5-kb enhancer trap insertion within the codon for amino acid L858 of the predicted protein. Very close linkage was demonstrated between the Ds insertion and the mutation, suggesting that the phenotype was caused by Ds insertion into, or very close to (within 0.9 cM), AtCesA4.
The wild-type IRX5 cDNA sequence was determined by RT-PCR. Primer pairs corresponding to the presumptive coding sequence (from P1 clone MRH10.14) were used to amplify the 5′ and 3′ halves of the gene. These fragments were cloned before sequencing, with two independent clones being sequenced. This cDNA sequence has been deposited in GenBank (accession no. AF458083). To identify the mutations causing the defect in cellulose production in EMS-generated alleles of irx5, RT-PCR was used to amplify the 5′ and 3′ regions of the mutated cDNAs (irx5-3 5′ was amplified from genomic DNA because of difficulty in amplifying this region from cDNA). Two independent clones were sequenced for each region, and the presence of the mutation in the corresponding region of genomic DNA was confirmed. irx5-2 was caused by a G-to-A nucleotide substitution. This substitution results in the replacement of W995 with a stop codon, resulting in a protein lacking 61 aa. irx5-3 had a C-to-T nucleotide substitution. This substitution results in the replacement of Q263 with a stop codon, resulting in a protein lacking 792 aa. Thus, three independent alleles of irx5 contain mutations in this gene, confirming that defects in this gene are responsible for the cellulose-deficient phenotype of irx5 plants.
Structure of the IRX5 Gene.
The position of the 12 introns determined by comparison of the cDNA and genomic sequences differs between the cDNA and the sequence predicted from the genomic sequence, presumably because of incorrect predictions from the genomic sequence. (This results in the Ds insertion actually being in the codon for L870 rather than L858, as predicted from the genomic sequence). The cDNA sequence encodes a predicted protein of 1,055 aa with a molecular mass of 120 kDa and an isoelectric point of 8.2. In common with other plant cellulose synthases, IRX5 is predicted to be a membrane protein, with a cytosolic N terminus followed by two membrane-spanning domains. The central portion of the protein is predicted to be cytosolic with six membrane-spanning domains at the C terminus. The central region also contains four motifs that have been identified as being conserved in all processive β glycosyl transferases. AtCesA4 contains regions of homology to all Arabidopsis CesA proteins (overall identity: 64% to AtCesA8 and 65% to AtCesA7).
IRX5 Is Expressed in the Same Cells as IRX1 and IRX3.
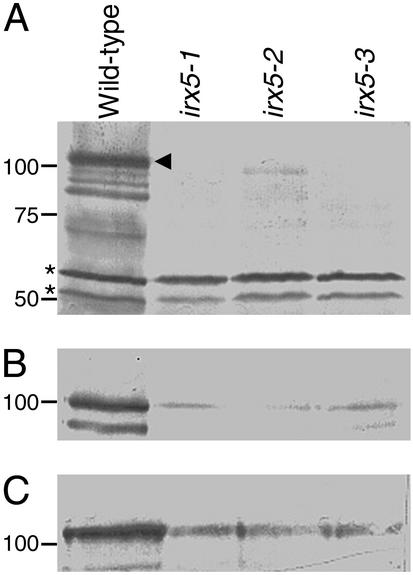
To detect IRX5, a specific polyclonal antibody was raised to a hypervariable region (amino acids 96–175) from IRX5. Fig. 2A shows protein gel blots demonstrating the specificity of this antibody. The IRX5 antibody recognizes a band in wild-type extracts corresponding to the correct size for IRX5, indicated by an arrowhead, in addition to bands showing a higher mobility, which are presumed to be degradation products. Rapid degradation of other CesA proteins has been described (10). No band corresponding to IRX5 is seen in irx5-1, which has a 5-kb Ds insertion in IRX5 and presumed to be a null allele, or in irx5-3, which contains a stop codon after 263 aa. A truncated protein is detected in irx5-2, consistent with this protein lacking 61 aa at the C terminus. These results suggest that the anti-IRX5 antibody is specific for IRX5. In all of these extracts, bands at 50 and 55 kDa are present (Fig. 2A, denoted by asterisks), indicating that they were caused by recognition of unrelated proteins and not degradation products of IRX5. IRX1 and IRX3 (recognized by specific polyclonal antibodies; ref. 10) are detectable in each of these extracts (Fig. 2 B and C) at reduced levels compared with wild-type.
