Abstract
Background
Pandemic Vibrio parahaemolyticus has undergone rapid changes in both K- and O-antigens, making detection of outbreaks more difficult. In order to understand these rapid changes, the genetic regions encoding these antigens must be examined. In Vibrio cholerae and Vibrio vulnificus, both O-antigen and capsular polysaccharides are encoded in a single region on the large chromosome; a similar arrangement in pandemic V. parahaemolyticus would help explain the rapid serotype changes. However, previous reports on "capsule" genes are controversial. Therefore, we set out to clarify and characterize these regions in pandemic V. parahaemolyticus O3:K6 by gene deletion using a chitin based transformation strategy.
Results
We generated different deletion mutants of putative polysaccharide genes and examined the mutants by immuno-blots with O and K specific antisera. Our results showed that O- and K-antigen genes are separated in V. parahaemolyticus O3:K6; the region encoding both O-antigen and capsule biosynthesis in other vibrios, i.e. genes between gmhD and rjg, determines the K6-antigen but not the O3-antigen in V. parahaemolyticus. The previously identified "capsule genes" on the smaller chromosome were related to exopolysaccharide synthesis, not K-antigen.
Conclusion
Understanding of the genetic basis of O- and K-antigens is critical to understanding the rapid changes in these polysaccharides seen in pandemic V. parahaemolyticus. This report confirms the genetic location of K-antigen synthesis in V. parahaemolyticus O3:K6 allowing us to focus future studies of the evolution of serotypes to this region.
Background
V. parahaemolyticus is a naturally occurring marine bacterium that has been recognized as an important food borne pathogen since a large outbreak occurred in Japan in 1950[1]. Before 1996, no particular serotype of V. parahaemolyticus was associated with outbreaks. In that year, there was a major outbreak in Kolkata, India caused by strains with increased virulence and more than half of the patient isolates were serotype O3:K6 [2]. These isolates quickly spread to other countries in Asia, followed by South America, Africa and the United States affecting tens of thousands people and resulting in the first known V. parahaemolyticus pandemic [2,3]. Strains from early in the pandemic were all serotype O3:K6 [4,5]. However, the pandemic strains have rapidly evolved to more than 20 serovariants including O3:K6, O4:K68, O1:K25, O1:KUT (K-untypable) and others [2]. The pandemic isolates are closely related (clonal) as shown by pulse-field gel electrophoresis, ribotyping, and multilocus sequence typing (MLST). Therefore, new serotypes seem to have arisen from the original pandemic O3:K6 strain by changes occurring in both the K- (capsule) and the O-antigen. Understanding the mechanism underlying rapid serotype conversion may help us develop improved diagnostics for identifying isolates with pandemic potential.
Eleven O and 65 K serotypes are recognized in V. parahaemolyticus. The lipopolysaccharide (LPS) of most Gram-negative bacilli consists of lipid A, core polysaccharide and the highly variable O side chain (O-antigen). The capsular or K-antigen is composed of high molecular weight polysaccharide and forms a dense, high molecular weight coat outside of the bacterial cells. Encapsulated pathogens can become invasive and cause septicemia due to their increased resistance to phagocytosis and complement-mediated killing. K- and O- antigens are generally encoded in discreet loci; but, in limited studies in V. cholerae and V. vulnificus isolates, O-antigen and K-antigen have been shown to be co-located [6-8]. A third form of polysaccharide, the exopolysaccharide, is a loose slime outside the cell that forms an intercellular matrix in biofilms. In V. cholerae, this exopolysaccharide is expressed by cells that display a rugose (wrinkled) colony phenotype [9].
Genetic study of surface polysaccharides in V. parahaemolyticus is limited and controversial. Guvener et al. have proposed a locus on chromosome II, VPA1403-VPA1412, for capsular polysaccharide biosynthesis, but have not shown a correlation with the K-antigen [10]. Comparison of restriction fragment polymorphisms of different serotypes led Okura et al to suggest a different locus, around VP214-VP237on chromosome I, for K-antigen genes and a region with homology to the LPS-core polysaccharide region for the O-antigen, but have not experimentally confirmed the function of these regions [11]. To resolve this controversy, we have investigated these putative K-antigen genetic determinants in an epidemic O3:K6 isolate by construction of gene deletions.
Results
Polysaccharide gene clusters in V. parahaemolyticus O3:K6
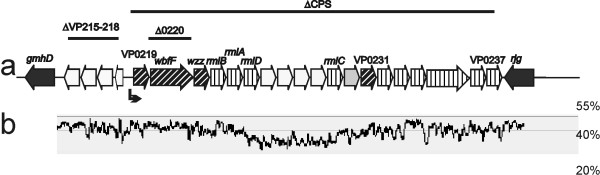
From the genome of V. parahaemolyticus RIMD2210633, we identified four gene clusters that may relate to surface polysaccharide synthesis judging by their homologs in V. cholerae and V. vulnificus (Figure 1). Region A includes genes VP0190-0214. Border genes in region A, i.e. VP0190-0191 and VP0211-0214 are homologous to genes in the other species that synthesize lipid A, Kdo or heptoses, which are all signature components of lipid A or core components in LPS. VP0214 is a homolog of gmhD, an ADP-L-glycero-D-manoheptose-6-epimerase, which has never been successfully deleted in the other species suggesting that its deletion was possibly lethal. Since there is good homology with known lipid A/core regions and mutations in their genes may be lethal, we have not attempted to delete this region in this study. Region B (VP0215-0237) lies between genes gmhD (VP0214) and rjg (VP0238), which define the regions for O-antigen biosynthesis in V. cholerae serogroups O1, O22, O31, O37 and O139 [7,12-16]. Besides O-antigen, this region also defines the capsule genes for non-O1 V. cholerae O31 and O139 [7,13]. In V. vulnificus, O-antigen and capsule genes are both located between gmhD and rjg as well [6]. Previous studies have found similar restriction fragment length polymorphism patterns in region B of strains with the same K serotype suggesting this region may contain the capsule genes [11]. However, region C (VPA1403-VPA1412) in chromosome II was previously identified as the capsule gene region [10]. We deleted genes in region B and C (table 1) to clarify this discrepancy and to answer the question if O- polysaccharide and capsule polysaccharide share the same genes in V. parahaemolyticus, as is the case in both V. cholerae and V. vulnificus.
