Abstract
Elevated levels of acute-phase serum amyloid A (A-SAA) cause amyloidosis and are a risk factor for atherosclerosis and its clinical complications, type 2 diabetes, as well as various malignancies. To investigate the genetic basis of A-SAA levels, we conducted the first genome-wide association study on baseline A-SAA concentrations in three population-based studies (KORA, TwinsUK, Sorbs) and one prospective case cohort study (LURIC), including a total of 4,212 participants of European descent, and identified two novel genetic susceptibility regions at 11p15.5-p13 and 1p31. The region at 11p15.5-p13 (rs4150642; p = 3.20×10−111) contains serum amyloid A1 (SAA1) and the adjacent general transcription factor 2 H1 (GTF2H1), Hermansky-Pudlak Syndrome 5 (HPS5), lactate dehydrogenase A (LDHA), and lactate dehydrogenase C (LDHC). This region explains 10.84% of the total variation of A-SAA levels in our data, which makes up 18.37% of the total estimated heritability. The second region encloses the leptin receptor (LEPR) gene at 1p31 (rs12753193; p = 1.22×10−11) and has been found to be associated with CRP and fibrinogen in previous studies. Our findings demonstrate a key role of the 11p15.5-p13 region in the regulation of baseline A-SAA levels and provide confirmative evidence of the importance of the 1p31 region for inflammatory processes and the close interplay between A-SAA, leptin, and other acute-phase proteins.
Author Summary
An elevated level of acute-phase serum amyloid A (A-SAA), a sensitive marker of the acute inflammatory state with high heritability estimates, causes amyloidosis and is a risk factor for atherosclerosis and its clinical complications, type 2 diabetes, as well as various malignancies. This study describes the first genome-wide association study on baseline A-SAA concentrations. In a meta-analysis of four genome-wide scans totalling 4,212 participants of European descent, we identified two novel genetic susceptibility regions on chromosomes 11 and 1 to be associated with baseline A-SAA concentrations. The chromosome 11 region contains the serum amyloid A1 gene and the adjacent genes and explains a high percentage of the total estimated heritability. The chromosome 1 region is a known genetic susceptibility region for inflammation. Taken together, we identified one region, which seems to be of key importance in the regulation of A-SAA levels and represents a novel potential target for the investigation of related clinical entities. In addition, our findings indicate a close interplay between A-SAA and other inflammatory proteins, as well as a larger role of a known genetic susceptibility region for inflammatory processes as it has been assumed in the past.
Introduction
Serum amyloid A (SAA) is a sensitive marker of the acute inflammatory state. Its isoforms are expressed constitutively (C-SAA) and show a rapid (up to 1000-fold) increased expression in response to inflammatory stimuli such as trauma, infection, injury, and stress during the acute phase (A-SAA) [1]. The high inductive capacity along with a high conservation of genes and proteins throughout evolution of vertebrates and invertebrates suggests that A-SAA plays a key role in pathogen defence and probably functions as an immune-effector molecule [1]. Acute inflammation has mainly beneficial effects in restoring homeostasis. However, in recent years, clinical and epidemiological studies have gathered substantial evidence that A-SAA is associated with obesity [2] and that prolonged and recurrent chronic infection as well as inflammation is causally involved in the pathogeneses of amyloidosis [1]. Furthermore, it induces, promotes, or influences susceptibility to several chronic diseases such as atherosclerosis and its clinical complications [3]–[9], type 2 diabetes [10], [11], and various malignancies [12]. The SAA gene family is located within 150 kb at chromosome 11 and comprises of four genes: SAA1 and SAA2, the bona fide acute-phase SAA isoforms, SAA3, a pseudogene in humans, and SAA4, a low level expressed gene coding for C-SAA [13], [14]. A-SAA expression is regulated by a variety of stimuli, including the pro-inflammatory cytokines TNF-α and IL-6, as well as glucocorticoids [15], [16]. Like other acute-phase proteins, A-SAA is expressed primarily by the liver [17]. However, extra-hepatic expression has been reported for different cell lines like epithelial cells, monocyte and macrophage cells, most endothelial cells, adipocytes, atherosclerotic lesions, and smooth muscle cells [17]. Twin studies suggest a substantial genetic contribution to baseline A-SAA concentrations with heritability estimates of 59% (95% confidence interval, 49–67%) [18]. The identification of genetic variants that are predisposed to elevated levels of A-SAA could provide important clues to the immune response pathways involved in the regulation of A-SAA levels which might also be of relevance for related clinical entities. In the past, association analyses between genetic variants and A-SAA levels were limited and restricted to allelic variants of SAA genes and protein concentrations [19]–[21].
We therefore conducted the first genome-wide association study on baseline A-SAA concentrations. In a meta-analysis of four genome-wide scans (KORA S4, LURIC, TwinsUK and Sorbs) we included 4,212 participants of European descent. Additionally, in order to account for known gender-specific differences in the regulation of A-SAA [22], [23] we stratified the analysis by gender.
