Abstract
Hybrid sterility of the heterogametic sex is one of the first postzygotic reproductive barriers to evolve during speciation, yet the molecular basis of hybrid sterility is poorly understood. We show that the hybrid male sterility gene Odysseussite homeobox gene (OdsH) encodes a protein that localizes to evolutionarily dynamic loci within heterochromatin and leads to their decondensation. In D. mauritiana X D. simulans male hybrids, OdsH from D. mauritiana (OdsHmau) acts as a sterilizing factor by associating with the heterochromatic Y chromosome of D. simulans, whereas D. simulans OdsH (OdsHsim) does not. Characterization of sterile hybrid testes revealed that OdsH abundance and localization in the premeiotic phases of spermatogenesis differs between species. These results reveal that rapid heterochromatin evolution affects the onset of hybrid sterility.
Keywords: D. mauritiana, D. simulans, Dobzhansky-Muller incompatibilities, heterochromatin, speciation
One facet of speciation studies is focused on mechanisms by which populations become reproductively isolated (1). Intrinsic postzygotic isolation is a reproductive isolating event resulting in the sterility or lethality of the F1 hybrid offspring following a successful fertilization event and the formation of the zygote (2). The Dobzhansky-Muller model (reviewed in ref. 2) proposes that such reproductive barriers occur due to incompatibilities between genetic loci arising as a by-product of divergence between two populations. The identification of loci involved in F1 hybrid sterility in the heterogametic sex (XY males or ZW females) is of particular interest, as this defect is postulated to be the earliest postzygotic isolation event to arise between incipient species (2, 3). Yet, the biological basis of the defects that result in hybrid sterility remain largely unknown.
Crosses between D. simulans and D. mauritiana, which separated ~250,000 years ago (9), produce sterile F1 hybrid males and fertile females. A series of introgressions of the D. mauritiana genome into a D. simulans genomic background revealed that the interaction of the D. mauritiana X chromosome-encoded OdsH (OdsHmau) protein with the male D. simulans genome resulted in F1 male hybrid sterility (4, 8, 10, 11). Considerable amino acid divergence was observed between OdsHsim and OdsHmau, especially within the putative DNA-binding homeodomain (16 non-synonymous and 3 synonymous changes) (4). Homeodomains are characteristic of a well-conserved family of transcription factors regulating early developmental patterning (12). OdsH evolved approximately 25 million years ago from a gene-duplication of unc-4 (13), which encodes a transcription factor that has somatic function in Drosophila (15).
OdsH expression in the testes (8) and its evolutionary descent from unc-4 (13) led to the proposal that OdsH encodes a transcription factor whose introduction into the hybrid background causes mis-expression of meiotic genes, and, therefore, hybrid sterility (16). However, this model fails to account for how the protein-DNA interaction interface may drive the changes observed in the OdsH homeodomain between Drosophila species. Ablation of the OdsH gene in D. melanogaster had only modest effects on male fertility (8) contrary to expectations that a deletion of OdsH would affect male fertility due to the misregulation of meiotic genes.
An alternative model suggests that evolutionary labile satellite DNAs – found in pericentric, telomeric and other heterochromatic regions – may result in the divergence of speciation genes (17, 18). Under this model, satellite DNAs and their expansions are perpetuated by female meiotic drive, but affect fitness through reductions in male fertility, which is evident in plant and animal species (19, 20). The evolution of satellite DNA-binding proteins is predicted to be one way to mitigate cost to male fertility and ensure species survival (17, 18). Therefore, we considered the alternative possibility that hybrid sterility genes like OdsH encode proteins that bind to satellite DNA repeats in pericentric or telomeric regions. Under this model, hybrid sterility could result from an inability to correctly package and condense heterochromatin.