Figure 1.
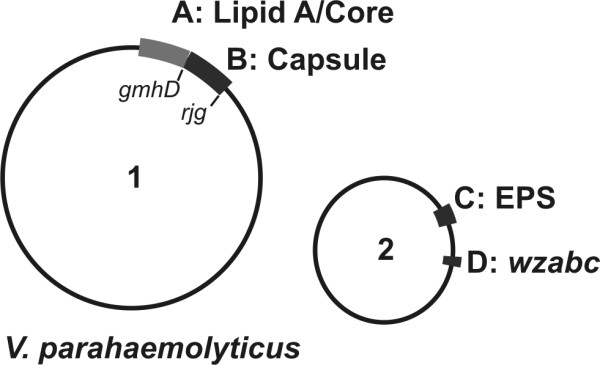
Gene clusters related to polysaccharide in Vibrio parahaemolyticus O3:K6. Two circles to represent two chromosomes. Function of each region is indicated. A (VP0190-0214), putative lipid A/core region; B (VP0215-0237), K-antigen/capsule region (CPS); C (VPA1403-1412), exopolysaccharide region (EPS); D (VPA1602-1604), putative polysaccharide exportation genes wza, b, c.
Table 1.
V. parahaemolyticus strains used in this study
| Strain | Description* |
|---|---|
| VP53 | Wild type |
| ∆0220 | Deletion of VP0220 (wbfF) in region B |
| ∆0220 plus complementation | ∆0220 trans-complemented with VP0220 |
| ∆CPS | Deletion of VP0219-0237 in region B |
| ∆VP215-218 | Deletion of VP0215-0218 in region B |
| ∆EPS | Deletion of VPA1403-1406 in region C |
| ∆EPS plus complementation | ∆EPS trans-complemented with VPA1403-1406 |
| ∆EPS plus empty vector | ∆EPS with pBBR1-MCS2 empty vector |
| ∆wzabc | Deletion of wza, wzb and wzc (VPA1602-1604) genes in region D |
*All mutants were derived from VP53.
Region B determines capsule (K-antigen)
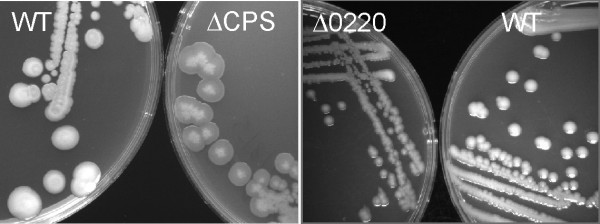
According to the annotation in GenBank [17], region B in V. parahaemolyticus encodes four hypothetical proteins that are upstream of gmhD and transcribed in the same direction, followed by an operon-like structure of 19 open reading frames in the opposite direction (Figure 2, Table 2). To investigate if region B is related to either O-antigen/K-antigen biogenesis in V. parahaemolyticus, we deleted the entire 21 kb operon of 19 open frames, VP0219-0237, and replaced it with a Cm cassette (Figure 2). The resulting mutant, ∆CPS, displayed a translucent phenotype consistent with loss of capsule expression, in contrast to an opaque phenotype in the wild type (Figure 3) [18].
Figure 2.
Capsule (K-antigen) genes in V. parahaemolyticus O3:K6. a) Bars with mutant names above indicate regions deleted in each mutant. Bent arrow indicates promoter. Design patterns of open reading frames indicate different classes of genes: vertical lines, pathway genes; diagonal lines, processing and transportation genes; grey box, glycosyltransferase; white box, functions not clear. b) GC percentage of the sequence in 120 bp windows, aligned to the genes in a.
Table 2.
K-antigen/Capsule genes of V. parahaemolyticus O3:K6
| Gene | Symbol | Putative function |
|---|---|---|
| VP0214 | gmhD | ADP-L-glycero-D-manoheptose-6-epimerase |
| VP0215 | hypothetical protein | |
| VP0216 | hypothetical protein | |
| VP0217 | putative regulator protein | |
| VP0218 | hypothetical protein | |
| VP0219 | hypothetical protein | |
| VP0220 | wbfF | capsule assembly protein |
| VP0221 | wzz | polysaccharide chain length determinant |
| VP0222 | rmlB | dTDP-glucose 4,6 dehydratase |
| VP0223 | rmlA | D-glucose-1-phosphate thymidylyltransferase |
| VP0224 | rmlD | dTDP-4-dehydrorhamnose reductase |
| VP0225 | hypothetical protein | |
| VP0226 | glycosyltranferase | |
| VP0227 | hypothetical protein | |
| VP0228 | hypothetical protein | |
| VP0229 | rmlC | dTDP-4-dehydrorhamnose 3,5-epimerase |
| VP0230 | glycosyltranferase | |
| VP0231 | UDP-galactose phosphate transferase | |
| VP0232 | similar to carbamoyl phosphate synthase | |
| VP0233 | hypothetical protein | |
| VP0234 | amino transferase | |
| VP0235 | putative epimerase | |
| VP0236 | UDP-glucose 6-dehydrogenase | |
| VP0237 | UTP-glucose-1-phosphate uridylyltransferase | |
| VP0238 | rjg | hypothetical protein |
Figure 3.