Figure 2.
Specificity of IRX5 antibody. Protein gel blot of wild-type and irx5-1, irx5-2, and irx5-3 extracts. (A) Blot probed with anti-IRX5 antibody. Arrowhead denotes full-length IRX5; asterisks represent nonspecific cross-reacting bands. (B) Blot probed with anti-IRX1 antibody. (C) Blot probed with anti-IRX3 antibody. Molecular mass markers are shown at left in kDa.
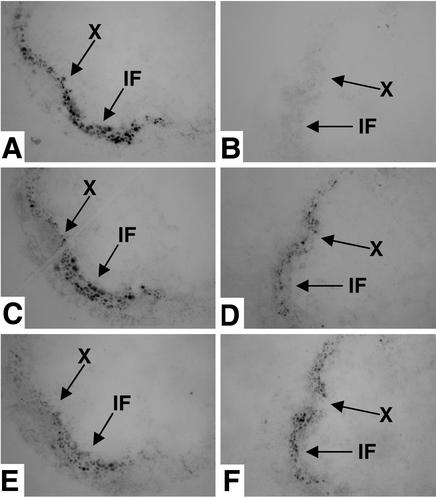
Tissue prints of stem sections were used to determine the expression of IRX5 and IRX1 in specific cells. IRX1 and IRX3 have been shown to be expressed in exactly the same cells (19). Fig. 3 A, C, and E shows that probing serial prints made from the same wild-type stem with antibodies recognizing IRX5, IRX1, and IRX3 reveals that these proteins are expressed in exactly the same cells at the same time. To control for recognition of the unrelated proteins of 50 and 55 kDa by the anti-IRX5 antibody, tissue prints were also made from irx5-1 plants, which lack detectable IRX5 protein. No specific labeling is detected when the anti-IRX5 antibody (Fig. 3B) is used, whereas a signal was detectable when the anti-IRX1 and anti-IRX3 antibodies (Fig. 3 D and F) are used, indicating that the labeling in the xylem and interfascicular region in prints from wild-type plants (Fig. 3A) is caused by specific recognition of IRX5. Tissue prints probed with anti-IRX5 antibody were the first prints made from the fresh cut face to ensure detection of IRX5. The expression patterns of IRX1 and IRX3 are unchanged in the irx5-1 mutant background (Fig. 3 D and F).
Figure 3.
Colocalization of IRX5, IRX1, and IRX3 as shown by tissue printing. Tissue prints of wild-type (A, C, and E) and irx5-1 (B, D, and F) stems probed with anti-IRX5 antibody (A and B), anti-IRX1 antibody (C and D), or anti-IRX3 antibody (E and F). X, xylem; IF, interfascicular region. Arrows indicate the same points in serial sections.
Interaction Among IRX5, IRX1, and IRX3.
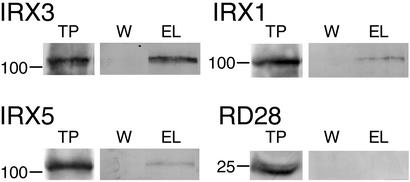
IRX1 and IRX3 have been shown to associate in the same protein complex, and a hexahistidine tagged IRX3 has been described (10) that may be used for the purification of IRX3 and interacting proteins by using immobilized metal affinity chromatography. IRX5 was solubilized in 2% Triton X-100 (data not shown), the same conditions used to solubilize IRX1 and IRX3 (10). Solubilized stem extract from the hexa-His tagged IRX3 line was bound to nickel resin to purify the tagged IRX3. Fig. 4 shows protein blots probed with antibodies recognizing IRX1, IRX3, and IRX5. Blots probed with the anti-IRX3 antibody (10) demonstrate that IRX3 binds to the nickel resin and is specifically eluted with imidazole after extensive washing to remove nonspecifically bound proteins, consistent with this binding being caused by the hexahistidine tag. An identical blot was probed with anti-IRX5 antibody, demonstrating that IRX5 was also purified, remaining bound to the resin until elution with imidazole. This was not caused by nonspecific interactions with the matrix, as performing the same experiment with wild-type plants (i.e., plants without tagged IRX3) did not retain IRX5 on the resin (data not shown). IRX1 is also retained on the nickel resin until elution with imidazole (Fig. 4), which is consistent with previous data (10). To determine that the association of IRX5 with IRX3 was not caused by nonspecific binding of membrane proteins, we used an antibody (RD28) that recognizes another abundant plasma membrane protein, aquaporin, as a control (20). Fig. 4 shows that aquaporin does not copurify with IRX3. Similar experiments have shown that IRX1, IRX3, and IRX5 are all coimmunoprecipitated by using antibodies specific to IRX1. Identical results are obtained when antibodies specific to either IRX3 or IRX5 are used (see below). These results all indicate that IRX1, IRX3, and IRX5 associate and are all components of the same protein complex.