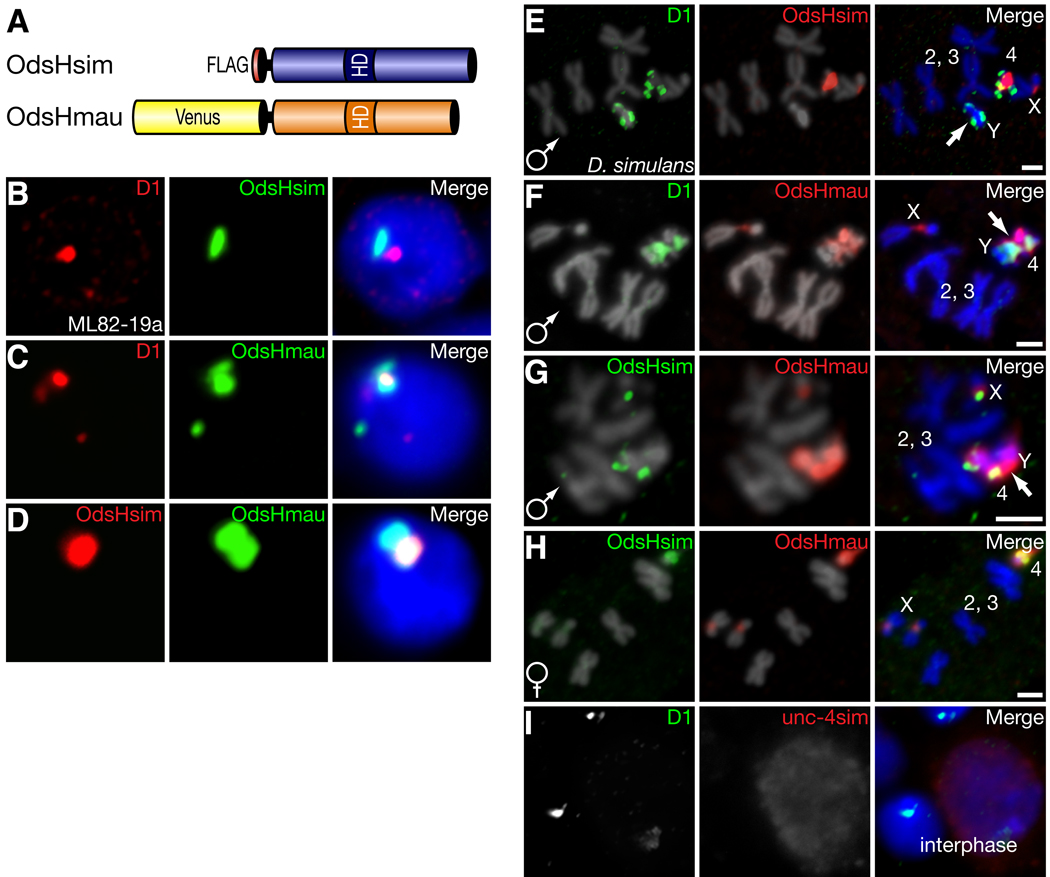
To distinguish between euchromatic versus heterochromatic localization, we expressed OdsHsim fused to a 3XFLAG epitope in a D. simulans embryonic cell culture line (Fig. 1A, B). We observed a punctate localization pattern of OdsHsim in interphase cells —reminiscent of the D1 satellite-binding protein (21). In D. simulans, D1 predominantly localizes to repetitive satellite sequences on the Y and 4th chromosomes (Fig. S1) providing a cytological marker relative to OdsH localization. On this basis OdsHsim localized adjacent to D1 in D. simulans cells (Fig. 1B). However, the localization of OdsHmau protein (fused to Venus, yellow fluorescence protein) partially overlapped with D1 (Fig. 1A, C). Co-expression of the OdsHsim and OdsHmau fusion proteins revealed that the two proteins localize to a common site, but that OdsHmau has additional localization (Fig. 1D).
Fig. 1. OdsH proteins differ in their localization to D. simulans heterochromatin.
We use D1 staining as (Fig. S1). (A) FLAG-OdsHsim or Venus-OdsHmau epitope-tagged proteins were expressed in transiently transfected D. simulans cultured cells (B–D) or in transgenic D. simulans larval neuroblasts (E–H) under the control of a heat-shock promoter. (B,C) D1 staining (red) is a cytological landmark for localization of OdsHsim (B) and OdsHmau (C) proteins (both shown in green) to D. simulans heterochromatin. DNA staining by DAPI shown in blue in merge. (D) Co-expression of OdsHsim (red) and OdsHmau (green). (E) OdsHsim (red) localizes to the X chromosome and adjacent to D1 staining (green) on the 4th chromosome but not to the Y chromosome. (F) OdsHmau (red) localization has additional localization to the Y chromosome. (G) Co-expression of OdsHsim (green) and OdsHmau (red) on male mitotic chromosomes versus (H) female mitotic chromosomes. Arrows in E–G highlight Y chromosomes. (I) GFP-Unc-4 (red) staining in interphase larval neuroblast cells of D. simulans is diffuse and does not overlap specifically with D1 (green). Scale bars represent 2 µm.
