Abstract
The pathway involving Bre1-dependent monoubiquitination of histone H2B lysine 123, which leads to Dot1-dependent methylation of histone H3 lysine 79 (H3K79me2), has been implicated in survival after exposure to ionizing radiation in Saccharomyces cerevisiae. We found that depletion of mammalian homologs of Bre1 compromises the response to ionizing radiation, leading to increased radiosensitivity and a G2/M checkpoint defect. The deficiency in Bre1a/b function was also associated with increased sensitivity to crosslinking drugs and defective formation of Rad51 foci in mouse cells, suggesting a defect in homologous recombinational repair analogous to that seen in Saccharomyces. In budding yeast, H3K79me2 is important for the recruitment of the checkpoint signaling protein Rad9 to sites of double-strand breaks (DSBs). However, in mammalian cells, 53BP1 (the Rad9 ortholog) in addition to H3K79me2 recognizes a different residue, H4K20me2, and some studies argue that it is H4K20me2 and not H3K79me2 that is the preferred target for 53BP1. We show here that depletion of Bre1b specifically reduced dimethylation of H3K79 without affecting dimethylation of H4K20. Thus our data suggest that the observed defects in the radiation response of Bre1a/b-deficient cells are associated with reduced H3K79me2 and not with H4K20me2.
INTRODUCTION
It has become increasingly clear in recent years that several types of post-translational histone modification including phosphorylation, acetylation, ubiquitination and methylation play significant roles in DNA repair mechanisms. A particular pathway that was first studied in Saccharomyces involves cross-histone modifications in which ubiquitination of histone H2B on residue K123 affects methylation of the K79 residue on histone H3 (1, 2). This pathway is widely conserved in eukaryotes and has been shown to influence radiation sensitivity and radiation-induced cell cycle checkpoints in yeast (3–8). Specifically, H2BK123 is ubiquitinated in Saccharomyces by the Rad6 ubiquitin conjugase in conjunction with the Bre1 ubiquitin ligase (9–11), and the H3K79 residue is uniquely methylated by the Dot1 methyltransferase (12–14). In the absence of H2BK123 ubiquitination, Dot1 remains able to monomethylate H3K79 (15), but the di- and trimethylation of this residue that occurs in wild-type Saccharomyces is largely abrogated. In addition, methylation of H3K4, which is mediated in wild-type Saccharomyces by the SET1 complex (COMPASS), is abrogated in bre1 Δ strains and other mutants that lack H2BK123 ubiquitination (1, 16, 17).
Deleting the DOT1 gene in Saccharomyces confers multiple phenotypes, including reduced telomeric silencing, increased radiation sensitivity, and abrogation of the radiation-induced G1 (but not G2/M) checkpoint (4– 6, 14, 18, 19). A subtle defect in G2/M was detected in dot1 mutants by Grenon et al. (7). Deleting the BRE1 gene confers radiation sensitivity equivalent to that of dot1null strains (6), indicating that it is di- and/or trimethylation of H3K79 rather than monomethylation that is important for wild-type radiation resistance. Epistasis analysis indicates that recombinational repair is affected in the absence of adequate H3K79 methylation, since double mutants involving dot1Δ or bre1Δ with the recombinational repair mutant rad51Δ show X-ray sensitivity that is equivalent to that of rad51Δ strains alone (6). In contrast, deleting DOT1 or BRE1 adds substantial sensitivity to mutants such as rad18Δ and rad5Δ that are involved in postreplication repair (6). Grenon et al. (7) reported that the Saccharomyces Rad9 protein binds via its tudor domain to methylated H3K79 after irradiation as part of the cell’s response to radiation. Abrogation of this binding may be responsible for the radiation sensitivity seen in mutants such as dot1Δ and rad9tudor (7). Further evidence for the importance of H3K79me2 comes from a recent study showing that both Dot1 and the Rad9 DNA damage checkpoint adaptor are required for efficient sister chromatid recombination in budding yeast (20). However, in Schizosaccharomyces pombe, H3K79 is not methylated, and the tudor domain of the Crb2 protein, which is closely related to Saccharomyces Rad9, binds after irradiation to dimethylated H4K20 residues (21, 22), implying significant pathway divergence. H4K20 is methylated in S. pombe by Set9, and SET9 deletion mutants demonstrate increased radiation sensitivity and defects in the G2/M-phase checkpoint (21, 23), whereas in Saccharomyces, H4K20 is not methylated (23, 24). H3K79 is not methylated in S. pombe, and although H3K4 is methylated in S. pombe (25), it is not dependent on ubiquitination of H2BK119 (the analog of H2BK123 in S. cerevisiae). To our knowledge no data point to dependence of H4K20 methylation on ubiquitination of H2BK119 in S. pombe.