Results
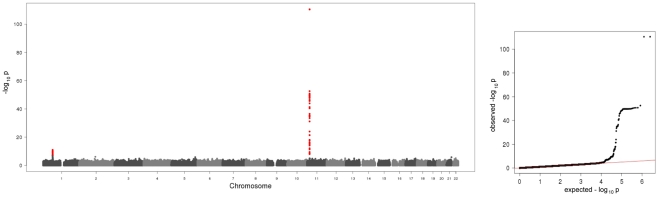
In the present meta-analysis of four genome wide scans 106 SNPs distributed across two regions showed genome-wide significant associations with p-values below the threshold of 5×10−8 (Figure 1, Table S1). Table 1 shows study specific results for the top hits within the two regions and three identified subregions (see below) of the meta-analysis as well as an additional region for men in the gender stratified analysis. Genotypic mean levels are provided in Table S2. Results of the single genome-wide studies were consistent across all four studies regarding the direction and magnitude of the effects. In addition, results were consistent between different genotyping technologies (Table S3). No deviations from the Hardy-Weinberg-Equilibrium were observed. The variable of inter-study heterogeneity (I2) showed homogeneity at the 1p31 locus. At the 11p15.5-p13 locus we observed I2 values that indicated a more distinct heterogeneity. This reflects the relatively large and varying beta values and differences in the minor allele frequency (Table S1). However, taking into account that this locus was clearly significantly associated with A-SAA in all studies included in the meta-analysis, results of the meta-analysis are reported based on a fixed effect model.
Figure 1. Manhattan plot and quantile-quantile plot of the results of the meta-analysis on baseline A-SAA levels.
The Manhattan plot on the left hand side displays all analyzed SNPs with their calculated p-values (p-values below the threshold of genome-wide significance are coloured red). The quantile-quantile plot on the right hand side points out the observed significant associations beyond those expected by chance.
Table 1. Study-specific results for the hits within the regions/subregions.
| SNP | chr | region/subregion | study | effect allele | other allele | imp | effect allele freq | n | beta | se(beta) | p |
| rs4150642 | 11 | 11p15.5-p13 | KORA S4 | G | C | I | 0.200 | 1785 | 0.529 | 0.032 | 1.92E-58 |
| region | LURIC | G | C | I | 0.212 | 961 | 0.569 | 0.075 | 6.51E-14 | ||
| Sorbs | G | C | I | 0.212 | 883 | 0.524 | 0.042 | 1.22E-32 | |||
| TwinsUK | G | C | I | 0.109 | 550 | 0.358 | 0.050 | 8.27E-13 | |||
| combined | G | C | - | 0.193 | 4179 | 0.501 | 0.022 | 3.20E-111 | |||
| validation | G | C | G | 0.217 | 2082 | 0.473 | 0.029 | 7.41E-58 | |||
| rs4638289 | 11 | SAA1 | KORA S4 | A | T | I | 0.347 | 1785 | 0.323 | 0.027 | 4.95E-31 |
| subregion | LURIC | A | T | I | 0.358 | 961 | 0.235 | 0.063 | 2.17E-04 | ||
| Sorbs | A | T | I | 0.320 | 883 | 0.405 | 0.044 | 1.71E-19 | |||
| TwinsUK | A | T | I | 0.214 | 514 | 0.206 | 0.041 | 5.70E-07 | |||
| combined | A | T | - | 0.327 | 4143 | 0.305 | 0.020 | 2.77E-53 | |||
| validation | A | T | G | 0.332 | 2091 | 0.292 | 0.025 | 1.61E-30 | |||
| rs4353250 | 11 | HPS5/GTF2H1 | KORA S4 | T | C | I | 0.339 | 1785 | 0.278 | 0.027 | 1.06E-24 |
| subregion | LURIC | T | C | I | 0.351 | 961 | 0.364 | 0.060 | 2.21E-09 | ||
| Sorbs | T | C | I | 0.413 | 883 | 0.310 | 0.035 | 3.21E-18 | |||
| TwinsUK | T | C | I | 0.256 | 577 | 0.188 | 0.035 | 9.01E-08 | |||
| combined | T | C | - | 0.346 | 4206 | 0.272 | 0.018 | 1.68E-51 | |||
| validation | T | C | G | 0.352 | 2125 | 0.345 | 0.025 | 5.82E-42 | |||
| rs2896526 | 11 | LDHA/LDHC | KORA S4 | G | A | I | 0.177 | 1785 | 0.265 | 0.033 | 3.99E-15 |
| subregion | LURIC | G | A | I | 0.181 | 961 | 0.271 | 0.079 | 6.58E-04 | ||
| Sorbs | G | A | I | 0.163 | 883 | 0.226 | 0.048 | 3.00E-06 | |||
| TwinsUK | G | A | I | 0.156 | 582 | 0.123 | 0.042 | 3.27E-03 | |||
| combined | G | A | - | 0.172 | 4211 | 0.221 | 0.023 | 4.12E-22 | |||
| validation | G | A | G | 0.187 | 2097 | 0.216 | 0.031 | 8.18E-12 | |||
| rs12753193 | 1 | 1p31 (LEPR) | KORA S4 | A | G | I | 0.613 | 1785 | 0.103 | 0.027 | 1.94E-04 |
| region | LURIC | A | G | I | 0.601 | 961 | 0.094 | 0.064 | 1.44E-01 | ||
| Sorbs | A | G | I | 0.599 | 883 | 0.177 | 0.038 | 4.15E-06 | |||