We mapped the chromosomal localization of tagged OdsHsim and OdsHmau proteins with N-terminal 1XFLAG or Venus, respectively. Expression and immunofluorescent detection of OdsHsim in mitotic larval neuroblast cells confirmed that OdsHsim was associated with repeat-rich regions of the D. simulans genome, namely the X pericentric region and the 4th chromosome (Fig. 1E). OdsHmau localized similarly on the D. simulans X and 4th chromosomes but showed gross localization to the D. simulans Y chromosome (Fig. 1F). Co-expression of OdsHsim and OdsHmau showed the additional localization of OdsHmau to the Y chromosome in male cells (Fig. 1G) whereas these two proteins localized identically in female cells (Fig. 1H). In Drosophila, both the Y and 4th chromosomes are principally heterochromatic, gene-poor and repeat-rich (22). On the basis of these results we conclude that both OdsHsim and OdsHmau encode heterochromatin-binding proteins but that they have different localization specificities resulting in altered localization to the D. simulans Y chromosome.
Because OdsH shares ancestry with a transcription factor, unc-4, we expressed a green fluorescence protein (GFP) tagged unc-4 protein to contrast its localization with that of the OdsH protein. As expected for a transcription factor, unc-4 showed diffuse staining in interphase neuroblast cells (Fig. 1I). Hence, the OdsH protein may have gained specificity for localization to heterochromatin since it diverged from unc-4.
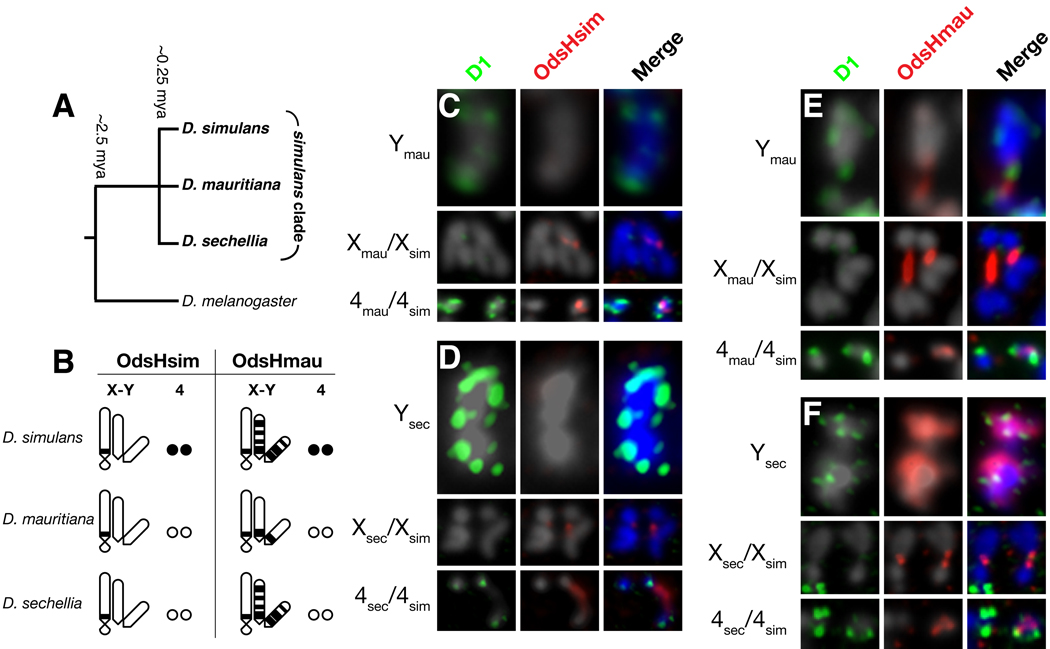
We investigated whether divergence of the underlying satellite-DNAs were associated with changes in OdsH binding specificity. We identified targets of OdsH binding in sibling species D. mauritiana or D. sechellia (Figs. 2A, S2) by crossing transgenic D. simulans lines expressing fusion proteins of either OdsHsim or OdsHmau, to these species. By examining localization in male and female hybrids, we found altered localization of OdsHsim and OdsHmau on the Y and 4th chromosomes of sister species (Figs. 2B–F, S3). The D. sechellia and D. simulans Y chromosomes were enriched for OdsHmau binding, whereas OdsHmau localization to its own Y chromosome was restricted (Fig. 2E, F). In contrast, OdsHsim did not associate with any of the three Y chromosomes (Fig. 2C, D). Furthermore, OdsHsim and OdsHmau bound to the 4th chromosome from D. simulans, but not from D. sechellia or D. mauritiana (Fig. 2B–F). Thus, the localization of OdsH proteins differed on homologous chromosomes from different Drosophila species, suggesting that there has been a reorganization or wholesale replacement of OdsH-binding sites within heterochromatin on both the Y and 4th chromosomes within the past 250,000 years (summarized in Fig. 2B). These rapid changes in heterochromatin mirror the rapid evolution of OdsH’s homedodomain during this span (4).