V. parahaemolyticus mutants ∆CPS and ∆0220 display translucent phenotype. Wild type (WT), ∆CPS and ∆0220 have grown on LB agar at 37°C for 24 hours.
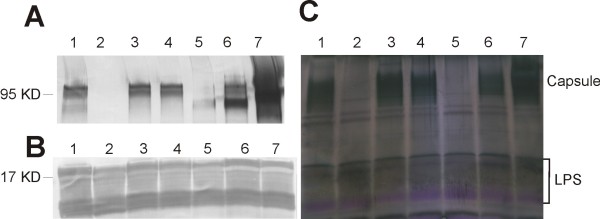
We then investigated the immunogenicity of wild type and ∆CPS mutant by immuno-blotting. Whole cell lysate treated with DNase, RNase and pronase was separated on SDS gels, stained with stains-all/silver stain; or blotted to PVDF membrane and probed with O3 or K6 specific antiserum. With the O3:K6 wild type, gels stained with stains-all/silver-stain showed low molecular weight bands circa 17 kDa and high molecular weight bands circa 95 kDa (Figure 4). Immuno-blot developed with O3 antiserum only detected the low molecular weight bands. The low molecular weight is consistent with the LPS/O-antigen. The O3 antiserum bound in the same amount and pattern in ∆CPS mutant as in wild type (Figure 4) indicating that the major operon between gmhD and rjg, i. e. VP0219-0237, is not involved in O antigen synthesis. Immunoblots developed with K6 antiserum only detected the high molecular weight polysaccharide (Figure 4) in the wild type O3:K6. The high molecular weight of the K-antigen is consistent with capsular polysaccharide. Binding of K6 antiserum was lost in the ∆CPS mutant indicating that region B is required for K antigen biosynthesis. Stains-all/Silver-stain also showed that the high molecular weight capsular polysaccharide was lost in the ΔCPS mutant (Figure 4).
Figure 4.
Immunoblots and stains-all/silver-stain of V. parahaemolyticus. Whole cells lysate treated with DNase, RNase and pronase was separated on polyacrylamide gel, transferred to PVDF membrane and probed with K6 specific antiserum (A), or O3 specific antiserum (B). Total polysaccharides were visualized by stains-all/silver-stain on polyacrylamide gel (C). lane 1, wild type VP53; lane 2, ∆CPS mutant; lane 3, ∆EPS mutant; lane 4, ∆wzabc mutant; lane 5, ∆0220 mutant; lane 6, ∆0220 mutant with trans-complementation; lane 7, ∆VP215-218 mutant.
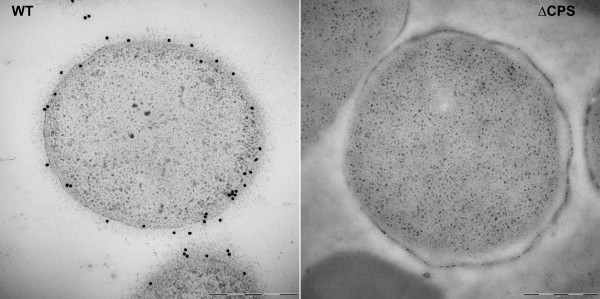
We further investigated the surface structural change in the ∆CPS mutant by immuno-gold EM using K6 antiserum (Figure 5). The EM image of wild type O3:K6 showed gold particles localized around the exterior of the cell consistent with a capsule-like structure surrounding the cell. This capsule structure was absent from ∆CPS mutant and there was no specific gold particle binding to the cell.
Figure 5.
Immuno-gold labeling TEM of V. parahaemolyticus with K6 antiserum. Thin sections samples were labeled with K6 antiserum, followed by gold attached secondary antibodies. Left, Wild type VP53 (WT), right, ∆CPS mutant. Bar equals to 500 nm.
K-antigen processing genes
In order to have some understanding of the capsule/K-antigen biosynthesis pathway, we investigated the polysaccharide processing and assembly genes in the genome of V. parahaemolyticus. We identified a small region outside of the K-antigen genes that contains wza, wzb, and wzc genes (Region D, Figure 1). Wza, b and c together constitute an important exportation system in group 1 and group 4 capsules in E. coli. A wza gene is present in the capsule gene region in both V. vulnificus and encapsulated non-O1 V. cholerae [7,19]. The wza gene in V. parahaemolyticus shares 75% and 64% amino acid identity to the V. vulnificus and V. cholerae wza respectively. To investigate the function of this system in V. parahaemolyticus O3:K6, we deleted all three genes in region D from V. parahaemolyticus to generate mutant Δwzabc. Δwzabc mutant did not show obvious phenotypic differences to the wild type. Immunoblots and stains-all/silver-stain with whole cell lysate indicated that neither the K antigen nor the O antigen production was affected in the Δwzabc mutant (Figure 4). To further investigate if the capsular polysaccharide accumulated in the cell, as would be anticipated if the exportation of capsule were interrupted, immunoblots and stains-all/silver stain with different cell fractions were performed (Figure 6). There was no difference in K-antigen present outside or inside the cells between the Δwzabc mutant and the wild type. Therefore, our results suggested that the wza, wzb and wzc exportation system was not required by either K6-antigen or O3-antigen production in V. parahaemolyticus O3:K6.