| TwinsUK | A | G | G | 0.628 | 583 | 0.133 | 0.033 | 5.86E-05 | |||
| combined | A | G | - | 0.609 | 4212 | 0.125 | 0.018 | 1.22E-11 | |||
| validation | A | G | G | 0.606 | 2127 | 0.085 | 0.025 | 8.60E-04 | |||
| rs549485 | 11 | 11p14 (SERGEF) | KORA S4 | T | C | G | 0.335 | 871 | 0.155 | 0.040 | 9.77E-05 |
| males only | region | LURIC | T | C | G | 0.368 | 691 | 0.105 | 0.076 | 1.17E-01 | |
| Sorbs | T | C | G | 0.309 | 361 | 0.272 | 0.144 | 3.18E-05 | |||
| combined | T | C | - | 0.342 | 1923 | 0.173 | 0.031 | 2.76E-08 | |||
| vlidation* | C | G | G | 0.349 | 1049 | 0.091 | 0.035 | 8.50E-03 |
*In the validation analysis rs549485 was replaced by rs493767 (r2 = 0.961, 3rd lowest p-value within this region in the gender stratified meta-analysis) for technical reasons.
The first region (193.3 kb of length) resides at 11p15.5-p13 and includes SAA1 one of the structure genes of A-SAA. Within this region the strongest association was found for two highly correlated intronic polymorphisms of the general transcription factor 2 H1 (GTF2H1) gene, rs4150642 (p = 3.20×10−111) and rs7103375 (p = 3.26×10−111) (Figure 2A). These two top hits show modest correlation (r2≤0.376) with other significantly associated SNPs within this region. When the structure of correlation and explained variances within the region were analysed three mostly independent subregions were identified (Table S4, Figure 2B–2D). The first subregion encloses the 5′ end of SAA1 (Figure 2B) with strongest association for rs4638289 (p = 2.77×10−53). The other two subregions harbour the genes Hermansky-Pudlak Syndrome 5 (HPS5) and GTF2H1 (Figure 2C) and lactate dehydrogenase A and C (LDHA and LDHC) (Figure 2D) with strongest associations for rs4353250 (p = 1.68×10−51) and rs2896526 (p = 4.12×10−22), two intronic polymorphisms of HPS5 and LDHA, respectively.
Figure 2. Regional plots of the genetic susceptibility regions/subregions.
The regional plots present gene regions and block structures of the region at 11p15.5-p13 (A), the SAA1 subregion (B), the HPS5/GTF2H1 subregion (C), the LDHA/LDHC subregion (D), and the region at 1p31 (E) and picture the probability values of the significantly associated SNPs, the colour representing the degree of correlation with the top hit of the respective region/subregion.
The second region was detected at 1p31 (Figure 2E). All 38 significantly associated variants cluster around the 3′ end of the leptin receptor gene (LEPR). The most significantly associated SNP, rs12753193, (p = 1.22×10−11) is located downstream of LEPR.
All associations were consistent in the KORA S4 validation analyses (Table 1 for the top hits, Table S5 for all SNPs selected for validation).
The entire regression model (including the top SNPs of the two genomic regions (rs4150642 and rs12753193), age, gender and BMI) explains 19.32% of the total variation of A-SAA in our data. With an explained variance of 10.84% for the top SNP (rs4150642) of the 11p15.5-p13 locus (5.57% for rs4638289 of the SAA1 subregion, 5.34% for rs4353250 of the HPS5/GTF2H1 subregion, and 2.37% for rs2896526 of the LDHA/LDHC subregion; Table S4) and 0.93% for the top SNP (rs12753193) of the 1p31 locus the identified genomic regions account for a major part of such variance.
When the analysis was stratified by gender, one additional SNP (rs549485) located about 350 kb apart from the SAA1 subregion at 11p14 in the secretion regulating guanine nucleotide exchange factor (SERGEF) gene showed a borderline significant association with A-SAA levels in men (p = 2.76×10−8) in the meta-analysis. In the validation analysis the association between two highly correlated SNPs within this region (rs493767 and rs550659, r2 = 0.961) and A-SAA levels was also borderline significant (p = 8.50×10−3 and p = 1.65×10−2, respectively). No significant differences between men and women were found within the regions identified in the overall meta-analysis (data not shown).