Fig. 2. OdsH-binding sites are evolutionary labile in sibling species.
(A) A schematic phylogeny of the recently diverged D. simulans, D. sechellia and D. mauritiana species. (B) Summary of localization studies (C–F) highlighting the differences in genomic localization of OdsHsim and OdsHmau to the Y and 4th chromosomes. (C) Localization of FLAG-OdsHsim fusion protein on neuroblast mitotic chromosomes from D. mauritiana/D. simulans and (D) D. sechellia/D. simulans hybrid larvae (Fig. S2). (E) Localization of Venus-OdsHmau fusion protein on neuroblast mitotic chromosomes from D. mauritiana/D. simulans and (F) D. sechellia/D. simulans hybrid larvae. In all cases OdsH staining is red and D1 staining green. Only relevant X, Y, and 4th chromosomes are shown.
Localization of OdsH appears to dramatically affect local chromosome condensation. For instance, in D. simulans neuroblast nuclei expressing OdsHmau the general decondensation and intercalation of Y and 4th chromosomes obscured our ability to distinguish these chromosomes from each other (Figs. 1F, G, S4). This is in contrast to nuclei expressing only OdsHsim in which the Y chromosome is condensed (Fig. 1E). Decondensation of the X and 4th chromosomes was also associated with the localization of either OdsHsim or OdsHmau (Fig. 2D–F). Decondensation was seen for both OdsHsim and OdsHmau and in all Drosophila genomes assayed (Fig. S4). It appears that the retention of an ancestral unc-4-like transcriptional function coupled with its localization to heterochromatic regions that are otherwise condensed, has resulted in OdsH-mediated chromosome decondensation in hybrids.
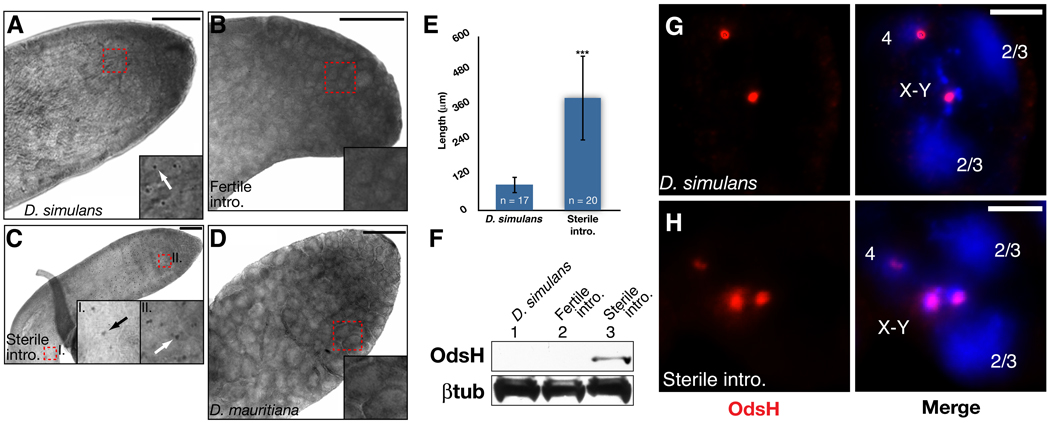
To confirm the localization of OdsH in D. simulans cell culture and mitotic chromosomes, we characterized endogenous OdsH localization in the D. simulans testis with an antibody raised to the C-terminus of OdsHsim (Fig. S5). Whole-mount immunohistochemistry (IHC) of OdsH in wildtype D. simulans testes showed that OdsH is restricted to developing post-mitotic primary spermatocytes in the G2 phase (Figs. 3A, S7, S8). Within these spermatocytes, immunofluorescence revealed two punctate dots, consistent with OdsHsim localization to only two major loci within the D. simulans genome (Fig. 3G). At this stage of spermatogenesis, homologous chromosomes arrange into individual chromatin domains associated with the nuclear membrane (23). Typically, two large hazy domains represent the associated 2nd and 3rd chromosomes, respectively, while the other two domains represent the associated X-Y and 4th chromosomes (23). The presence of OdsH in the latter chromatin domains suggests that endogenous OdsH binds to loci on the X and 4th chromosomes, consistent with our observations that OdsHsim binds to the X and 4th chromosomes of D. simulans.
