Abstract
In this study, we isolated a porcine circovirus 2 (PCV2) strain from piglets co-infected with porcine reproductive and respiratory syndrome virus (PRRSV). The complete genome of this strain was sequenced, phylogenetic and polymorphic analyses were carried out. BLAST searches revealed the highest sequence identity (99.5% nt and 99.3% aa) to Guangxi strain EF675230. The phylogenetic tree showed that clustering of the isolates didn't strongly correlate to geographical distribution. Polymorphic analyses demonstrated that the amino acids at most of the polymorphic sites in Open Reading Frame 1(ORF1) and 2 (ORF2)belong to the same amino acid group according to chemical or structural properties, and revealed that highly polymorphic regions overlapped with the known immunoreactive epitopes of ORF2.
1 Introduction
Postweaning multisystemic wasting syndrome (PMWS), characterized by growth retardation, paleness of the skin, dyspnea, and increased mortality rates [1,2], was first described in Canada in 1991, and is now widespread throughout swine production areas of the world [3,4]. The genome of PCV is a single-stranded circular DNA of about 1.76 kb. ORF1 encodes the Rep proteins involved in virus replication and is highly conserved among isolates [5]. ORF2 encodes a 234 amino acid (aa) Cap protein, which is the main structural protein and also the major antigen inducing neutralizing immune responses [6]. The ORF3 protein is involved in PCV2-induced apoptosis by the caspase-8 and caspase-3 pathways [7]. However, healthy pigs experimentally inoculated with PCV2 developed only mild clinical symptoms [8,9], suggesting that other concomitant factors may be needed for the development of typical clinical PMWS [10,11]. Experimental studies on co-infection with PRRSV and PCV2 resulted in the microscopic lesions associated with PMWS and/or porcine dermatitis and nephropathy syndrome (PDNS), and lead to the development of severe disease [12].
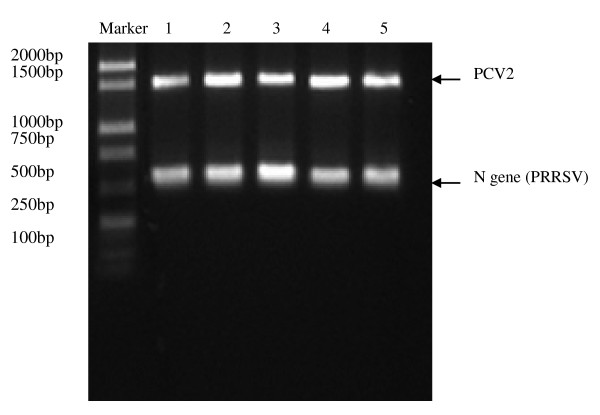
In may 2008, severe disease, known as ''high fever'' occurred in several pig farms in shanghai, leading to a 57% death rate. PRRSV was detected in all the diseased piglets, genome sequence blast showed the strain belongs to genotype 2, 99.4% homologous to the PRRS virus strain JXA1 isolated in China, which has been proved to cause porcine high fever disease with high morbidity and mortality[13]. There was no Porcine Parvovirus (PPV) detected in all the samples, but we detected PCV2 from all the PRRS infected piglets (Figure 1). To investigate the genetic relationship of this newly identified Shanghai PCV2 isolate with existing viruses isolated from other parts of the world, we sequenced the complete genome of the sh0901 strain and carried out phylogenetic and polymorphic analyses.
Figure 1.
Dection of PCV2 and PRRSV in five diseased pigs. Blood samples from five diseased pigs were collected, viruses DNA and RNA were purified, respectively. After RT-PCR and PCR amplification, the amplified products run 1% agrose gel.
2. Materials and methods
2.1 Primer design and synthesis
Two pairs of primers were designed according to the published PCV2 genome sequence of strain AY691679, using the Primer 5.0 software. The sequences of the primers were as follows: upper primers, F1- 5'-AGAATTCAACCTTAACCTTTCT-3' and F2- 5'-AATCCTTCCGAAGACGAGCGCA-3'; reverse primers, R1- 5'-AGAATTCTGGCCCTGCTCC-3' and R2- 5'-TGGGCTCCACTGCTGTTATTC-3'. Primers were synthesized by the Shanghai Sangon Biological Engineering Technology & Services Co. Ltd. (Shanghai, PR China).