Discussion
Based on a meta-analysis of four genome-wide association studies including 4,212 participants of European descent two novel genetic susceptibility regions were identified to be associated with baseline A-SAA concentrations. With 11.68% explained variance in our data, which makes up 19.76% of the total estimated heritability of 59%, these two regions seem to have a major impact on baseline A-SAA concentrations. The region at 11p15.5-p13 accounts for most of the explained variance. Its SAA1 subregion contains part of a highly conserved region between the two bona fide acute-phase structure genes SAA1 and SAA2, which consist of at least 5 and 2 allelic variants, respectively [1], [24]. These two genes are concurrently induced during the acute-phase [1], and cluster within 18 kb of each other in a head to head arrangement [25]. This study is the first presenting the complex genetic architecture of A-SAA levels at this locus. In the identified region, there has been evidence of regulatory elements like C/EBPalpha and C/EBPbeta (http://genome.ucsc.edu), which are necessary for the full responsiveness to IL-1β and IL-6 either alone or in combination [1]. Our finding underlines the high functional potential for this region.
The adjacent GTF2H1 is a basal transcription factor involved in nucleotide excision repair of DNA and RNA transcription by RNA polymerase II [26]. HPS5 encodes a protein, which is probably involved in organelle biogenesis associated with melanosomes and platelet dense granule, its mutations lead to a homonymous clinical entity [27]. And LDHA and LDHC, which are expressed in muscle tissue and in testes, respectively, encode for lactate dehydrogenase, an enzyme that catalyzes the interconversion of lactate and pyruvate [28].
Variants of the GTF2H1 gene have been recently found to be associated with lung cancer in a Chinese population [29]. Furthermore, it was demonstrated that LDHA is involved in tumour genecity and its reduction causes bioenergetic and oxidative stress leading to cell death [30]–[32]. Finally, Kosolowski et al. [33] found LDHC to be expressed in several types of tumour cell lines. It is thought, that recurrent or persistent chronic inflammation may play a role in carcinogenesis by causing DNA damage, inciting tissue reparative proliferation and/or by creating an environment that is enriched with tumour-promoting cytokines and growth factors [12]. Furthermore, SAA synthesis could be found in human carcinoma metastases and cancer cell lines [17].
As the approach taken in this study is observational in nature it is not possible to draw causal inferences. For that reason, it could be possible that not genes, but small regulatory elements may be responsible for the findings. This is most likely the case as the identified region contains one structure gene and the adjacent region. In any case, the major impact on baseline A-SAA concentrations demonstrates a key role of the 11p15.5-p13 region in the regulation of inflammation. Therefore, the identification of causal variants and their impact on diseases related to elevated baseline A-SAA concentrations represent promising targets for future functional and epidemiological studies.
The second region was found on chromosome 1p31, harbouring the LEPR gene locus. Leptin, an important circulating signal for the regulation of body weight, was found to be correlated with SAA concentrations independently of BMI, and both were expressed in adipose tissue [34]. In the KORA F3 study (Text S1) a moderate but significant correlation was found between circulating A-SAA and leptin concentrations in blood in 181 participants with measurements of both proteins (Spearman correlation = 0.25, p = 7×10−4). So far it is unclear whether leptin influences SAA expression directly or via the leptin stimulated cytokines, IL-6 and TNF-α [34]. LEPR is a single transmembrane receptor of the cytokine receptor family most related to the gp130 signal-transducing component of the IL-6 receptor, the granulocyte colony-stimulating factor (GCSF) receptor, and the Leukaemia Inhibitory Factor (LIF) receptor, all of which are thought to play an essential role in the inflammatory process [35], [36]. Previous studies have provided evidence of an association of the LEPR gene locus with CRP and fibrinogen [37]–[40], which were both correlated with A-SAA in the KORA S4 study (Text S1) (CRP: Spearman correlation = 0.58, p = 3.22×10−155, and fibrinogen: Spearman correlation = 0.31, p = 3.89×10−41; N = 1734). The finding gives confirmative evidence of the importance of the LEPR gene locus for inflammatory processes and the close relationship between leptin, A-SAA, CRP and fibrinogen.
Furthermore, in the gender stratified analysis one region containing SERGEF was identified to be presumably associated with A-SAA in men. The adjacency of this identified region to the SAA gene family suggests that regulatory elements may be responsible for this signal. However, the association with A-SAA levels was only borderline significant in our study and therefore awaits replication.
Two limitations of our study have to be mentioned. Firstly, due to the restrictions in laboratory methods our analyses were confined to the A-SAA isoforms and did not capture the constitutively expressed C-SAA isoform which might also be of interest, especially when analyzing baseline SAA levels. Secondly, the number of studies with genome-wide data and measured A-SAA levels was limited compared to other genome-wide association studies. Nevertheless, the study had enough power to detect two novel genetic susceptibility regions for A-SAA which explain 19.76% of the total estimated heritability already. Furthermore, results were consistent across all four studies and within different genotyping platforms, the regions are biologically highly plausible, and the results may contribute to future research on the regulation of inflammatory response and its role in related clinical entities.