Figure 6.
Immuno blot and stains-all/silver-stain of cell fractions. Outer membrane (OM) and cytoplasmic (CP) fractions were separated on polyacrylamide gel, then were either transferred to PVDF membrane and probed with K6 specific antiserum (A), or stained with stains-all/silver stain (B). Lane1, wild type CP; lane 2, ∆wzabc CP; lane 3, ∆EPS CP; lane 4, wild type OM; lane 5, ∆wzabc OM; lane 6, ∆EPS OM.
However, a K-antigen processing system similar to the O-antigen/capsule polysaccharide genes in V. cholerae O139 [13,20,21] is present in V. parahaemolyticus. VP0219-0221 are homologous to wbfE, wbfF and wzz genes in V. cholerae O139, sharing 49%, 69% and 54% amino acid identities. Therefore a similar capsule processing mechanism may exist for both taxa. We generated an in frame deletion of VP0220, the wbfF homolog. Mutant ∆0220 displayed an intermediate level of translucence. Immunoblots indicated that deletion of VP0220 did not affect O3 antigen synthesis (Figure 4). However, the midpoint of the K-antigen band shifted in this mutant, suggesting a role of VP0220 in the later stage of the K-antigen processing. Complementation of ∆0220 with over expressed wild type VP0220 gene restored mostly the pattern of the wild type K antigen (Figure 4). However, there was more reactive material away from the midpoint of the K-antigen band in the complemented mutant than the wild type (Figure 4), possibly due to the over expression of VP0220 or other reasons that remain unclear.
Other K-antigen region features
A complete set of genes of the rhamnose pathway rmlBADC are present in the K-antigen genes of V. parahaemolyticus. However, four open reading frames, VP0225-0228, are inserted between the rmlD and rmlC genes. Analysis of the GC percentage revealed that the average GC percentage in VP0225-0228 is lower than the rest of the genes in this operon (Figure 2). The unusual arrangement of the rhamnose gene order and the mosaic GC percentage pattern indicated that there was a recent recombination event in the K antigen genes.
Between gmhD and the K-antigen operon like genes, there are four genes (VP0215-0218) transcribed to the opposite direction (Figure 2). In frame deletion of these four genes led to the over expression of K-antigen polysaccharides (Figure 4), suggesting these genes may have a regulatory role in capsule expression.
Region C is the exopolysaccharide gene cluster (EPS)
Region C on chromosome II (VPA1403-1412) contains 10 genes related to polysaccharide biogenesis suggested by Guvener et al to responsible for capsular polysaccharide synthesis in V. parahaemolyticus [10]. However, we found that the first 4 genes were similar to exopolysaccharide genes encoding the rugose phenotype in V. cholerae [9], sharing the same gene order and 31-54% amino acid identity to their V. cholerae homologs. We also compared region C in V. parahaemolyticus O3:K6 and O4:K68 (GenBank accession number ACFO00000000) and found that sequences in this region were almost identical in the different serotypes of V. parahaemolyticus and thus unlikely to be involved in synthesis of either O- or K-antigen. To clarify the function of this gene cluster, we deleted genesVPA1403-1406 to generate mutant ∆EPS. The ∆EPS mutant displayed an opaque phenotype similar to the wild type on LB agar, and immunoblots showed that neither the K6 nor the O3 antigens were affected in the ∆EPS mutant (Figure 4). Wild type V. parahaemolyticus displays phase variation in the colony morphology under certain conditions. Growth in APW#3 media, which induced the rugose phenotype in V. cholerae [22], also resulted a rugose colony morphology in V. parahaemolyticus with a raised and wrinkled central area (Figure 7). Unlike the wild type, the ∆EPS mutant lost the ability to become rugose after incubation in APW#3 media. Complementation of the ∆EPS mutant by wild type VPA1403-1406 restored the ability to the rugose phase variation (Figure 7). Therefore, we believe that genes in region C, previously referred to as "capsule genes" are not the genes defining the K-antigen, but in fact, are more appropriately designated exopolysaccharide genes.
Figure 7.
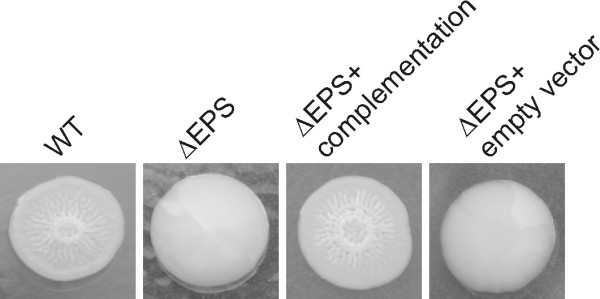
Colony morphology of V. parahaemolyticus. Wild type (WT) V. parahaemolyticus displayed rugose phenotype when incubated in APW#3 media followed by 48-72 hours incubation on LB agar. Mutant ∆EPS only displayed smooth phenotype under the same conditions. Complementation of ∆EPS by the EPS genes restored the rugose phenotype while the ∆EPS mutant with empty vector remained smooth.