Fig. 3. Endogenous OdsH localization to G2 spermatocyte nuclei.
(A) Whole-mount immunohistochemistry with an OdsH antibody on adult male testes from D. simulans wildtype, (B) D. simulans fertile introgression, (C) D. simulans sterile introgression, and (D) D. mauritiana. Scale bars represent 50 µm. An 8X zoom on the regions identified by the red dashed boxes is shown in the insets along with arrows pointing to OdsH protein. (E) OdsH protein expression was significantly expanded in the sterile line, measured by the length of detectable protein expression within the testes (p < 0.001). (F) Western blot from D. simulans wildtype, fertile introgression, and sterile introgression testes immunoblotted for OdsH (top panel) and β-tubulin control (bottom panel). (G) OdsH (red) detected by immunofluorescence in G2 primary spermatocyte nuclei from the adult testes of D. simulans wildtype and (H) D. simulans sterile introgression. DNA staining in blue. Scale bars represent 5 µm.
The antibody we raised to OdsHsim also recognizes OdsHmau with high specificity (Fig. S5). Therefore, we investigated OdsH localization using whole-mount IHC on the testes of a D. simulans male sterile line, containing an introgression of the OdsH-containing region from the D. mauritiana X chromosome (Figs. 3C, S6) (4). Immunofluorescence of OdsH confirmed the localization of OdsHmau to three loci corresponding to the X, Y, and 4th chromosomes within these primary spermatocytes (Fig. 3H). We also observed an expansion of OdsH protein localization in late-G2 primary spermatocytes of the sterile introgression line as compared to wild type lines (Fig. 3E) suggesting an increase in protein expression or stability. Western blot analysis confirmed that OdsH protein levels are increased in the sterile introgression testes (Fig. 3F). Despite the presence of OdsH protein at this late stage of primary spermatocyte development, we did not observe OdsH protein in spermatocytes that have progressed from G2 to meiotic prophase or a delay in the onset of meiosis (Fig. S8).
A fertile introgression line differing from the sterile line in that exons 3 and 4 of OdsH are derived from D. simulans (Fig. S6) (4) showed no OdsH expression, both by IHC and by Western analyses (Fig. 3B, F). In addition, we observed no OdsHmau expression in the testes of D. mauritiana males (Fig. 3D). Our characterization of endogenous OdsH levels and localization reveal differences between both sibling Drosophila species and the sterile and fertile D. simulans introgression lines. While it is still unclear how the additional binding capacity of OdsHmau adversely affects premeiotic stages of sperm development to cause male sterility, there is precedent for premeiotic defects manifesting in post-meiotic dysfunction in Drosophila and mice (24, 25, 26).
Since the studied introgression lines only differ at their OdsH locus, OdsHmau is unambiguously linked to the hybrid male sterility phenotype (8). Our findings reveal the genetic architecture underlying OdsH-mediated hybrid sterility. Both D. mauritiana and fertile introgressed D. simulans males express no OdsH protein, yet are completely fertile, consistent with the fact that OdsH function is not required for male fertility in D. melanogaster (8). Thus OdsHmau-mediated hybrid sterility involves both the unusual expression and retention of OdsHmau protein in the D. simulans testis, as well as an unusual localization and possibly decondensation of the D. simulans Y chromosome. We conclude on the basis of these data that hybrid male sterility is caused by a gain-of-function interaction between OdsHmau and some component of the D. simulans Y chromosome heterochromatin, with this protein-DNA interaction representing the Dobzhansky-Muller incompatibility.
OdsH shares similarities with the hybrid sterility genes Prdm9 (or Meisetz) in mouse (7) and Overdrive (Ovd) in Drosophila (5); all of which encode proteins with putative DNA binding domains. Satellite DNAs have also been implicated in hybrid inviability, including a pericentric satellite locus (Zhr) (27, 28) and a gene encoding a heterochromatin-binding protein (Lhr) (29). Thus, rapidly evolving repetitive DNA elements driven by genetic conflict may represent a major evolutionary force driving sequence divergence of speciation genes that would ultimately result in hybrid incompatibilities (17, 18).