Taken together, the present meta-analysis is the first whole genome approach to identify genetic variants that are associated with baseline A-SAA concentrations. Two novel genetic susceptibility regions were identified to be associated with baseline A-SAA concentrations. The findings demonstrate a major impact of the 11p15.5-p13 gene region on the regulation of inflammation and suggest a close interplay between leptin, A-SAA, and other acute-phase proteins as well as a larger role of the LEPR gene locus in inflammatory processes as it has been assumed in the past.
Materials and Methods
Participating studies
The present meta-analysis combined data from four genome-wide scans: one survey of the Cooperative Health Research in the Region of Augsburg (KORA S4), the Ludwigshafen Risk and Cardiovascular Health study (LURIC), the UK Adult Twin Register (TwinsUK) and a self-contained population from Eastern Germany (Sorbs) (). Approval was obtained by each of the local Ethic Committees for all studies and written informed consent was given by all study participants. In total, the meta-analysis included 4,212 individuals (1,928 males, 2,284 females) of European ancestry with measured baseline A-SAA concentrations. For validation analyses we used data of 2,136 participants of the KORA S4 sample, which were not included in the meta-analysis (Text S1). Sample sizes and characteristics of the study participants of the four genome-wide scans and the validation sample are displayed in Table S6.
Measurement of A-SAA concentrations
In all four studies, study participants were fasting and EDTA plasma samples were analyzed by immunonephelometry on a BNAII device from Siemens, Germany, and well-validated automated microparticle capture enzyme immunoassays [10], [41]. The inter-assay coefficients of variation were below 7% in all four studies.
Genome-wide genotyping and imputation
For genotyping different platforms as the Affymetrix 500K GeneChip array (Sorbs), Affymetrix 6.0 GeneChip array (KORA S4, LURIC, Sorbs), Illumina HumanHap300 BeadChip (317K) (TwinsUK) and Illumina Human 610K BeadChip (TwinsUK) were used. Quality control before imputation was undertaken in each study separately. Detailed information on genotyping and imputation is reported in Table S7. Imputation based on the HapMap Phase 2 CEU population was performed using IMPUTE [42] in all studies. After imputation all genotype data had to meet the following quality criteria: a minor allele frequency ≥0.01, a call rate per SNP ≥0.9, and r2.hat metrics ≥0.40 for imputed SNPs. In total, 2,593,456 genotyped or imputed autosomal SNPs were analyzed in the meta-analysis.
For validation and comparison of genotyping platforms, we selected 27 of the most significantly associated SNPs. Genotyping of these SNPs was performed with the MassARRAY system using the iPLEX technology (Sequenom, San Diego, CA) in the KORA S4 study. The allele-dependent primer extension products were loaded onto one 384-element chip using a nanoliter pipetting system (SpectroCHIP, Spectro-POINT Spotter; Sequenom), and the samples were analyzed by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (Bruker Daltonik, Leipzig, Germany). The resulting mass spectra were analyzed for peak identification via the SpectroTYPER RT 3.4 software (Sequenom). To control for reproducibility, 9.8% of samples was genotyped in duplicate with a discordance rate of less than 0.5%.
Genome-wide association analyses and meta-analysis
In each study, linear regression models for all available SNPs have been calculated on ln-transformed A-SAA levels in mg/l. The genetic effect has been assumed to be additive. Adjustment has been made for age, gender, BMI, and study specific covariates, i.e. the Friesinger Score in the LURIC population [43] and a genotyping batch variable in the TwinsUK population. Additionally, this analysis was undertaken stratified by gender. The genome-wide scans were calculated with the analysis software SNPTEST (http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html) (KORA S4, LURIC) QUICKTEST (http://toby.freeshell.org/software/quicktest.shtml) (Sorbs) and Merlin (http://www.sph.umich.edu/csg/abecasis/Merlin/) (TwinsUK).
The results of all four genome-wide scans were meta-analysed using a fixed-effects model applying inverse variance weighting with the METAL software (www.sph.umich.edu/csg/abecasis/metal). Study specific results were corrected for population stratification using the genomic control method. For the overall meta-analysis, the inflation factor was 1.009. No further correction was applied.
P-values below the threshold of p = 5×10−8, which corresponds to a Bonferroni correction for the estimated number of one million tests for independent common variants in the human genome of European individuals [44], were considered to be significant.
As a measure for between study heterogeneity I2 was calculated [45]. Deviations from Hardy-Weinberg-Equilibrium were tested for all identified SNPs by means of the exact Hardy Weinberg test. For the calculation of explained variances, we subtracted the multiple R2 value of the covariate model from those of the full model including covariates and top hits of the loci in every single study and assessed the weighted mean (KORA S4, LURIC, and the Sorbs). We tested adjacent regions for independency by analyzing the significance of their top SNPs in a joint model.