2.2 DNA extraction
Five homogenized Lung samples were collected from deceased pigs, DNA was extracted using a DNA mini kit (Tiangen Inc., PR China) according to the manufacturer's instructions.
2.3 Whole genome amplification
The thermocycling conditions were: 3 min at 94°C, then 35 cycles consisting of 30 s at 94°C, 35 s at 60°C and 2 min at 72°C, with a final extension at 72°C for 10 min. The PCR products were subjected to 1% (w/v) agarose gel electrophoresis, and visualized by ethidium bromide staining under UV illumination. Five PCR products were then subjected to DNA sequencing by the Shanghai Generay Biotech Co. Ltd. (Shanghai, PR China) with the same primers used for PCR.
2.4 Phylogenetic analysis of ORF2
Sequence comparisons were made by aligning the sequence of the virus isolate with those of other isolates using the algorithm CLUSTALW (version 1.8) method in the program MEGALIGN (DNASTAR, Lasergene version 7) at both the nucleotide (nt) and deduced aa levels. To analyze the homology and evolutionary relatedness, prototype genes for PCV2 were obtained from GenBank at the National Center for Biotechnology Information, USA (http://www.ncbi.nlm.nih.gov). Phylogenetic analyses were conducted using MEGA version 4.0 and elaborated with both parsimony and distance methods, supplying statistical support with bootstrapping over 1000 replicates.
3. Results
3.1 Genome sequence of the isolated strain
PCR amplification showed the five diseased pigs co-infected with PCV2 and PRRSV (Figure 1). The genome of the isolated viruses was 1767 nucleotide (nt) in length. The genome sequence was assembled, aligned using Seqman software (DNASTAR, Lasergene version 7), submitted to GenBank and assigned the accession number, GU124593. The genome sequence was confirmed to be PCV2 by BLAST searches against the GenBank database.
3.2 Sequence and phylogenetic analysis of ORF2
The ORF2 DNA sequence of the virus isolate shared 99.4% nt identity with the Guangxi isolate (accession number EF675230) and 94.0% with the Jiangsu isolate (accession number AY691679). The results indicated that the ORF2 sequence was not distinct in different geographical areas.
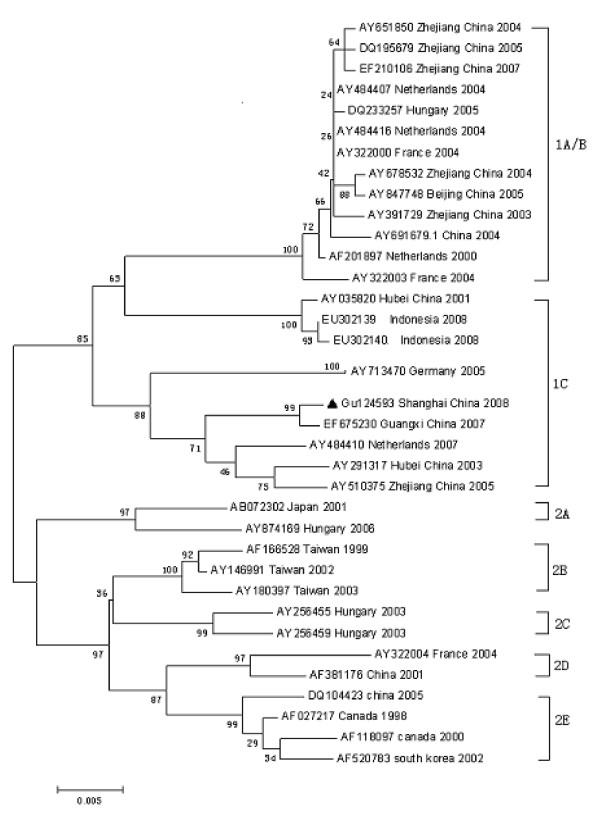
To determine the evolutionary relatedness of the new isolate, the ORF2 gene sequence was aligned with selected PCV2 isolates acquired from GenBank and performed phylogenetic analysis. The data showed that the virus isolate was classified into group 1 cluster C with the Guangxi strain (Figure 2).