Discussion
The genetic region encoding the capsular polysaccharide, or K antigen in V. parahaemolyticus has been controversial, with two different investigators suggesting different loci [10,11]. In our study, construction of gene deletions with confirmation of loss of binding K6-specific antiserum in immunoblots provided solid evidence that the region between genes gmhD and rjg (VP0215-0237) on chromosome I was the genetic determinant of the K6-antigen in the pandemic V. parahaemolyticus O3:K6 serotype. This antigen consists of high molecular weight polysaccharide that is located on the surface of the cell. Loss of this antigen resulted in a translucent colony morphology. These data are consistent with the K6 antigen being a typical vibrio capsular polysaccharide. Our study supports the location suggested by Okura et al as encoding the K-antigen [11]. Although O-antigen genes have been identified in a conserved locus between genes gmhD and rjg in both V. cholerae and V. vulnificus, our study found that this locus in V. parahaemolyticus was not involved in O-antigen biosynthesis. We also showed that gene cluster referred to as "capsule" genes by Guvener et al (VPA1403-VPA1412) was not related to either K-antigen capsule polysaccharide or O-antigen but was instead related to exopolysaccharide production, which causes rugose phase variation. We suggest reserving the term "capsule" for K-antigen polysaccharides and referring to the rugose related polysaccharide exopolysaccharide.
Our understanding of the major surface polysaccharides in V. parahaemolyticus had been limited, in part, due to our limited ability to perform genetic manipulations in this species. Genetic manipulation of genes in V. parahaemolyticus was previously achieved by first cloning the DNA of interest into a suicide plasmid that cannot replicate in V. parahaemolyticus, propagating the plasmid in an E. coli host, then transferring the plasmid from E. coli to V. parahaemolyticus by conjugation, followed by counter selection against the E. coli host and screening for mutants of V. parahaemolyticus [23]. The procedure is tedious and time consuming. There are few reports using electroporation in V. parahaemolyticus and no report of successful chemical transformation [24,25]. We tested electroporation on V. parahaemolyticus and had limited success with plasmid DNA but no success with linear DNA (data not shown). Chemical transformation was also not successful. Therefore we sought alternative methods for targeted gene deletion in V. parahaemolyticus. Meibom et al. reported that V. cholerae became competent and took up foreign DNA when cultured with chitin [26]. The chitin based transformation method was later successfully adapted for V. vulnificus [27]. We modified the chitin based transformation technique and developed a rapid method to mutate genes in V. parahaemolyticus. On average, 150 mutants were obtained from each transformation. Since only one mutant is needed in most cases, this transformation efficiency will satisfy most deletion applications in V. parahaemolyticus.
Capsule biogenesis in E. coli is classified into 4 groups. Exportation of group 1 and 4 capsules rely on Wza proteins, while group 2 and 3 may rely on CPSM and CPST proteins [28]. Previous research has shown that capsules in V. cholerae O31 and V. vulnificus have similarities to E. coli group 1- or group 4 capsules; with a wza gene inside the capsule gene cluster [6,7,19]. Genomic analysis also revealed that a wza gene was present in the putative capsule regions in the other published genomes of V. vulnificus and non-O1, non-O139 V. cholerae [29]. In contrast, the wza gene was present in V. parahaemolyticus, but was not within the capsular polysaccharide region. Furthermore, mutagenesis of this gene showed it was not required for K antigen biosynthesis. Deletion of wbfF changed the pattern of the K-antigen seen on immunoblots, suggesting that this gene may play a role in assembly of the capsule. However, the processing of K-antigen by the wbfF gene and possibly the adjacent wzz gene, and the regulation role of the upstream genes will require further investigation.
In both V. cholerae and V. vulnificus the capsule and O-antigen genes lie in a region similar to the O-antigen region of enteric, such as E. coli, and that specific genes may be shared by both biosynthetic pathways [6,7]. Pandemic V. parahaemolyticus has changed rapidly in both O and K types, leading to the hypothesis that the genetic determinants of O and K also share the same genetic locus thus allowing a single genetic event to alter the structure of both antigens. However, our finding is not consistent with this hypothesis. Our experiments clearly demonstrated that genes determining the K-antigen in pandemic V. parahaemolyticus were located in the region determining both surface polysaccharides in the other vibrios, but that the O-antigen genes are located elsewhere. From our data and Okura et al's observations on polysaccharide genes, we speculate that the region with homology to LPS core regions may be playing the role of O antigen. This speculation is consistent with the finding that the LPS in V. parahaemolyticus are rough type [30]. Since the core genes are adjacent to the capsule genes, they could still be replaced in the same recombination event and give rise to both new O- and K-antigens. Analysis of putative O and K antigen genes in a different serotype O4:K68 revealed that these regions are distinct from those of O3:K6 serotype despite their highly similar genetic backbones [11] and suggested both the O and K regions were replaced during the serotype conversion (Chen et al: Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence, submitted).
Conclusion
Understanding of the genetic basis of O- and K-antigens is critical to understanding the rapid changes in these polysaccharides seen in pandemic V. parahaemolyticus. This is also important in understanding the virulence of V. parahaemolyticus as the O- and K-antigens represent major surface antigens responsible for protective immunity. In this study, we found the O and K genes were separated in V. parahaemolyticus but their locus maybe adjacent. This report also confirms the genetic location of K-antigen synthesis in V. parahaemolyticus O3:K6 allowing us to focus future studies of the evolution of serotypes to this region.