The current study examined the role of the Bre1-dependent pathway of H3K79 methylation in the mammalian DNA damage response. In human cells, the conserved modification pathway involves ubiquitination of H2B on residue K120, mediated by a ubiquitin ligase complex encoded by the BRE1A and BRE1B genes (26, 27). As in yeast, this facilitates methylation of H3K79 by the homologous methyltransferase hDOT1L. It is known that Dot1L deficiency leads to increased radiation sensitivity in mouse embryonic fibroblasts and in 293 cells (28) and that deletion of Dot1L leads to embryonic lethality in mice (29). With respect to radiation response, mammalian cells may include features analogous to both S. cerevisiae and S. pombe. Conflicting data have been reported from different cell systems [reviewed in ref. (30)]. Huyen et al. (31) reported that the 53BP1 protein, which is closely related to the Saccharomyces Rad9 and S. pombe Crb2 proteins, binds methylated H3K79 via residues in the tudor domain that were also required for the recruitment of 53BP1 to DSB sites. In addition, inhibition of DOT1L in human U2OS cells inhibited 53BP1 recruitment to DSB sites (31). However, Botuyan et al. (32) found normal binding of 53BP1 to DSB sites in Dot1-deficient mouse embryonic fibroblast cells and found evidence instead that 53BP1 binds to dimethylated H4K20 residues and is dependent on H4K20 methylation for its recruitment to foci after irradiation. In mice, loss of H4K20 methylation was found to be associated with a defect in G1/S transition but not with the G2/M (entry into mitosis) checkpoint defect or with increased sensitivity to radiation (33).
In contrast to studies of phenotypes related to down-regulation of H3K79 methylation, there appears to be little direct information concerning the role of H2BK120 monoubiquitination in radiation resistance or cell cycle checkpoints in mammals. Given the complex roles of different histone modifications in radiation responses and the fact that H2BK120 ubiquitination has major impacts on H3K4 methylation (1, 2, 16, 17) in addition to its role in H3K79 methylation and on transcription (27, 34, 35), we wished to determine how down-regulating H2B ubiquitination would affect DNA repair and cell cycle checkpoints. We therefore constructed shRNA knockdown cell lines involving mouse Bre1a and Bre1b and human BRE1A and BRE1B, and here we report the effects of these knockdowns on sensitivity to radiation and chlorambucil, formation of Rad51 foci, and the G2/M checkpoint. H2BK120 ubiquitination is thought to facilitate methyltransferase access to H3K79, and because H3K79 and H4K20 are located close to each other on a nucleosomal surface (36), it seemed possible that the defects in the radiation response of Bre1-deficient cells could result from reduced methylation of H4K20. However, while we confirmed that Bre1 knockdown reduces H3K79 dimethylation, we found no effect on H4K20, and our data support a role for H2B ubiquitination in mammalian recombinational repair mediated by H3K79 methylation and analogous to that found in Saccharomyces cerevisiae.
MATERIALS AND METHODS
Cell Culture and Reagents
Radiation-induced mouse fibrosarcoma (RIF-1) cells were cultured in Dulbecco’s modified Eagle medium with glutamate (Gibco BRL) with 15% tetracycline-free fetal bovine serum (Clontech). The lentivirus-based tetracycline-inducible shRNA-expressing system was obtained from Dr. Eric Campeau, University of Massachusetts (37).
Antibodies
The following antibodies were used in this study: anti-H3K79me2 (Abcam), anti-H3K79me1 (Abcam), anti-H4K20me2 (Upstate), antipanH3 (Upstate). Polyclonal antibody recognizing Bre1b (Rnf40) was custom made by Zymed Laboratories (Invitrogen, CA). The antibody was raised in rabbits against synthetic peptide (CQRVYSRGDSEAPGE) conjugated to KLH under a proprietary PolyQuick regimen, and affinity purified using a Sulfolink kit (Pierce, IL).
Short Hairpin RNA (shRNA) Transduction
The shRNA sequences directed against GFP (5′-GAAGCAGCACGACTTCTTC-3′), mBre1a (sequence “Bre1a shRNA1” 5′-ACATCCGCATCATCCTTAA-3′, sequence “Bre1a shRNA2” 5′-GTGAAGTCCTAAGGTATAA-3′), or mBre1b (sequence “Bre1b shRNA1” 5′-ACATATGGAGAGTGATGAA-3′, sequence “Bre1b shRNA2” 5′-CAAGATAAAGTGACATCGA-3′) were cloned into lentiviral vectors conferring puromycin or hygromycin resistance (37). Lentiviruses were created as described in ref. (37). RIF-1 cells were transduced with shRNA-encoding lentiviruses and selected in corresponding antibiotic for 7–10 days. To create RIF-1 cells with inducible expression of shRNAs, cells were cotransduced with lentivirus-containing expression vector for Tet repressor protein and were grown for 2 weeks to select for stably transduced heterogeneous or clonal cell populations. shRNA expression was induced by treatment with 2 µg/ml doxycycline for 3–6 days before assessment of target gene knockdown.