Accession numbers
The OMIM (http://www.ncbi.nlm.nih.gov/omim) accession numbers for genes mentioned in this article are 104750 for SAA1, 607521 for HPS5, 189972 for GTF2H1, 150000 for LDHA, 150150 for LDHC, 601007 for LEPR, and 606051 for SERGEF.
Supporting Information
List of all significantly associated SNPs of the meta-analysis.
(0.05 MB PDF)
Genotypic mean levels.
(0.02 MB PDF)
Comparison between different genotyping technologies in the KORA study.
(0.09 MB PDF)
Analysis of the structure of the chromosome 11 region.
(0.04 MB PDF)
Results of the validation analyses.
(0.04 MB PDF)
Study characteristics of the four studies of the meta-analysis and the validation sample.
(0.04 MB PDF)
Study specific information on genotyping and imputation.
(0.06 MB PDF)
Study descriptions.
(0.08 MB PDF)
Acknowledgments
The KORA authors acknowledge the contribution of Peter Lichtner, Gertrud Eckstein, Guido Fischer, Norman Klopp, Nicole Spada, and all members of the Helmholtz Zentrum München genotyping staff for generating the SNP data, as well as all members of field staffs who were involved in the planning and conduction of the KORA Augsburg studies. The KORA authors also want to thank Jens Baumert and Eva Lattka from the Helmholtz Zentrum München for their assistance.
The TwinsUK authors acknowledge the support of the National Eye Institute via an NIH/CIDR genotyping project (PI: Terri Young) and the staff from the Genotyping Facilities at the Wellcome Trust Sanger Institute led by Leena Peltonen and Panos Deloukas for sample preparation, quality control, and genotyping; Le Centre National de Génotypage, France, led by Mark Lathrop, for genotyping; Duke University, North Carolina, USA, led by David Goldstein, for genotyping; and the Finnish Institute of Molecular Medicine, Finnish Genome Center, University of Helsinki, led by Aarno Palotie.
The Sorbs authors would like to thank Knut Krohn from the Microarray Core Facility of the Interdisciplinary Centre for Clinical Research (IZKF), University of Leipzig, Germany, for his excellent genotyping support and Nigel William Rayner from the Wellcome Trust Centre for Human Genetics, University of Oxford, UK, as well as John Broxholm from the Bioinformatics Core Unit of the Wellcome Trust Centre for Human Genetics for their excellent bioinformatics support.
Footnotes
The authors have declared that no competing interests exist.
The KORA research platform was initiated and financed by the Helmholtz Center Munich, German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Part of this work was financed by the German National Genome Research Network (NGFN-2, NGFNPlus 01GS0823, and NGFNPlus 01GS0834) and through additional funds from the University of Ulm. Our research was supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ and by a grant from the BMBF to the German Center for Diabetes Research (DZD e.V.), as well as from the BMBF funded German Network for Mitochondrial Disorders (mitoNET 01GM0862) and Systems Biology of Metabotypes (SysMBo 0315494A). Furthermore, the study received funding from the European Community's Seventh Framework Programme (FP7/2007–2013), ENGAGE project, grant agreement HEALTH-F4-2007-201413. The LURIC part of this study has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 201668; AtheroRemo. It was further supported by the Centre of Excellence “Metabolic disorder” Baden-Württemberg, Ulm, University Ulm, Germany. The Sorbs part of this study was supported by grants from the Interdisciplinary Centre for Clinical Research at the University of Leipzig (B27; N06) and from the German Diabetes Association as well as by a grant from the DFG (KFO 152). Furthermore, the authors received funding through the European Community's Seventh Framework Programme (FP7/2007–2013), ENGAGE project, grant agreement HEALTH-F4-2007- 201413. The TwinsUK study was funded by the Wellcome Trust, the EU MyEuropia Marie Curie Research Training Network, the European Community's Seventh Framework Programme (FP7/2007–2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and (FP7/2007–2013), ENGAGE project grant agreement HEALTH-F4-2007-201413, and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also received support from the Dept of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London and by the WT Grant n. 091746/Z/10/Z. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (G20234). Genotyping was in part performed by CIDR as part of an NEI/NIH project grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, He X, Shi X, Huang C, Liu J, et al. Association between serum amyloid A and obesity: a meta-analysis and systematic review. Inflamm Res. 2010;59:323–334. doi: 10.1007/s00011-010-0163-y. [DOI] [PubMed] [Google Scholar]