Methods
Bacterial strains and growth condition
At the time of this study, we didn't have access to the sequenced strain RIMD 2210633 and numerous studies showed that the pandemic strains of V. parahaemolyticus O3:K6 are highly clonal and homogenous in their genomes. In particular, the polysaccharide genes are almost identical in RIMD2210633 and two O3:K6 pandemic strains sequenced (Chen et al: Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence, submitted), and primers based on RIMD2210633 sequence successfully amplified target DNA from VP53, an O3:K6 pandemic strain isolated from a patient in Kolkata, India in 1996 [5], and subsequent sequence was confirmed to be identical to RIMD2210633. Thus all mutants were generated from V. parahaemolyticus VP53. Unless otherwise stated, bacteria were cultured in LB broth or LB agar at 37°C. Antibiotics were added in the following concentration when needed: chloramphenicol at 10 μg/ml, and Kanamycin at 50 μg/ml for Escherichia coli and 100 μg/ml for V. parahaemolyticus.
To induce rugose phenotype, a single colony was inoculated into 2 ml APW#3 broth [22], incubated at 37°C statically for 48 hours. Then 1 μl of culture was spotted on LB agar plate and incubated at 30°C for 48-72 hours. Pictures were taken when colony size reached about half centimeter.
Construction of Mutants
Genetic regions to be targeted and primer sequences were determined based on the annotation of V. parahaemolyticus genome RIMD2210633 (GenBank Accession BA000031 and BA000032). Several mutants, including a mutation deleting the entire K-antigen structural gene operon on chromosome I (VP0219-0237), several partial deletion mutations in the region on chromosome I (VP0215-0218 and VP0220 gene), and a deletion mutation of exopolysaccharide region in chromosome II (VPA1403-1406) as well as a deletion mutation in a separate region containing polysaccharide transport genes wza, wzb, and wzc were constructed (Table 1). Polymerase Chain Reaction (PCR) was performed using Taq DNA polymerase (Thermo Fisher, Waltham, MA). PCR products were purified on Qiagen PCR purification columns (Qiagen, Valencia, CA). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA).
DNA was prepared for crossover recombination by overlapping PCR. First, three DNA fragments were amplified by PCR separately, including a fragment (500-1000 bp) upstream of targeted gene in V. parahaemolyticus, a fragment (500-1000 bp) downstream of targeted gene in V. parahaemolyticus and a chloramphenicol resistant gene (Cm) in pKD3 [31]. The 3' end of the reverse primer in the upstream DNA was complementary to the forward primer of Cm, and the 5' end of the forward primer of downstream DNA was complementary to the reverse primer of Cm. Then the three fragments were mixed and assembled into one piece in a second PCR reaction where the product was amplified by primers at the two extremes. Genes deleted and primers used are listed in (Table 3). Two to four micrograms of PCR product were used to transform V. parahaemolyticus VP53.
Table 3.
Primers used in this study
| Target | Location | Primer sequence |
|---|---|---|
| Cm cassette | pKD3 forward 1 | GTGTAGGCTGGAGCTGCTTC |
| pKD3 reverse 1 | CATATGAATATCCTCCTTA | |
| pKD3 forward 2 | ACCTGTGACGGAAGATCACTTCG | |
| pKD3 reverse 2 | AGGAACTTCATTTAAATGGCGCG | |
| flanking sequence of VP0220 (wbfF) gene | upstream forward* | CCCAGCCATAACTAACACTAACCCGT |
| upstream reverse | GAAGCAGCTCCAGCCTACACGATAATTCGCTATTTAAATCGAGAGTTAAA | |
| downstream forward | TAAGGAGGATATTCATATGGGAACGACAAGATCATTCCAATCAG | |
| downstream reverse* | TAGGCTAAGTTCTGAGAGGTTTCCG | |
| flanking sequence of CPS region VP0219-0237 | upstream forward | AATACTAGTGAGCTGTGTTCTTCATTATTAATCCT |
| upstream reverse | CGAAGTGATCTTCCGTCACAGGTGCAGTGAATGTCTGTTAACTCT | |
| downstream forward | CGCGCCATTTAAATGAAGTTCCTCAGGCTCGTTACCAATGTGCT | |
| downstream reverse | GCCAATTATCCTAGACTCACCACT | |
| flanking sequence of VP0215-0218 | upstream forward | CACCAGCATTGATCTGGTTATTCA |
| upstream reverse | CGCGCCATTTAAATGAAGTTCCTTATTTAAGGGAGCTTCGGCTCCCT | |
| downstream forward | CGAAGTGATCTTCCGTCACAGGTAGAGCTGTGTTCTTCATTATTAATCC | |
| downstream reverse | TCGGCATTAGAGTAGCTCACTAACG | |
| flanking sequence of EPS genes VPA1403-1406 | upstream forward $ | CCACTACCCACAGAACCGCTTTGT |
| upstream reverse | CGAAGTGATCTTCCGTCACAGGTATGACCTAGTTTCCCTTCTAGCA | |
| downstream forward | CGCGCCATTTAAATGAAGTTCCTAGCCAGGTTTAACCAACATATTGA | |
| downstream reverse$ | ACCACTCCAAAGGGTAGTGGTGA | |
| flanking sequence of wza, wzb, wzc genes | upstream forward | CAGGGAATCAAGCATACGTTGAA |
| upstream reverse | CGAAGTGATCTTCCGTCACAGGTATTCATCTGACGTAAAGAGCGA | |
| downstream forward | CGCGCCATTTAAATGAAGTTCCTATCTAGATCGCTAATTTGACCAAA | |
| downstream reverse | GAGCAGCAAAGCTGCAGATTGA | |
*Primers to amplify DNA for ∆0220 complementation.
$Primers to amplify DNA for ∆EPS complementation.