RT-PCR Analysis
Total RNA extracts were prepared using an RNeasy kit (Qiagen). cDNAs were synthesized with M-MLV reverse transcriptase (Invitrogen). Both oligo-(dT) and random-hexamer primers were used to obtain cDNAs. The sequences of primers specific to Bre1a, Bre1b or GAPDH were: Bre1aF, 5′-GCTTCGGGAACACATTGAAAAG-3′; Bre1aR, 5′-CTGTGAGTAGGTCCCCCAAAC-3′; Bre1bF, 5′-ACACTCCTCATCGTCAACCG-3′; Bre1bR, 5′-GGGATCACATCACGGGTCAG-3′; GAPDHF, 5′-TGACCACAGTCCATGCCATC-3′; and GAPDHR, 5′-GACGGACACATTGGGGGTAG-3′.
Western Blot Analysis
Total cellular protein extracts were prepared by lysing cell pellets in buffer containing 50 mM Tris, pH 7.5, 1% SDS, 10% glycerol, 0.15M NaCl, 0.5 mM EDTA, 0.2 mM PMSF, and 100× protease inhibitor cocktail (Sigma-Aldrich). Twenty or 50 µg of protein samples was separated on 4–12% NuPAGE Bis-Tris polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes. Protein blots were probed with antibodies, washed and incubated with corresponding secondary antibodies conjugated with alkaline phosphatase (Vector Laboratories). Protein bands were detected using a Storm scanner (Molecular Dynamics).
Radiation and Drug Treatment
After irradiation (MARK-1 cesium γ-ray source, J. L. Shepherd and Associates, dose rate 11.49 Gy min) or 1 h of drug treatment, cells were immediately plated for colony formation. Cells were plated in triplicate for each treatment condition. After 10–14 days dishes were rinsed with 0.9% NaCl and stained with crystal violet, and visible colonies containing more than 50 cells were counted.
G2 Cell Cycle Delay
Cells were irradiated with 4 Gy of γ rays and fixed for FACS analysis 16 h later.
G2/M Cell Cycle Checkpoint Analysis
Cells were irradiated with 3 Gy and 1 h after irradiation were fixed for FACS analysis. FITC-conjugated antibody against phospho-H3Ser10 (Upstate) was used for visualization of mitotic cells.
Immunofluorecsence
Cells were grown on cover slips, irradiated with 6 Gy, and fixed at different times postirradiation using 2% (w/v) paraformaldehyde in PBS and permeabilized using 0.1% Triton X-100 solution. Cells were incubated with anti-Rad51 antibody (H-92, Santa Cruz Biotechnologies, 1:200) and detected with secondary anti-rabbit Alexa594 antibody (Invitrogen, 1:400). To quantify Rad51 foci, we counted cells with more than 10 distinct bright foci. At least 200 nuclei were counted for each sample.
RESULTS