Figure 4.
Copurification of IRX5, IRX3, and IRX1 as shown by protein gel blots. Total protein (TP), pooled washes (W), and eluate (EL) from NHisIRX3 probed with anti-IRX3, IRX5, IRX1, and RD28 (aquaporin) antibodies. Molecular mass markers are shown at left in kDa.
Association of IRX1, IRX3, and IRX5 in Mutant Backgrounds.
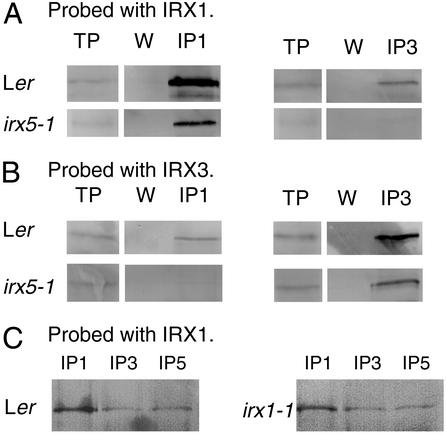
Immunoprecipitation was used to determine the interactions of IRX1 and IRX3 when the third component of the complex, IRX5, was absent. Fig. 5A shows that in solubilized extracts from wild-type plants antibodies specific to IRX3 coimmunoprecipitate IRX1. Similarly the IRX1 specific antibody is capable of coimmunoprecipitating IRX3 from wild-type extracts (Fig. 5B). However, when extracts from irx5-1 plants (in which there is no detectable IRX5 protein) is used, IRX1 is no longer precipitated by the anti-IRX3 antibody at a detectable level (Fig. 5A). Similarly, IRX3 is no longer detectable in the proteins precipitated by the IRX1 antibody (Fig. 5B). Control immunoprecipitations demonstrate that IRX1 and IRX3 are both present in the irx5-1 extracts (Fig. 5 A and B).
Figure 5.
Association of IRX1 and IRX3 in wild type, irx5-1, and irx1-1. Detergent solubilized extracts from wild type, irx5-1, and irx1-1 immunoprecipitated with anti-IRX1 antibody (IP1), anti-IRX3 antibody (IP3), or anti-IRX5 antibody (IP5). TP, total protein; W, final wash. (A and C) Blots probed with anti-IRX1 antibodies. (B) Blots probed with anti-IRX3 antibodies.
All irx5 alleles that we have isolated result in premature termination of the proteins. No alleles in which there is a full- length, mutated protein are available to test the association of the other subunits in the presence of mutant IRX5. It is possible, however, to test the association of IRX3 and IRX5 in the presence of mutated IRX1. The irx1-1 allele is caused by a point mutation that alters a highly conserved aspartate residue (10). Although this protein is assumed to have no activity, this mutation is likely to have little effect on the structure of the protein, and levels of IRX1 protein in irx1-1 plants are comparable to wild type (10). Coimmunoprecipitation experiments on irx1-1 extracts show that antibodies specifically recognizing IRX3 and IRX5 are able to coprecipitate IRX1, in a manner identical to that seen in wild-type (Fig. 5C). Thus, the presence of a mutated form of IRX1 does not affect the interactions of these three proteins.
Discussion
Results presented here show that the cellulose deficiency in irx5 is caused by mutations in the cellulose synthase gene AtCesA4. irx5 affects exactly the same cells as irx1 and irx3, indicating that all three proteins are essential for cellulose synthesis.