Materials and Methods
Antibodies
A rabbit polyclonal antibody was raised to a peptide consisting of amino acids 119–136 of D1 (QCB, Hopkinton, MA) and affinity purified. A mouse polyclonal antibody was raised to a bacterially expressed glutathione S-transferase (GST) fusion protein of the C-terminal half of the OdsHsim protein (amino acids 210–370). The specificity of the OdsH antibody was determined by immunoblotting and cytology (Fig. S5).
D. simulans Cell Cytology
The D. simulans ML82-19a cell line derived from 4–19 hour embryos used in the cell culture experiments was obtained from the Drosophila Genomics Resource Center (DGRC, Indiana University, Bloomington, IN). OdsHsim and OdsHmau cDNAs were amplified from testes RNA isolated from wildtype D. simulans and D. mauritiana lines by reverse transcription-polymerase chain reaction (RT-PCR) (Superscript III One-Step RT-PCR Kit, Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The following primers were used to amplify OdsHsim: OdsSim1-F 5’-CACCATGCACGTGTCCGGCTGG-3’ and OdsSim2-RS 5’-TTACGATCCCAACAGGTATTTGATGC-3’. The same forward primer (OdsSim1-F) was used to amplify OdsHmau in combination with OdsMau2-RS 5’-CTATTCCACTTCCACTTCCATATCCTCG-3’. A cDNA clone of OdsHmel was obtained from the Berkeley Drosophila Genome Project (BDGP, Berkeley, CA) and amplified by PCR (Platinum Taq DNA Polymerase, Invitrogen) using the following primer set: OdsMel1-F 5’-CACCATGCAAGTGAGCGGCTGG-3’ and OdsMel2-RS 5’-TTACGATCCCAACAGGGATTCG-3’. The cDNAs were directionally TOPO-cloned into a pENTR vector (Invitrogen) and sequence verified. Subsequently, the cDNAs were recombined into either an N-terminal Venus (yellow fluorescent protein) or N-terminal 3XFLAG Gateway Destination vector (http://www.ciwemb.edu/labs/murphy/Gatewayvectors.html) via LR Recombinase according to manufacturer’s protocol (Invitrogen). ML82-19a cells were seeded onto a coverslip and 1–2 ug of plasmid vector DNA was transfected with Cellfection reagent (Invitrogen). Expression of the fusion protein was induced by a 45 minute heatshock at 37°C. Cells were allowed to recover at room temperature for 3 hours, swelled in hypotonic solution and fixed in 4% formaldehyde as before {Henikoff, Ahmad, PNAS, 2000}. Cells were blocked in PBG: 1XPBS + 0.2% fish gelatin and 0.5% BSA (Sigma-Aldrich, St. Louis, MO) for 45 minutes at room temperature. Primary antibodies were diluted in PBG as follows: rabbit anti-D1 1:1,000 and mouse anti-M2 FLAG 1:2,000 (Sigma-Aldrich). Washes were conducted in 1XPBS plus 0.3% Triton X-100 (PBST). Texas red anti-rabbit diluted 1:100 and anti-mouse AlexaFluor 568 (Invitrogen) diluted 1:1,000 were used for secondary detection. DNA was stained with DAPI and coverslips were mounted in Vectashield (Vector Labs, Burlingame, CA). Slides were observed under a Zeiss AxioPlan inverted scope, deconvolved, and processed with AxioVision software package (Zeiss, Thornwood, NY).