Depletion of Bre1a and Bre1b in Mouse Cells Leads to Increased Sensitivity to Ionizing Radiation
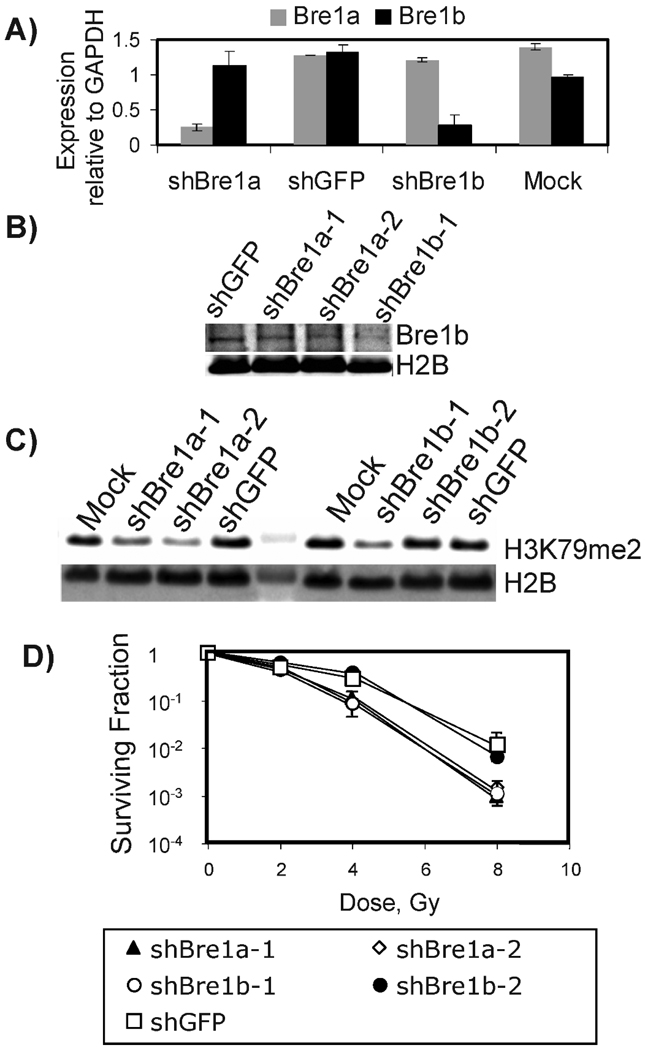
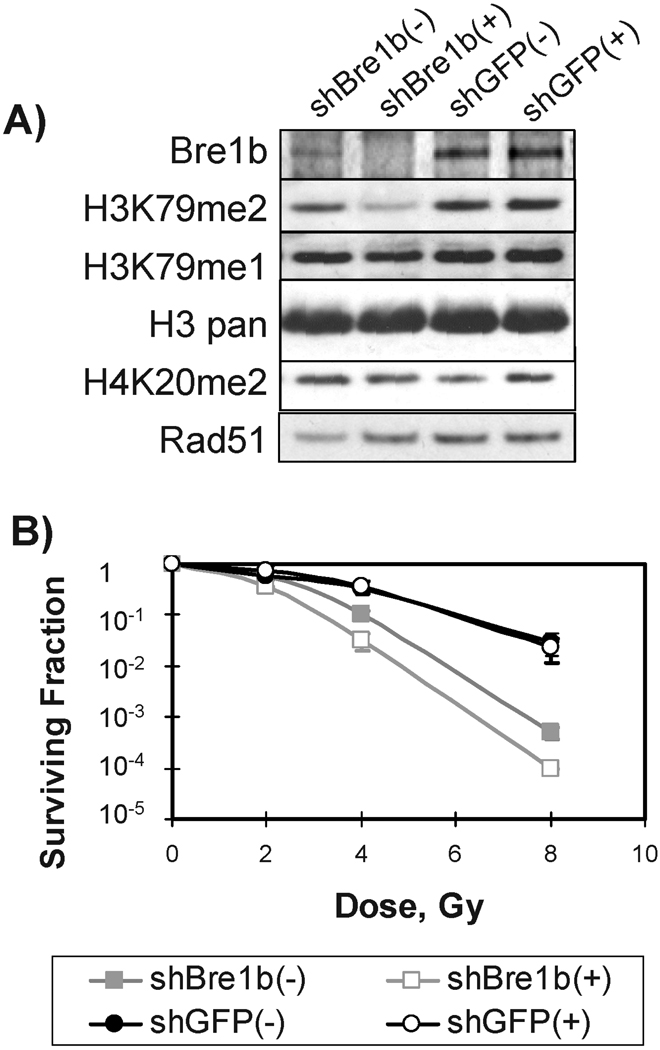
To establish whether Bre1 homologs contribute to radiation resistance in mammalian cells, we designed shRNA sequences uniquely targeting expression of BRE1a or BRE1b (Fig. 1A and B). BRE1-mediated monoubiquitination of histone H2B facilitates methylation of histone H3 residues K4 and K79 (26, 27). As expected, RNAi knockdown of either of the Bre1 mouse homologs Bre1a and Bre1b reduced dimethylation of histone H3K79 (Fig. 1C). The cells that displayed a reduction in methylation of H3K79 showed increased sensitivity to ionizing radiation (Fig. 1C and D). The cell line expressing construct Bre1b shRNA2 that failed to knock down Bre1b exhibited a survival comparable to that of the control GFP shRNA cells. Attempts to increase the level of knockdown, such as with double transduction with Bre1a and Bre1b shRNA or transduction with greater quantities of shRNA, were unsuccessful. Of note, the RIF-1 cells constitutively expressing Bre1a or Bre1b shRNAs grew significantly more slowly than mock-treated or control cells expressing GFP shRNA (data not shown). This observation and the fact that our attempts to increase knockdown of the proteins were unsuccessful suggest that cells need a critical level of Bre1a and Bre1b to survive. To avoid the problem of insufficient cell numbers, we created stable cell lines in which the knockdown of Bre1a and/or Bre1b could be induced by addition of doxycycline. Five days after addition of doxycycline, no Bre1b protein was detected in the Bre1b shRNA-inducible cells (Fig. 2A), while Bre1b protein was present in cells not treated with doxycycline or in GFP shRNA cells with or without doxycycline. However, the doxycycline-inducible Bre1b shRNA system was leaky, and partial knockdown of Bre1b occurred even when doxycycline was not present (Fig. 2A). The levels of H3K79me2 were proportional to the levels of Bre1b protein, while monomethylation of H3K79 was affected to a lesser degree (Fig. 2A). Moreover, the decrease in levels of H3K79 dimethylation strongly correlated with an increase in radiation sensitivity (Fig. 2B). We did not notice major changes in cell cycle distribution caused by depletion of Bre1b (data not shown and Fig. 3B). Depletion of Bre1b did not affect the levels of H4K20me2 and Rad51 (Fig. 2A).