Stems from strong alleles of irx1, irx3, and irx5 all contain approximately a third of wild-type cellulose levels (Table 1), and all three loci appear equally important in cellulose synthesis in the secondary cell wall. Because these measurements are from tissue also containing primary cell walls, the true reduction in secondary cell wall cellulose is likely to be greater, as indicated by irx3 plants having little or no cellulose in the secondary cell walls (21). This observation suggests that these three genes are not redundant with one another, implying that all three gene products IRX1 (AtCesA8), IRX3 (AtCesA7) and IRX5 (AtCesA4) are essential to make cellulose in most, if not all, lignified secondary cell walls. Consistent with this suggestion, IRX5 was shown to be expressed in exactly the same cells as IRX1 and IRX3 at the same time. Immobilized metal affinity chromatography on detergent solubilized extracts from plants containing epitope tagged IRX3 demonstrated that both IRX5 and IRX1 were specifically bound to IRX3, whereas another plasma membrane protein, the aquaporin recognized by antibody RD28, was not. These results demonstrate that IRX1, IRX3, and IRX5 are all part of a complex essential for cellulose synthesis in secondary cell walls.
Analysis of Arabidopsis mutants deficient in cellulose synthesis in the primary cell wall also supports the idea that three members of the CesA gene family are required. Mutations in AtCesA1 (rsw1) and AtCesA6 (prc1), both give similar radial swelling phenotypes in the root (2, 5). In addition, mutations in either AtCesA3 (ixr1) (7) or AtCesA6 (ixr2) (6) confer resistance to the cellulose synthase inhibitor isoxaben, suggesting that AtCesA3 is also essential. Furthermore, the mutant eli1 has a radial swelling phenotype that is similar to rsw1 and prc1 and is caused by a mutation in AtCesA3 (A. Cano Delgado and M. Bevan, personal communication), suggesting a role in primary cell wall cellulose synthesis. The implication of these findings is that three distinct classes of CesA proteins are present in rosettes in both primary and secondary cell walls.
It is apparent from data presented here that different CesA proteins are required to assemble the cellulose synthesizing rosette. The fact that interactions between IRX1 and IRX3 are at or below the limits of detection in the absence of IRX5 suggest that these proteins are assembled in a highly ordered manner to make a full size complex. We do not exclude the possibility that there is an undetectable level of interaction between IRX1 and IRX3 in the absence of IRX5, but a comparison with the level of interaction found in wild-type indicates that any interactions are greatly reduced. In addition, in the presence of a mutated form of IRX1 (in irx1-1 plants), the interactions among IRX1, IRX3, and IRX5 are indistinguishable from wild-type. It is possible that IRX1 differs from IRX5 in that it is not required for assembly of the other two subunits. The simplest and most likely explanation, however, is that all three proteins need to be present to assemble the complex correctly, and that the presence of a mutated form of one of the subunits does not affect this assembly.
It has been suggested that the rosettes are transported intact to the plasma membrane (22). Thus, assembly of the complex may occur within the endoplasmic reticulum (ER). There are several examples of oligomeric protein complexes in which the correct folding and assembly of protein subunits in the ER depends on the presence of other subunits of the complex (23, 24). An example of this is the H,K-ATPase in which the β subunit, a type II membrane protein, is essential for correct packing and insertion into the membrane of the polytopic α subunit (23). In this case the incorrectly assembled proteins are degraded. Less IRX1 and IRX3 are consistently found in irx5 mutants, and there appears to be a similar reduction in the level of both proteins. It is possible that these reductions are caused by the lack of interaction between IRX1 and IRX3 in irx5 plants and their subsequent degradation. This data must be interpreted with caution, however, because of the altered morphology of irx5 plants that tend to be slightly smaller and grow more slowly than the wild type.
Cellulose microfibrils are synthesized in a strictly controlled orientation that is essential for normal plant growth and development. This orientation may be altered during formation of the cell wall or in response to a variety of factors such as plant growth regulators. The control of the orientation of cellulose deposition is achieved through altering the direction of deposition from the cellulose synthesizing rosettes. These rosettes are elaborate structures which may contain 18–36 catalytic subunits and probably a large number of additional proteins. The versatile nature of cellulose deposition in higher plants may be due in some extent at least to the unique organization of these rosettes. The importance of CesA proteins in the assembly of these complexes is demonstrated by the rsw1 mutation in which a comparatively minor amino acid change in AtCesA1 causes disassembly of the rosettes (2).