- 3.Schillinger M, Exner M, Mlekusch W, Sabeti S, Amighi J, et al. Inflammation and Carotid Artery—Risk for Atherosclerosis Study (ICARAS). Circulation. 2005;111:2203–2209. doi: 10.1161/01.CIR.0000163569.97918.C0. [DOI] [PubMed] [Google Scholar]
- 4.Johnson BD, Kip KE, Marroquin OC, Ridker PM, Kelsey SF, et al. Serum amyloid A as a predictor of coronary artery disease and cardiovascular outcome in women: the National Heart, Lung, and Blood Institute-Sponsored Women's Ischemia Syndrome Evaluation (WISE). Circulation. 2004;109:726–732. doi: 10.1161/01.CIR.0000115516.54550.B1. [DOI] [PubMed] [Google Scholar]
- 5.Maier W, Altwegg LA, Corti R, Gay S, Hersberger M, et al. Inflammatory markers at the site of ruptured plaque in acute myocardial infarction: locally increased interleukin-6 and serum amyloid A but decreased C-reactive protein. Circulation. 2005;111:1355–1361. doi: 10.1161/01.CIR.0000158479.58589.0A. [DOI] [PubMed] [Google Scholar]
- 6.Fyfe AI, Rothenberg LS, DeBeer FC, Cantor RM, Rotter JI, et al. Association between serum amyloid A proteins and coronary artery disease: evidence from two distinct arteriosclerotic processes. Circulation. 1997;96:2914–2919. doi: 10.1161/01.cir.96.9.2914. [DOI] [PubMed] [Google Scholar]
- 7.Danesh J, Whincup P, Walker M, Lennon L, Thomson A, et al. Low grade inflammation and coronary heart disease: prospective study and updated meta-analyses. BMJ. 2000;321:199–204. doi: 10.1136/bmj.321.7255.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridker PM, Rifai N, Pfeffer MA, Sacks FM, Moye LA, et al. Inflammation, pravastatin, and the risk of coronary events after myocardial infarction in patients with average cholesterol levels. Cholesterol and Recurrent Events (CARE) Investigators. Circulation. 1998;98:839–844. doi: 10.1161/01.cir.98.9.839. [DOI] [PubMed] [Google Scholar]
- 9.Kinlay S, Schwartz GG, Olsson AG, Rifai N, Szarek M, et al. Myocardial Ischemia Reduction with Aggressive Cholesterol Lowering (MIRACL) Study Investigators. Inflammation, statin therapy, and risk of stroke after anacute coronary Syndrome in the MIRACL study. Arterioscler Thromb Vasc Biol. 2008;28:142–147. doi: 10.1161/ATVBAHA.107.151787. [DOI] [PubMed] [Google Scholar]
- 10.Müller S, Martin S, Koenig W, Hanifi-Moghaddam P, Rathmann W, et al. Impaired glucose tolerance is associated with increased serum concentrations of interleukin 6 and co-regulated acute-phase proteins but not TNF-α or its receptors. Diabetologia. 2002;45:805–812. doi: 10.1007/s00125-002-0829-2. [DOI] [PubMed] [Google Scholar]
- 11.Yang R-Z, Lee M-J, Hu H, Pollin TI, Ryan AS, et al. Acute-phase serum amyloid A: an inflammatory adipokine and potential link between obesity and its metabolic complications. PLoS Med. 2006;3:e287. doi: 10.1371/journal.pmed.0030287. doi: 10.1371/journal.pmed.0030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malle E, Sodin-Semrl S, Kovacevic A. Serum amyloid A: an acute-phase protein involved in tumour pathogenesis. Cell Mol Life Sci. 2009;66:9–26. doi: 10.1007/s00018-008-8321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sellar GC, Jordan SA, Bickmore WA, Fantes JA, van Heyningen V, et al. The human serum amyloid A protein (SAA) superfamily gene cluster: mapping to chromosome 11p15.1 by physical and genetic linkage analysis. Genomics. 1994;19:221–227. doi: 10.1006/geno.1994.1051. [DOI] [PubMed] [Google Scholar]
- 14.Sellar GC, Oghene K, Boyle S, Bickmore WA, Whitehead AS. Organization of the region encompassing the human serum amyloid A (SAA) gene family on chromosome 11p15.1. Genomics. 1994;23:492–495. doi: 10.1006/geno.1994.1530. [DOI] [PubMed] [Google Scholar]
- 15.Thorn CF, Lu ZY, Whitehead AS. Regulation of the human acute-phase serum amyloid A genes by tumour necrosis factor-α, interleukin-6 and glucocorticoids in hepatic and epithelial cell lines. Scand J Immunol. 2004;59:152–158. doi: 10.1111/j.0300-9475.2004.01369.x. [DOI] [PubMed] [Google Scholar]
- 16.Jensen LE, Whitehead AS. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem J. 1998;334:489–503. doi: 10.1042/bj3340489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol. 2000;7:64–69. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 18.MacGregor AJ, Gallimore JR, Spector TD, Pepys MB. Genetic effects on baseline values of C-reactive protein and serum amyloid a protein: a comparison of monozygotic and dizygotic twins. Clin Chem. 2004;50:130–134. doi: 10.1373/clinchem.2003.028258. [DOI] [PubMed] [Google Scholar]
- 19.Yamada T, Wada A, Itoh Y, Itoh K. Serum amyloid A1 alleles and plasma concentrations of serum amyloid A. Amyloid. 1999;6:199–204. doi: 10.3109/13506129909007327. [DOI] [PubMed] [Google Scholar]