Chitin based transformation
Transformations were performed using a modification of the chitin based method of Meibom [26], which has been successfully used in V. vulnificus [27]. V. parahaemolyticus was cultured to an optical density of 0.4 - 0.6, pelleted and resuspended to an optical density of 0.2 in sterile seawater. Two ml of this culture were added to pieces of sterile crab shell in a culture plate and incubated at 30°C overnight. The next day, culture supernatant was discarded and 2 ml of fresh sea water, along with 2-4 μg of DNA that was prepared as described above, were added. The mixture was returned to 30°C and incubated overnight. Bacteria were then released from crab shells by vortexing and plated on chloramphenicol agar. Deletions in mutants were confirmed by PCR and sequencing.
Complementation of mutants
DNA fragments deleted in the ∆0220 or ∆EPS mutants were amplified from the wild type genomic DNA (primers listed in Table 3) and cloned into the BamHI and XmaI site of plasmid pBBR1-MCS2 [32]. The resulting plasmids were propagated in E. coli S17pir and then mobilized d into V. parahaemolyticus mutants by conjugation as described previously[7]. All constructs were confirmed by restriction enzyme digestion patterns and sequencing.
Immunoblots
To prepare whole cell lysate for polysaccharide analysis, one ml of bacteria culture grown to an O.D. ~1.0 was pelleted, resuspended in 100 μl cell lysis solutions (200 μg/ml lysozyme, 50 μg/ml DNase, 100 μg/ml RNase A) per O.D. and incubated at 37°C for one hour. SDS was added to the solution to 0.5% final concentration and incubated for an additional 30 min. Pronase was then added to 100 μg/ml and samples were incubated at 37°C overnight. Samples were mixed with equal amount of sample buffer (Biorad), boiled for 10 min, separated in a 15% SDS polyacrylamide gel and then transferred to PVDF membranes (Bio-Rad, Hercules, CA). Cell fractions were prepared as described by Koga and Kawata [33]. Briefly, bacteria were treated with lysis buffer (0.6 M sucrose, 100 μg/ml lysozyme, 2.5 mM EDTA and 50 mM Tris-HCl, pH 8.0) at 37°C for 20 min, and then centrifuged at 8000 g for 15 min. The supernatant represented the outer membrane fraction and the pellet represented the cytoplasmic fraction. Cell fraction samples were then treated with DNase and RNase followed by pronase. Aliquots equal to 1 × 108 cells were separated and blotted as described above. The membranes were blocked with 3% skim milk, and incubated with O3 or K6 specific typing sera (Denka Seiken, Japan), followed by binding with a secondary goat anti-rabbit antibody conjugated with alkaline phosphatase (Bio-Rad). Alkaline phosphatase activity was detected by GAR-AP detection kit (Bio-Rad).
Stains-all/silver-stain
Polysaccharides were stained by a combination of stains-all/silver-stain method adapted from [34]. After electrophoresis, polyacrylamide gel was fixed following the fixative step as instructed by the silver stain plus kit (Biorad). The gel was then washed with water four times, 10 min each, to ensure the removal of SDS. The gel was stained for 2 hours with a solution containing 4 mg/ml stains-all (MP Biomedicals), 5% formamide, 25% isopropanol and 15 mM Tris-HCL, pH8.8. The gel was de-stained with water until background became clear (about 30 min). Silver stain was then performed following the staining and developing step as instructed by the silver stain plus kit.
Immuno-gold EM
Immuno-gold EM was performed in the Interdisciplinary Center for Biotechnology Research at the University of Florida. V. parahaemolyticus samples were treated by high-pressure freezing, followed by freeze-substitution, embedded in EPOXY resin and thin sectioned. Samples were then labeled with K6 antiserum, followed by gold-labeled secondary antibodies.
Authors' contributions
YC, JGM and JAJ conceived the study. YC and JD designed and performed the experimental works. YC and JAJ drafted the manuscript. All authors read and proved the final manuscript.
Contributor Information
Yuansha Chen, Email: yuansha.chen@pathology.ufl.edu.
Jianli Dai, Email: dai.jian.li@ufl.edu.
J Glenn Morris, Jr, Email: jgmorris@ufl.edu.
Judith A Johnson, Email: jajohnson@pathology.ufl.edu.
Acknowledgements
We thank G. Balakrish Nair and O. Colin Stine for their suggestions and supplying bacterial strains and Michael E. Kovach for providing plasmid pBBR1-MCS2. We also thank Paul Gulig for sharing his chitin based transformation protocol before publication and Lolia Fernandez for reading our manuscript.