At present, it is only possible to speculate on how these three classes of CesA interact. It is possible that each “lobe” of the hexameric rosette is made up of either a single type of CesA or a defined number of each of the three classes of CesA. A requirement of both of these models is the need for specific interactions between different CesA proteins in a regular structure. These specific interactions may allow the formation of a correctly assembled rosette, emphasizing the importance of the three distinct catalytic subunits in this process. Which of these models is correct will only be confirmed by defining the specific regions responsible for these interactions and determination of the stoichiometry of the different CesA proteins within the rosette. This information is essential for a proper understanding of how cellulose is synthesized, and the identification of a third cellulose synthase catalytic subunit essential for secondary cell wall cellulose synthesis has provided new tools for understanding the interactions between these proteins, and is an important step toward understanding cellulose synthesis.
Acknowledgments
We thank M. Bevan for sharing unpublished results. We are grateful to the Nottingham Arabidopsis Stock Centre for supplying transposon starter lines. This work was supported by the Biotechnology and Biological Sciences Research Council Grant P13259 (to N.G.T.) and European Union Grant FAIR-97-3072 (to R.M.H.).
Abbreviation
- EMS
ethylmethylsulphonate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF458083).
References
- 1.Brown R M. J Macromol Sci Pure Appl Chem. 1996;A33:1345–1373. [Google Scholar]
- 2.Arioli T, Peng L, Betzner A S, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 3.Kimura S, Laosinchai W, Itoh T, Cui X, Linder C R, Brown R M. Plant Cell. 1999;11:2075–2085. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richmond T A, Somerville C R. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Hofte H. Plant Cell. 2000;12:2409–2423. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desprez T, Vernhettes S, Fagard M, Refregier G, Desnos T, Aletti E, Py N, Pelletier S, Hofte H. Plant Physiol. 2002;128:482–490. doi: 10.1104/pp.010822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner S R, Somerville C R. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor N G, Scheible W-R, Cutler S, Somerville C R, Turner S R. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor N G, Laurie S, Turner S R. Plant Cell. 2000;12:2529–2539. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng L, Kawagoe Y, Hogan P, Delmer D. Science. 2002;295:147–150. doi: 10.1126/science.1064281. [DOI] [PubMed] [Google Scholar]
- 12.Edwards K, Johnstone C, Thompson C. Nucleic Acids Res. 1991;19:1349–1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-G, Mitsukawa N, Oosumi T, Whittier R F. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 14.Koneczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 17.Sundaresan V, Springer P, Volpe T, Haward S, Jones J D G, Dean C, Ma H, Martienssen R. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 18.Jones L, Ennos A R, Turner S R. Plant J. 2001;26:205–216. doi: 10.1046/j.1365-313x.2001.01021.x. [DOI] [PubMed] [Google Scholar]
- 19.Turner S R, Taylor N, Jones L. Plant Mol Biol. 2001;47:209–219. [PubMed] [Google Scholar]
- 20.Daniels M J, Mirkov T E, Chrispeels M J. Plant Physiol. 1994;106:1325–1333. doi: 10.1104/pp.106.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha M-A, MacKinnon I M, Sturcova A, Apperley D C, McCann M C, Turner S R, Jarvis M C. Phytochemistry. 2002;61:7–14. doi: 10.1016/s0031-9422(02)00199-1. [DOI] [PubMed] [Google Scholar]
- 22.Haigler C H, Brown R M. Protoplasma. 1986;134:111–120. [Google Scholar]
- 23.Beggah A T, Beguin P, Bamberg K, Sachs G, Geering K. J Biol Chem. 1999;274:8217–8223. doi: 10.1074/jbc.274.12.8217. [DOI] [PubMed] [Google Scholar]
- 24.Geering K. In: Membrane Protein Assembly. Heijne G, editor. New York: Springer; 1997. pp. 173–188. [Google Scholar]