FIG. 1.
Constitutive knockdown of Bre1a and Bre1b in mouse RIF-1 cells increases radiation sensitivity. Depletion of Bre1a or Bre1b reduces target gene expression (panels A, B) and dimethylation of histone H3K79 (panel C). RIF-1 cells were infected with lentiviral vectors expressing shRNA against Bre1a or Bre1b and GFP (control). Total RNA was prepared 7 days post-transduction and used for RT-PCR analysis. Cellular protein extracts were prepared 10 days post-transduction. Lane 5 in panel C shows a molecular weight protein marker. Panel D: Cells with reduced H3K79me2 show increased sensitivity to ionizing radiation. Shown is a representative graph from an experiment that was repeated twice or more for some conditions. Standard errors are shown when larger than symbols. “sh” before a gene name indicates shRNA.
FIG. 2.
Inducible knockdown of Bre1b increases radiation sensitivity of mouse RIF-1 cells. Panel A: RIF-1 cells expressing Bre1b shRNA or GFP shRNA were grown in the presence of doxycycline (2 µg/ml) for 5 days. Bre1b was down-regulated in the Bre1b shRNA cells even in the absence of doxycycline. Panel B: Inducible depletion of Bre1b in RIF-1 cells increases radiation sensitivity. (+) and (−) indicate incubation in the presence and absence of doxycycline, respectively. Note that the leaky phenotype of the Bre1b shRNA cell line (panel A) results in intermediate sensitivity in the absence of doxycycline [Bre1b shRNA(+)]. Shown are means and standard errors of data from three to five independent experiments. “sh” before a gene name indicates shRNA.
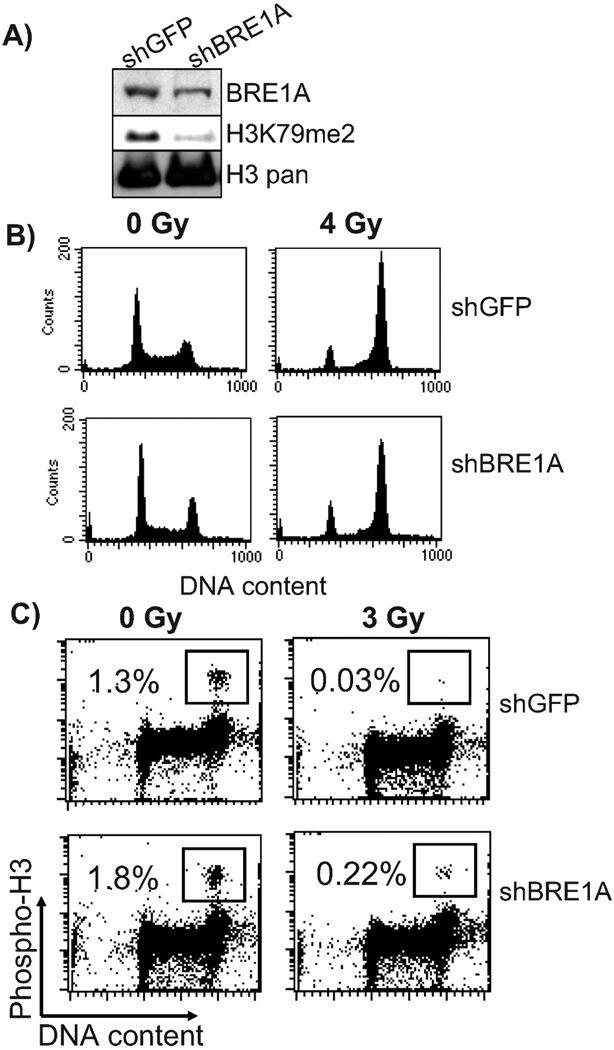
FIG. 3.
Depletion of BRE1A in the human U2OS cells deregulated the G2/M checkpoint without affecting radiation-induced G2 delay. Panel A: Depletion of BRE1A results in reduced dimethylated H3K79 in U2OS cells. Panel B: Depletion of BRE1A does not affect G2 delay after exposure to 4 Gy. Panel C: G2/M checkpoint function is defective in BRE1A-depleted cells irradiated with 3 Gy and fixed for FACS analysis 1 h later. Data shown are representative of two independent experiments. “sh” before a gene name indicates shRNA.