Transgenic Analyses
1XFLAG-OdsHsim, Venus-OdsHmau, Venus-OdsHmel, and Venus-unc-4 were amplified by PCR (Platinum Taq, Invitrogen) from the Destination vectors described above with the following primer pairs that include a 5’ NotI and 3’ XbaI restriction sites for downstream subcloning: (1XFLAG-OdsHsim) JJB428-FLAGSimOdsF 5’-GCGGCCGCATGGACTACAAAGATGACGACGATAAGATGCACGTGTCCGGCTGGTCATCC-3’ and JJB406-OdsSimRXba 5’-GCTCTAGATTACGATCCCAACAGGTATTTGATGC-3’; (Venus-OdsHmau) JJB405-YFPNotIF 5’-GCGGCCGCATGGTGAGCAAGGGCGAGGAGC-3’ and JJB414-OdsmauRX 5’-GCTCTAGACTATTCCACTTCCACTTCCATATCCTCG-3’; (Venus-OdsHmel) JJB405-YFPNotIF and JJB407-OdsMelRXba 5’-TCTAGATTACGATCCCAACAGGGATTCGATGC-3’; (Venus-unc-4) JJB405-YFPNotIF and JJB472-unc4XbaIR 5’-GCTCTAGATCACGTGTTGTTGAACAGCAAGGACTCG-3’. The PCR products were TOPO-cloned into the pCR2.1 vector (Invitrogen), sequence verified, then digested with NotI/XbaI for subcloning into a heatshock pCaSpeR P-element transformation vector. The constructs were injected into D. simulans w501 or D. melanogaster w1118 embryos by BestGene, Inc (Chino Hills, CA). Several transformants were recovered for each construct and tested for transgene expression. Larvae were heatshocked at 37°C for 50 minutes to induce expression of the transgene and allowed to recover at room temperature for 1 hour. After recovery, larval brains were dissected in 0.7% NaCl, incubated in 10−3 M colchicine for 1.5 hours, treated in hypotonic solution for 3 minutes, and fixed in 45% acetic acid/2% formaldehyde/PBS for 3 min. Samples were blocked in PBG as described above and incubated with 1:100 JL8 (anti-YFP, Invitrogen), 1:1,000 diluted anti-D1, and/or 1:500 diluted anti-M5 FLAG (Sigma-Aldrich) for 1 hour at room temperature. Washes were done in 1XPBS plus 0.1% Triton X-100. Anti-mouse AlexaFluor 568 and anti-rabbit AlexaFluor 488 secondary antibodies were used as described above. DNA was stained, slides mounted, and analyzed as described above.
Testes Cytology
Whole-mount immunohistochemistry
Testes were dissected in 0.7% NaCl solution from newly eclosed D. simulans w501, D. mauritiana, and D. simulans fertile and sterile introgression males. Anti-OdsH was diluted to 1:1,000 in PBG and conducted according to protocol {Henderson, Drosophila Cytogenetic Protocols, 2004} using the Vectastain-ABC anti-mouse Peroxidase kit (Vector Labs). Testes were mounted in 80% glycerol. Mounted testes were observed with a Zeiss AxioPlan inverted scope using phase contrast DIC. Final images are a composite of several smaller images taken with the same exposure time and constructed and processed in Adobe Photoshop CS3.
Since spermatocyte development follows a spatial-temporal program, we quantified the persistence of OdsH protein in the male testes in D. simulans w501 and D. simulans sterile introgression lines by measuring the length in µm of OdsH protein expression. The total number of testes analyzed was compiled over at least three independent experiments. Significance was determined by a Student Two-tailed t-test.
Whole-mount immunofluorescence
Testes were dissected from the genotypes described above and fixed in 3.7% formaldehyde in PBS for 30 min. and subsequently treated as above {Henderson, Drosophila Cytogenetic Protocols, 2004}. Testes were incubated overnight at 4°C in 1:2,000 diluted anti-OdsH and 1:200 diluted anti-phosphoH3 serine 10 (Millipore, Billerica, MA). Anti-rabbit AlexaFluor 488 and anti-mouse AlexaFluor 568 (Invitrogen), both diluted 1:1,000 were used for secondary detection. DNA was stained with DAPI. Testes were mounted in VectaShield (Vector Labs). Individual images were taken with a Zeiss AxioPlan inverted microscope and assembled in Adobe Photoshop CS3 to create a composite image. Despite maintaining the same exposure time between individual pictures, some variation in color was observed when the composite was assembled.
Spermatocyte immunofluorescence
As described above, testes were dissected from newly eclosed males and fixed according to Gunsalus et al. (J. Cell Biol. 131, 1243(1995)) and stained with 1:1,000 diluted anti-OdsH according to protocol 5.6 (Bonaccorsi, S. et al., 2000, Drosophila Protocols, Chapter 5 of the Blue Book). DNA was stained with DAPI. Samples were mounted and analyzed as described above.