Figure 2.
Phylogenetic tree of the ORF2 gene of PCV2 strains.
3.3 Polymorphic analysis of ORF1, ORF2, and ORF3
Sequence alignment of the 46 geographically distinct PCV2 isolates revealed four polymorphic aa sites within ORF1 (Table 1). The aa at positions 35 are acidic residues, the aa at positions 77 and 105 belong to the group of nonpolar aa, while the aa at position 121 belong to the polar, uncharged group. Among the 22 polymorphic sites in ORF2, there were two aa differences at 16 sites, among which each pair of aa at the 12 sites belonged to the same amino acids group according to their chemical structure. There are six polymorphic sites in ORF3, the amino acids at the polymorphic sites belong to different aa groups, except the aa at position 100, indicated high divergence in the chemical and structural properties of the aa of ORF3 in different PCV2 strains.
Table 1.
The distribution of amino acids at polymorphic sites in ORF1, ORF2, and ORF3, from 45 PCV2 strains
| Amino acid position | The distribution of amino acids at the polymorphic sites | ||||
|---|---|---|---|---|---|
| ORF1 | 33 | 8 D | 37 E | ||
| 77 | 23 F | 22 L | |||
| 105 | 18M | 27 I | |||
| 121 | 4 S | 41T | |||
| ORF2 | 2 | 14 F | 31Y | ||
| 30 | 39V | 6 L | |||
| 53 | 39F | 6I | |||
| 57 | 34V | 12 I | |||
| 59 | 15R | 7K | 23A | ||
| 63 | 25R | 11K | 5S | 4T | |
| 75 | 39N | 6 K | |||
| 76 | 40I | 5L | |||
| 77 | 37N | 8D | |||
| 87 | 8 T | 37 S | |||
| 89 | 31P | 14K | |||
| 90 | 13R | 13I | 19L | ||
| 91 | 22T | 13S | |||
| 92 | 23V | 12I | |||
| 134 | 9N | 36T | |||
| 151 | 28T | 17P | |||
| 169 | 10R | 33S | 2G | ||
| 190 | 19T | 14S | 12A | ||
| 191 | 5R | 13A | 27 G | ||
| 206 | 30I | 15K | |||
| 210 | 13E | 14D | |||
| 215 | 8I | 37V | |||
| ORF3 | 29 | 34T | 11A | ||
| 40 | 12 R | 10S | 23G | ||
| 88 | 41Q | 4E | |||
| 100 | 25F | 20L | |||
| 101 | 9Y | 9N | 6D | 21H | |
| 102 | 32Q | 13K | |||
4. Discussion
In the present study, the diseased piglets were infected with PRRSV and PCV2 viruses, detected by PCR and RT-PCR amplification, therefore, the levels of exposure to the infectious agents were theoretically identical for all animals. The severe lesions and clinical symptoms indicated that PRRSV could be a predisposing factor to the high mortality, as previously suggested [14]. Several hypothesis have been suggested to explain this epidemiological situation, PRRSV may interfere with PCV2 clearance, favor the persistence of the virus. However, the nature of the interaction between PRRSV and PCV2 has not been elucidated to date. The ORF2 protein has been considered as a major immunogenic capsid protein, able to stimulate a protective response in pigs. Five immunoreactive epitopes in ORF2 were identified by Mahe' et al using Pepscan analysis[15], these included residues 25-43, 65-87, 113-147,157-183 and 193-207. Larochelle et al (2002) also identified three major regions of aa heterogeneity among ORF2 sequences at residues 59-80, 121-136 and 180-191, and two of these regions corresponded to two of the immunoreactive epitopes demonstrated by Mahe' et al [16]. In this study, we identified six highly polymorphic regions, 26-50, 57-83, 90-120, 134-154, 169-191 and 206-215, by polymorphic analysis, overlapped with corresponded immunoreactive epitopes, thus demonstrated that polymorphic analysis could be applied to deduce the immunoreactive epitopes of viruses. Our analysis show that ORF1 is highly conserved with only four polymorphic sites, and the amino acids at each of the sites belong to the same groups regarding chemical structure, which indicate that these mutations have no big influence on the activity of the Rep protein. Polymorphic sites were clustered in the capsid protein, and overlapped with the immunoreactive epitopes, So ORF2 may play a major role in the varied pathogenicity of PCV2 isolates.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JZ carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. CQ carried out the immunoassays and participated in the sequence alignment. JZ participated in the design of the study and performed the statistical analysis. All authors read and approved the final manuscript.