Depletion of BRE1A in Human Cells is Associated with Defective G2/M Arrest
RIF-1 cell cultures were previously reported to contain a small fraction of tetraploid cells (38); therefore, to investigate the function of radiation-induced checkpoints in cells with Bre1 knockdown, we chose to use U2OS cells instead. As with the knockdown of the Bre1a/b complex in the RIF-1 cells, the knockdown of BRE1A led to a reduction in dimethylation of H3K79 (Fig. 3A). FACS analysis of U2OS cells exposed to 4 Gy of ionizing radiation and fixed 16 h after irradiation revealed an increase in the fraction of cells with G2/M content (from ~26% in unirradiated cells to ~77% in cells treated with 4 Gy), pointing to a normal function of the G2 delay checkpoints (Fig. 3B). Cells expressing BRE1A shRNA also displayed a delay in G2 (G2/M content increased from ~31% to ~71%) (Fig. 3B). Abrogation of 53BP1 has been reported to result in a defect in the G2/M cell cycle arrest detectable after exposure to low doses (3 Gy) of radiation (39). Because H3K79me2 is one of the two reported binding targets of the tudor domain of 53BP1, it seemed likely that reduction in H3K79me2 would result in defective G2/M cell cycle arrest. To assess the G2/M checkpoint in BRE1A-depleted cells, we irradiated cells with 3 Gy and fixed them 1 h postirradiation. As shown in Fig. 3C, hBRE1-deficient cells displayed a radiation-induced G2/M checkpoint defect, because more than fivefold more cells entered mitosis in BRE1A-depleted cells than in control cells expressing GFP shRNA.
Depletion of Bre1b in Mouse Cells Leads to Defects in Homologous Recombination
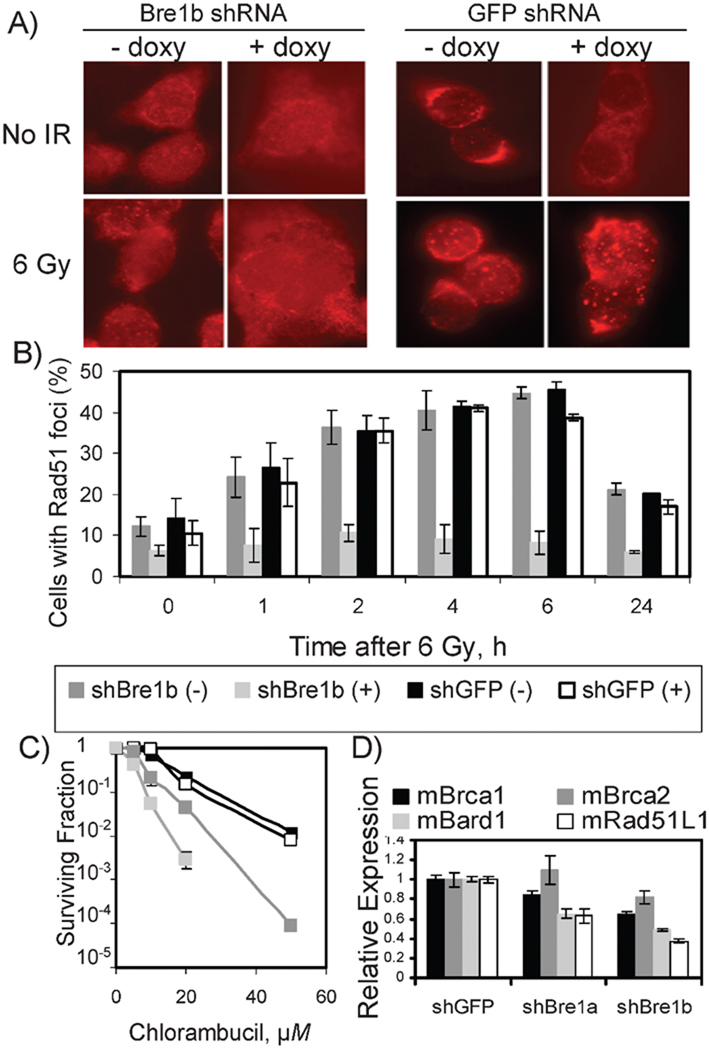
A defect in homologous recombination (HR) repair in the yeast bre1Δ mutant has been reported previously by Game et al. (6) on the basis of radiation sensitivity and epistasis analysis. Consistent with the defect in HR, the Bre1b knockdown cells in our study showed a drastic reduction in radiation-dependent formation of Rad51 foci (Fig. 4A and B) and increased sensitivity to the crosslinking agents chlorambucil and mitomycin C (Fig. 4C and our unpublished observations) and to ionizing radiation (Fig. 1D and 2B). The level of spontaneous Rad51 foci was also significantly lower in the Bre1b knockdown cells (Fig. 4B), suggesting that Bre1b-deficient cells could not efficiently resolve double-strand breaks occurring during replication. The HR defect associated with the loss of Bre1b in RIF-1 cells manifested itself as an overall reduction rather than a delay in Rad51 focus formation in response to radiation. Although the frequency of cells with Rad51 foci was similar between the Bre1b shRNA(−) cells and the control GFP shRNA(−/+) cells, the foci formed in the Bre1b shRNA(−) cells were smaller. The levels of Rad51 protein in the Bre1b knockdown cells were not affected by the loss of Bre1b (Fig. 1A), but the analysis of the gene expression array (to be reported elsewhere) suggested that at least three HR genes were affected by the depletion of Bre1a or Bre1b. Here we found that deficiency in either Bre1a or Bre1b resulted in suboptimal expression of the Brca1, Bard1 and Rad51L1 genes (Fig. 4D).