Western blot analysis
Protein extracts were prepared from 30 dissected testes from D. simulans w501, D. simulans sterile, and D. simulans fertile lines. Testes were directly lysed in 50 µL of 2X SDS loading buffer (100 mM Tris-HCl, pH 6.8; 4% SDS; 12% glycerol 0.02% bromophenol blue) plus 10% β-mercaptoethanol (Sigma-Aldrich). Protein lysates were incubated at 95°C for 5 minutes before approximately a third of the lysate was added to a 4%–12% gradient acrylamide gel for gel electrophoresis. The blot was incubated with 1:2,000 diluted anti-OdsH or 1:50 diluted anti-tubulin (Sigma-Aldrich). Primary antibodies were detected by incubating the blot in 1:5,000 diluted anti-mouse or anti-rabbit horseradish peroxidase conjugated secondary antibodies (GE Healthcare, Piscataway, NJ). Bound antibody was detected with the Super-Signal West Pico Chemiluminescent kit (Pierce, Rockford, IL).
Supplementary Material
Literature Cited
- 1.Mayr E. Systematics and the origin of species from the viewpoint of a zoologist. New York: Columbia University Press; 1942. [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland: Sinauer Associates; 2004. [Google Scholar]
- 3.Laurie CC. Genetics. 1997;147:937. doi: 10.1093/genetics/147.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ting CT, Tsaur SC, Wu ML, Wu CI. Science. 1998;282:1501. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 5.Phadnis N, Orr HA. Science. 2009;323:376. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masly JP, Jones CD, Noor MA, Locke J, Orr HA. Science. 2006;313:1448. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 7.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. Science. 2009;323:373. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 8.Sun S, Ting CT, Wu CI. Science. 2004;305:81. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- 9.Kliman RM, et al. Genetics. 2000;156:1913. doi: 10.1093/genetics/156.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez DE, Wu CI. Genetics. 1995;140:201. doi: 10.1093/genetics/140.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez DE, Wu CI, Johnson NA, Wu ML. Genetics. 1993;134:261. doi: 10.1093/genetics/134.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hueber SD, Lohmann I. Bioessays. 2008;30:965. doi: 10.1002/bies.20823. [DOI] [PubMed] [Google Scholar]
- 13.Ting CT, et al. Proc Natl Acad Sci U S A. 2004;101:12232. doi: 10.1073/pnas.0401975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabuchi K, Yoshikawa S, Yuasa Y, Sawamoto K, Okano H. Neurosci Lett. 1998;257:49. doi: 10.1016/s0304-3940(98)00799-x. [DOI] [PubMed] [Google Scholar]
- 16.Nei M, Zhang J. Science. 1998;282:1428. doi: 10.1126/science.282.5393.1428. [DOI] [PubMed] [Google Scholar]
- 17.Henikoff S, Ahmad K, Malik HS. Science. 2001;293:1098. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 18.Henikoff S, Malik HS. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- 19.Fishman L, Saunders A. Science. 2008;322:1559. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 20.Daniel A. Am J Med Genet. 2002;111:450. doi: 10.1002/ajmg.10618. [DOI] [PubMed] [Google Scholar]
- 21.Aulner N, et al. Mol Cell Biol. 2002;22:1218. doi: 10.1128/MCB.22.4.1218-1232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, Golic KG, Hawley RS. Drosophila : a laboratory handbook. ed. 2nd. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 23.Cenci G, Bonaccorsi S, Pisano C, Verni F, Gatti M. J Cell Sci. 1994;107:3521. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- 24.McKee BD. Curr Top Dev Biol. 1998;37:77. doi: 10.1016/s0070-2153(08)60172-6. [DOI] [PubMed] [Google Scholar]
- 25.Tomkiel JE. Genetica. 2000;109:95. doi: 10.1023/a:1026544402411. [DOI] [PubMed] [Google Scholar]
- 26.Forejt J. Trends Genet. 1996;12:412. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- 27.Sawamura K, Yamamoto MT, Watanabe TK. Genetics. 1993;133:307. doi: 10.1093/genetics/133.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferree PM, Barbash DA. PLoS Biology. 2009;7:e1000234. doi: 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brideau NJ, et al. Science. 2006;314:1292. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 30.We thank C-I. Wu for the D. simulans fertile and sterile introgression lines, C. Ting for scientific discussions and sharing data, G. Findlay for initial observations on OdsH cytology, and K. Ahmad, S. Biggins, N. Elde, S. Henikoff, N. Phadnis, T. Tsukiyama, and D. Vermaak for comments on the manuscript. Supported by NIH training grant PHS NRSA T32 GM07270 (JJB), and grants from the Mathers foundation and NIH R01-GM74108 (HSM). HSM is an Early-Career Scientist of the Howard Hughes Medical Institute.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.