References
- Fujino L, Okuno Y, Nakada D, Aoyama A, Fukai K, Mukai T, Uebo T. On the bacteriological examination of shirasu food poisoning. Med J Osaka Univ. 1953;4:299–304. [Google Scholar]
- Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20(1):39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair GB, Hormazabal JC. The Vibrio parahaemolyticus pandemic. Rev Chilena Infectol. 2005;22(2):125–130. doi: 10.4067/s0716-10182005000200002. [DOI] [PubMed] [Google Scholar]
- Chowdhury NR, Chakraborty S, Ramamurthy T, Nishibuchi M, Yamasaki S, Takeda Y, Nair GB. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg Infect Dis. 2000;6(6):631–636. doi: 10.3201/eid0606.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury NR, Stine OC, Morris JG, Nair GB. Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J Clin Microbiol. 2004;42(3):1280–1282. doi: 10.1128/JCM.42.3.1280-1282.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhamchik A, Wilde C, Rowe-Magnus DA. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect Immun. 2007;75(12):5550–5558. doi: 10.1128/IAI.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Bystricky P, Adeyeye J, Panigrahi P, Ali A, Johnson JA, Bush CA, Morris JG Jr, Stine OC. The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region. BMC Microbiol. 2007;7:20. doi: 10.1186/1471-2180-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Maneval D Jr, Panigrahi P, Joseph A, Levine MM, Kaper JB, Morris JG Jr, Johnson JA. The capsule and O antigen in Vibrio cholerae O139 Bengal are associated with a genetic region not present in Vibrio cholerae O1. Infect Immun. 1995;63(1):317–323. doi: 10.1128/iai.63.1.317-323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96(7):4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, McCarter LL. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J Bacteriol. 2003;185(18):5431–5441. doi: 10.1128/JB.185.18.5431-5441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M, Osawa R, Tokunaga A, Morita M, Arakawa E, Watanabe H. Genetic analyses of the putative O and K antigen gene clusters of pandemic Vibrio parahaemolyticus. Microbiol Immunol. 2008;52(5):251–264. doi: 10.1111/j.1348-0421.2008.00027.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Stine OC, Morris JG, Johnson JA. Genetic variation of capsule/LPS biogenesis in two serogroup O31 Vibrio cholerae isolates. FEMS Microbiol Lett. 2007;273(2):133–139. doi: 10.1111/j.1574-6968.2007.00808.x. [DOI] [PubMed] [Google Scholar]
- Comstock LE, Johnson JA, Michalski JM, Morris JG Jr, Kaper JB. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol. 1996;19(4):815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- Li M, Shimada T, Morris JG Jr, Sulakvelidze A, Sozhamannan S. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect Immun. 2002;70(5):2441–2453. doi: 10.1128/IAI.70.5.2441-2453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning PA, Heuzenroeder MW, Yeadon J, Leavesley DI, Reeves PR, Rowley D. Molecular cloning and expression in Escherichia coli K-12 of the O antigens of the Inaba and Ogawa serotypes of the Vibrio cholerae O1 lipopolysaccharides and their potential for vaccine development. Infect Immun. 1986;53(2):272–277. doi: 10.1128/iai.53.2.272-277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Shimizu T, Hoshino K, Ho ST, Shimada T, Nair GB, Takeda Y. The genes responsible for O-antigen synthesis of vibrio cholerae O139 are closely related to those of vibrio cholerae O22. Gene. 1999;237(2):321–332. doi: 10.1016/S0378-1119(99)00344-3. [DOI] [PubMed] [Google Scholar]
- Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A. et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361(9359):743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Panigrahi P, Morris JG Jr. Non-O1 Vibrio cholerae NRT36S produces a polysaccharide capsule that determines colony morphology, serum resistance, and virulence in mice. Infect Immun. 1992;60(3):864–869. doi: 10.1128/iai.60.3.864-869.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AC, Powell JL, Kaper JB, Morris JG Jr. Identification of a group 1-like capsular polysaccharide operon for Vibrio vulnificus. Infect Immun. 2001;69(11):6893–6901. doi: 10.1128/IAI.69.11.6893-6901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroeher UH, Manning PA. Genetics of Vibrio cholerae O1 and O139 surface polysaccharides. Boca Raton, Fl.: CRC Press; 1999. [Google Scholar]
- Stroeher UH, Parasivam G, Dredge BK, Manning PA. Novel Vibrio cholerae O139 genes involved in lipopolysaccharide biosynthesis. J Bacteriol. 1997;179(8):2740–2747. doi: 10.1128/jb.179.8.2740-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Rashid MH, Karaolis DK. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl Environ Microbiol. 2002;68(11):5773–5778. doi: 10.1128/AEM.68.11.5773-5778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Yamamoto K, Honda T, Ming X. Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh) J Bacteriol. 1994;176(15):4757–4760. doi: 10.1128/jb.176.15.4757-4760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Griffiths MW. Mg2+-free buffer elevates transformation efficiency of Vibrio parahaemolyticus by electroporation. Lett Appl Microbiol. 2009;48(3):349–354. doi: 10.1111/j.1472-765X.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- Hamashima H, Iwasaki M, Arai T. A simple and rapid method for transformation of Vibrio species by electroporation. Methods Mol Biol. 1995;47:155–160. doi: 10.1385/0-89603-310-4:155. [DOI] [PubMed] [Google Scholar]
- Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310(5755):1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- Gulig PA, Tucker MS, Thiaville PC, Joseph JL, Brown RN. USERTM friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl Environ Microbiol. 2009;75(15):4936–49. doi: 10.1128/AEM.02564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Lee JH. et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106(36):15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi T, Kondo S, Hisatsune K. Vibrio parahaemolyticus O serotypes from O1 to O13 all produce R-type lipopolysaccharide: SDS-PAGE and compositional sugar analysis. FEMS Microbiol Lett. 1995;130(2-3):287–292. doi: 10.1111/j.1574-6968.1995.tb07733.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1):175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Koga T, Kawata T. Isolation and characterization of the outer membrane from Vibrio parahaemolyticus. J Gen Microbiol. 1983;129(10):3185–3196. doi: 10.1099/00221287-129-10-3185. [DOI] [PubMed] [Google Scholar]
- Goldberg HA, Warner KJ. The staining of acidic proteins on polyacrylamide gels: enhanced sensitivity and stability of "Stains-all" staining in combination with silver nitrate. Anal Biochem. 1997;251(2):227–233. doi: 10.1006/abio.1997.2252. [DOI] [PubMed] [Google Scholar]