FIG. 4.
Depletion of Bre1b affects recombinational repair-related phenotypes in mouse REF-1 cells. Panel A: Rad51 focus formation is defective in Bre1b-deficient cells. Cells were irradiated with 6 Gy and fixed 2 h after irradiation. Panel B: Rad51 focus formation at different times after irradiation with 6 Gy. Means and standard deviations from four independent experiments are shown. Panel C: Bre1b-deficient cells are sensitive to chlorambucil. Note the intermediate sensitivity of the Bre1b shRNA cell line in the absence of doxycycline [Bre1b shRNA(+)]. Color is the same as in panel B. Means and ranges from two independent experiments are shown. Panel D: RT-PCR shows down-regulation of expression of homologous recombination proteins mBrca1, mBard1 and mRad51L1 in Bre1b-deficient RIF-1 cells. “sh” before a gene name indicates shRNA.
DISCUSSION
Using shRNA to suppress the function of the Bre1a/b (BRE1A/B) complex, we examined the role of these proteins in the radiation response of mammalian cells. We found that knockdown of either Bre1a or Bre1b in mouse cells resulted in a moderate but significant increase in radiation sensitivity. In addition, depletion of BRE1A resulted in defective G2/M checkpoint arrest in human cells. We found that compromised function of the Bre1a/b complex was associated with defects in homologous recombination, manifested as increased sensitivity to crosslinking agents and abnormal recruitment of Rad51 protein to the ionizing radiation-induced foci. Our results indicate that the defects observed in the Bre1a/b knockdowns are not due to altered methylation of H4K20 but rather are associated with reduced dimethylation of H3K79.
Monoubiquitination of H2B mediated by Bre1 is known to facilitate methylation of histone H3 residues K4 and K79 in yeast and mammals (1, 2, 26, 27). Extensive evidence points to a significant role of H3K79 methylation in radiation sensitivity in yeast, while methylation of H3K4 is not thought to have an impact, because several yeast mutants defective in H3K4 are not sensitive to ionizing radiation (6). It is believed that the H3K79 methylation plays a role in radiation response through binding of the Rad9 (or its ortholog 53BP1) tudor domain to dimethylated H3K79. Yeast cells mutant in the Rad9 tudor domain show a radiation sensitivity similar to that of dot1 deletion mutants (and less than that of the rad9 deletion mutant) (7), and deletion of DOT1 does not add further sensitivity to rad9 tudor domain (7) or rad9 deletion (6) mutants. In mammalian cells, recruitment of 53BP1-GFP fusion protein to sites of DNA damage is abolished by mutations in the tudor domain of 53BP1 and by suppression of methylation of H3K79 in the DOT1L knockdown cells (31). Knockdown of Dot1L has been shown to result in increased radiation sensitivity in mammalian cells (28). However, it remains to be seen whether mutation in the tudor domain of 53BP1 would lead to increased sensitivity to radiation in mammalian cells as it does with Rad9 in Saccharomyces (7).
Similarly, H3K79 methylation has also been shown to participate in the activation of G1 and intra-S checkpoints in response to DNA damage in Saccharomyces (3, 4, 6). In these studies the bre1Δ strains largely showed the same cell cycle checkpoint defects as dot1Δ, rad9 tudor domain and H3K79 mutants, implying that the checkpoint role of Bre1 was mediated by the H3K79 dimethylation it regulates. These studies showed no obvious defect in the G2/M mitotic checkpoint in cells arrested by nocodazole before irradiation. Note that the “G2/M” checkpoint measured in nocodazole-arrested cells should be distinguished from the G2/M mitotic entry checkpoint, because nocodazole arrests cells in prometaphase of mitosis, and the G2/M checkpoint controls the transition from G2 to M. None of the above studies examined the function of the G2/M transition checkpoint known to be defective in 53BP1-deficient cells (39).
The controversy still remains unresolved as to which methylated residue (H3K79me2 or H4K20me2) is preferred by the damage sensor 53BP1 in mammalian cells [see ref. (30) for review]. Our data show that H4K20 dimethylation is not affected by the Bre1a/b status in mouse RIF-1 cells. Therefore, based on available evidence, we attribute the defects in the radiation response of the Bre1a or Bre1b knockdowns to changes in Bre1-dependent dimethylation of H3K79. Separate evidence supporting the roles of each of H3K79me2 and H4K20me2 is strong (31, 32), so it seems that there is a role for both those modifications in radiation response in mammalian cells, with one of the two modifications probably being more important for a specific situation depending on the cell cycle stage or cell type. Thus, although the defect in the G2/M checkpoint we observed in BRE1A-deficient cells can be explained by suboptimal H2B ubiquitination-regulated transcription of cell cycle control genes (our unpublished observations), it may also be caused by abnormal recruitment of 53BP1 to the sites of double-strand breaks resulting from impaired H3K79 dimethylation.