Contributor Information
Jianzhong Yi, Email: yijianzhong@yahoo.com.
Chengqian Liu, Email: liuchengqian306@sohu.com.
Acknowledgements
This research was supported by research funds from the Shanghai Pujiang Program, project no. 07pj14074 and Technology Commission of Shanghai Municipality(Project No.2007-11-1),.
References
- Tischer I, Glederblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- Harding JCS, Clark EG. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS) J Swine Health Prod. 1997;5:201–203. [Google Scholar]
- Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Investig. 1999;11:3–14. doi: 10.1177/104063879901100101. [DOI] [PubMed] [Google Scholar]
- Harms PA, Sorden SD, Halbur PG, Bolin SR, Lager KM, Morozov I, Paul PS. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol. 2001;38:528–539. doi: 10.1354/vp.38-5-528. [DOI] [PubMed] [Google Scholar]
- Olvera A, Cortey M, Segalés J. Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology. 2007;357(2):175–85. doi: 10.1016/j.virol.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen I, Kwang J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol. 2005;79:8262–8274. doi: 10.1128/JVI.79.13.8262-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell C, Segale's J, Ramos-Vara JA, Folch JM, Rodrı'guez-Arrioja GM, Duran CO, Balasch M, Plana-Dura'n J, Domingo M. Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet Rec. 2000;146:40–43. doi: 10.1136/vr.146.2.40. [DOI] [PubMed] [Google Scholar]
- Mankertz A, Mankertz J, Wolf K, Buhk H-J. Identification of a protein essential for replication of porcine circovirus. J Gen Virol. 1998;79:381–384. doi: 10.1099/0022-1317-79-2-381. [DOI] [PubMed] [Google Scholar]
- Cheung AK. Comparative analysis of the transcriptional patterns of pathogenic and nonpathogenic porcine circoviruses. Virology. 2003;310:41–49. doi: 10.1016/S0042-6822(03)00096-5. [DOI] [PubMed] [Google Scholar]
- Choi J, Stevenson GW, Kiupel M, Harrach B, Anothayanontha L, Kanitz CL, Mittal SK. Sequence analysis of old and new strains of porcine circovirus associated with congenital tremors in pigs and their comparison with strains involved with postweaning multisystemic wasting syndrome. Can J Vet Res. 2002;66:217–224. [PMC free article] [PubMed] [Google Scholar]
- Rovira A, Balasch M, Segale's J, Garcı'a L, Plana-Dura'n J, Rosell C, Ellerbrok H, Mankertz A, Domingo M. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J Virol. 2002;76:3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian K, Yu Y, Zhao T. et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in china and molecular dissection of the unique hallmark. PLoS ONE. 2007;6:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan GM, Kennedy S, McNeilly F, Foster JC, Ellis JA, Krakowka SJ, Meehan BM, Adair BM. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol. 1999;121:1–11. doi: 10.1053/jcpa.1998.0295. [DOI] [PubMed] [Google Scholar]
- Mahe' D, Blanchard P, Truong C, Arnauld C, Le Cann P, Cariolet R, Madec F, Albina E, Jestin A. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J GenVirol. 2000;81:1815–1824. doi: 10.1099/0022-1317-81-7-1815. [DOI] [PubMed] [Google Scholar]
- Larochelle R, Magar R, D'Allaire S. Genetic characterization and phylogenetic analysis of porcine circovirus type 2 (PCV2) strains from cases presenting various clinical conditions. Virus Res. 2002;90:101–11. doi: 10.1016/S0168-1702(02)00141-7. [DOI] [PubMed] [Google Scholar]