- 20.Booth DR, Booth SE, Gillmore JD, Hawkins PN, Pepys MB. SAA1 alleles as risk factors in reactive systemic AA amyloidosis. Amyloid. 1998;5:262–265. doi: 10.3109/13506129809007299. [DOI] [PubMed] [Google Scholar]
- 21.Bakkaloglu A, Duzova A, Ozen S, Balci B, Besbas N, et al. Influence of Serum Amyloid A (SAA1) and SAA2 gene polymorphisms on renal amyloidosis, and on SAA/C-reactive protein values in patients with familial mediterranean fever in the Turkish population. J Rheumatol. 2004;31:1139–1142. [PubMed] [Google Scholar]
- 22.Sjöholm K, Lundgren M, Olsson M, Eriksson JW. Association of serum amyloid A levels with adipocyte size and serum levels of adipokines: differences between men and women. Cytokine. 2009;48:260–266. doi: 10.1016/j.cyto.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Thorand B, Baumert J, Döring A, Herder C, Kolb H, et al. Sex differences in the relation of body composition to markers of inflammation. Atherosclerosis. 2006;184:216–224. doi: 10.1016/j.atherosclerosis.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Sipe J. Revised nomenclature for serum amyloid A (SAA). Nomenclature Committee of the International Society of Amyloidosis. Part 2. Amyloid. 1999;6:67–70. doi: 10.3109/13506129908993291. [DOI] [PubMed] [Google Scholar]
- 25.Kluve-Beckerman B, Song M. Genes encoding human serum amyloid A proteins SAA1 and SAA2 are located 18 kb apart in opposite transcriptional orientations. Gene. 1995;159:289–290. doi: 10.1016/0378-1119(95)00027-4. [DOI] [PubMed] [Google Scholar]
- 26.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 27.Huizing M, Hess R, Dorward H, Claassen DA, Helip-Wooley A, et al. Cellular, molecular and clinical characterization of patients with Hermansky-Pudlak syndrome type 5. Traffic. 2004;5:711–722. doi: 10.1111/j.1600-0854.2004.00208.x. [DOI] [PubMed] [Google Scholar]
- 28.Cori CF. The glucose-lactic acid cycle and gluconeogenesis. Curr Top Cell Regul. 1981;18:377–387. [PubMed] [Google Scholar]
- 29.Wu W, Liu H, Lei R, Chen D, Zhang S, et al. Genetic variants in GTF2H1 and risk of lung cancer: a case-control analysis in a Chinese population. Lung Cancer. 2008;63:180–186. doi: 10.1016/j.lungcan.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Xie H, Valera VA, Merino MJ, Amato AM, Signoretti S, et al. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol Cancer Ther. 2009;8:626–635. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koslowski M, Türeci O, Bell C, Krause P, Lehr HA, et al. Multiple splice variants of lactate dehydrogenase C selectively expressed in human cancer. Cancer Res. 2002;62:6750–6755. [PubMed] [Google Scholar]
- 34.Lappalainen T, Kolehmainen M, Schwab U, Pulkkinen L, Laaksonen DE, et al. Serum concentrations and expressions of serum amyloid A and leptin in adipose tissue are interrelated: the Genobin Study. Eur J Endocrinol. 2008;158:333–341. doi: 10.1530/EJE-07-0598. [DOI] [PubMed] [Google Scholar]
- 35.Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 36.Tartaglia LA. The leptin receptor. J Biol Chem. 1997;272:6093–6096. doi: 10.1074/jbc.272.10.6093. [DOI] [PubMed] [Google Scholar]
- 37.Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YY, Gottardo L, Mlynarski W, Frazier W, Nolan D, et al. Genetic variability at the leptin receptor (LEPR) locus is a determinant of plasma fibrinogen and C-reactive protein levels. Atherosclerosis. 2007;191:121–127. doi: 10.1016/j.atherosclerosis.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 39.Ridker PM, Pare G, Parker A, Zee RY, Danik JS, et al. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am J Hum Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkins J, Gallimore JR, Tennent GA, Hawkins PN, Limburg PC, et al. Rapid automated enzyme immunoassay of serum amyloid A. Clin Chem. 1994;40:1284–1290. [PubMed] [Google Scholar]
- 42.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 43.Friesinger GC, Page EE, Ross RS. Prognostic significance of coronary arteriography. Trans Assoc Am Physicians. 1970;83:78–92. [PubMed] [Google Scholar]
- 44.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 45.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all significantly associated SNPs of the meta-analysis.
(0.05 MB PDF)
Genotypic mean levels.
(0.02 MB PDF)
Comparison between different genotyping technologies in the KORA study.
(0.09 MB PDF)
Analysis of the structure of the chromosome 11 region.
(0.04 MB PDF)
Results of the validation analyses.
(0.04 MB PDF)
Study characteristics of the four studies of the meta-analysis and the validation sample.
(0.04 MB PDF)
Study specific information on genotyping and imputation.
(0.06 MB PDF)
Study descriptions.
(0.08 MB PDF)