A defect in HR is one of the consequences of the Bre1a/b depletion that could contribute directly to the increased radiation sensitivity. HR repair has been shown to be defective in the yeast bre1Δ mutant (6). In that study the radiation sensitivity phenotypes of single and multiple mutants involving bre1Δ, lge1Δ, rtf1Δ, dot1Δ and rad51Δ suggested that each gene affected radiation resistance through the same RAD51-dependent mechanism mediated by H3K79 methylation. Consistent with this, a recent study showed that both Dot1 and Rad9 are required for efficient sister chromatid recombination in Saccharomyces cerevisiae (20). The observation (7) that the dot1Δ rad9 tudor domain double mutant strain shows radiation sensitivity equivalent to that of the single mutants provides support for the argument that the observed binding of Rad9 to H3K79me2 is involved in this Rad51-dependent repair. In mammalian cells, the formation of Rad51 foci has been reported to be unaffected by 53BP1 status (39). Therefore, the defect in Rad51 focus formation we observed may be a consequence of suboptimal expression of HR genes [(35) and our observations] due to downregulation of H2B ubiquitination-regulated transcription.
In summary, we found that mammalian Bre1 confers radiation resistance and is important for homologous recombination and for the normal function of the G2/M checkpoint. We also found that loss of mammalian Bre1 does not affect dimethylation of H4K20. Thus our finding of a radiation sensitivity phenotype in the Bre1-deficient cells supports a role for H3K79me2 in mammalian repair. The defects we observed in Bre1-depleted mammalian cells are reminiscent of those ones caused by the deletion of Bre1 in Saccharomyces cerevisiae, pointing to an evolutionary conservation of the pathway in eukaryotes.
ACKNOWLEDGMENTS
We are indebted to Dr. Eric Campeau for supplying vectors that were used in this study as part of an inducible H1 lentiviral RNA interference system. This work was supported by National Institutes of Health grants CA-118202 and CA-067166 awarded to JMB.
REFERENCES
- 1.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: transhistone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 2.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 3.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 4.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Game JC, Williamson MS, Baccari C. X-ray survival characteristics and genetic analysis for nine Saccharomyces deletion mutants that show altered radiation sensitivity. Genetics. 2005;169:51–63. doi: 10.1534/genetics.104.028613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Game JC, Williamson MS, Spicakova T, Brown JM. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics. 2006;173:1951–1968. doi: 10.1534/genetics.106.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grenon M, Costelloe T, Jimeno S, O’Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast. 2007;24:105–119. doi: 10.1002/yea.1441. [DOI] [PubMed] [Google Scholar]
- 8.Game JC, Chernikova SB. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair. 2009;8:470–482. doi: 10.1016/j.dnarep.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 10.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 11.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 12.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 13.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 15.Dehe PM, Pamblanco M, Luciano P, Lebrun R, Moinier D, Sendra R, Verreault A, Tordera V, Geli V. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 2005;353:477–484. doi: 10.1016/j.jmb.2005.08.059. [DOI] [PubMed] [Google Scholar]
- 16.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 17.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 18.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conde F, San-Segundo PA. Role of Dot1 in the response to alkylating DNA damage in Saccharomyces cerevisiae: regulation of DNA damage tolerance by the error-prone polymerases Polzeta/Rev1. Genetics. 2008;179:1197–1210. doi: 10.1534/genetics.108.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 22.Greeson NT, Sengupta R, Arida AR, Jenuwein T, Sanders SL. Di-methyl H4 Lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J. Biol. Chem. 2008;283:33168–33174. doi: 10.1074/jbc.M806857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Reinberg D. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 24.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 25.Zofall M, Grewal SI. HULC, a histone H2B ubiquitinating complex, modulates heterochromatin independent of histone methylation in fission yeast. J. Biol. Chem. 2007;282:14065–14072. doi: 10.1074/jbc.M700292200. [DOI] [PubMed] [Google Scholar]
- 26.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Lin YH, Kakadia PM, Chen Y, Li YQ, Deshpande AJ, Buske C, Zhang KL, Zhang Y, Xu GL, Bohlander SK. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood. 2009;114:651–658. doi: 10.1182/blood-2009-03-209395. [DOI] [PubMed] [Google Scholar]
- 29.Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Chen T. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem. Soc. Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 31.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 32.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, Jenuwein T. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Oren M. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PloS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twentyman PR, Brown JM, Gray JW, Franko AJ, Scoles MA, Kallman RF. A new mouse tumor model system (RIF-1) for comparison of end-point studies. J. Natl. Cancer Inst. 1980;64:595–604. [PubMed] [Google Scholar]
- 39.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]