Abstract
Rationale
Congenital heart defects (CHD) represent the most common human birth defects. Even though the genetic cause for these syndromes has been linked to respective candidate genes, the underlying molecular mechanisms are still largely unknown. Disturbance of neural crest cell (NCC) migration into the derivatives of the pharyngeal arches and pouches can account for many of the developmental defects.
Objective
To investigate the function of miRNA in NCCs and cardiovascular system.
Methods and Results
we deleted Dicer from neural crest cell (NCC) lineage and showed that Dicer conditional mutants exhibit severe defects in multiple craniofacial and cardiovascular structures, many of them are observed in human NCCS patients. We found that cranial NCCs require Dicer for their survival and deletion of Dicer led to massive cell death and complete loss of NCC-derived craniofacial structures. In contrast, Dicer and miRNAs were not essential for the survival of cardiac NCCs. However, the migration and patterning of these cells were impaired in Dicer knockout mice, resulting in a spectrum of cardiovascular abnormalities, including Type B Interrupted Aortic Arch (IAA-B), Double Outlet Right Ventricle (DORV) and Ventricular Septal Defect (VSD). We show that Dicer loss-of-function was, at least in part, mediated by miR-21 and miR-181a, which in turn, repressed the protein level of Sprouty 2, an inhibitor of Erk1/2 signaling.
Conclusion
Our results uncovered a central role for Dicer and miRNAs in neural crest cells survival, migration and patterning in craniofacial and cardiovascular development which, when mutated, lead to congenital neuro-craniofacial-cardiac defects.
Keywords: microRNAs, neural crest cells, Dicer, cardiovascular development
Introduction
Congenital heart diseases are present in about 1 percent of live births and are the leading cause of human congenital birth defects. The pathogenesis of congenital heart disease is varied and the genetic cause of disease remains to be completely understood. Intriguingly, phenotypic overlap between genetic disorders in cardiovascular and neuronal-craniofacial defects, including DiGeorge Syndrome (DGS), Noonan syndrome, LEOPARD syndrome, cardio-facio-cutaneous (CFC) syndrome, and Costello syndrome, has been described1, 2. DGS is caused by a deletion of 3 million base pairs on one copy of chromosome 223, 4. Genetic studies in mouse have shown that heterozygous deletion of Tbx1, a gene located in the deleted region of 22q11, causes a DGS-like phenotype5–7. However, signals downstream of Tbx1 which mediate DGS phenotype remain unclear. Genetic studies have linked ~50% of Noonan syndrome to mutations in PTPN11 gene, which encodes SHP-2 protein, an SH2 domain-containing, non-receptor tyrosine phosphatase (PTPase) essential for cellular proliferation, differentiation and migration8. Numerous human genetic studies revealed that mutations in the components of the RAS/RAF/MEK/ERK signaling cascade, which are key elements of a major intracellular signal transduction pathway involved in essential biological processes, were associated with many of the above-mentioned neuronal-craniofacial-cardiovascular syndromes9. Genetic studies in animal models in which the components of the signaling cascade were deleted, conventionally or in a spatiotemporal-specific manner, further support this view10.
The development of outflow tract and aorticpulmonary arteries is a complex process which involves precise regulation of cell proliferation, differentiation, migration and apoptosis of cells derived from multiple sources. Perturbation of this developmental process is often associated with congenital cardiovascular defects11. Neural crest cells (NCCs) are a transient, migratory population of progenitor cells that give rise to many cell types, including vascular smooth muscle cells, chondrocytes, melanocytes and neurons12. Cranial NCCs, a subset of NCCs, which migrate into 1st and 2nd pharyngeal arches, give rise to cranial ganglia, the maxilla, the mandible as well as muscle and cartilage of the head and neck13. Cardiac NCCs, another subset of NCCs that migrate into the 3rd, 4th and 6th pharyngeal arches, populate the aorticpulmonary septum and conotruncal cushion. They also contribute to the development of smooth muscle of great vessels14. Cardiac NCCs have long been known to be essential for proper development and patterning of great arteries and for outflow tract septation. Ablation of NCCs during early embryogenesis resulted in severe defects in cardiovascular structures11. However, signals that are necessary for the proper NCC development, patterning, survival and migration are not completely understood15, 16.
miRNAs are a class of ~22 nucleotide non-coding RNAs that regulate gene expression post-transcriptionally17. Despite the identification of large number of miRNAs18, the biological roles of most miRNAs and the molecular mechanisms underlying their function remain largely unknown. Dicer is an endonuclease required for miRNA biogenesis and the maturation of miRNAs19. Our previous studies revealed an essential role of Dicer and miRNAs in normal heart formation and function20. Recent reports documented that NCC-expressed miRNAs are essential for the development of craniofacial structure 21, 22. In the present study, we generated neural-crest-cell-specific Dicer mutant mice. Targeted deletion of Dicer in NCCs resulted in severe defects in craniofacial structures and malformation of cardiovascular system, including ventricular septal defects, double outlet right ventricle and type B interrupted aortic arch. Our studies demonstrated that Dicer is required for the survival of cranial NCCs, whereas deletion of Dicer in cardiac NCCs altered their migration, patterning and the morphogenesis of aortic arch arteries and outflow tract.
Results
Neural-crest-cell-specific Dicer mutant mice are perinatal lethal
To investigate the role of Dicer and miRNAs in the development of neural crest cell (NCC) lineage, we deleted Dicer in NCCs. We bred the Dicer-floxP mice, in which the 22nd and 23rd exons of Dicer gene are flanked by two loxP sites23, with a Wnt1-Cre transgenic mouse line (Supplemental Figure IA). The Wnt1-cre displayed robust Cre recombinase activity in NCC lineage as early as embryonic day 9.5 (E9.5), as evidenced by the Rosa-LacZ indicator mice (Supplemental Figure IB), consistent with a previous report24. Mice with heterozygous deletion of Dicer in NCCs (Wnt1-Cre/Dicerflox/+) were fertile and exhibited no apparent abnormalities. However, homozygous mutant mice (Wnt1-Cre/Dicerflox/flox) were found dead shortly after birth with no exception. Genotyping of 228 embryos at different developmental stages identified the Mendelian ratio for all four expected genotypes (Supplemental Table I), indicating that NCC-specific deletion of Dicer did not result in embryonic lethality. qRT-PCR and Western blot analyses using samples from pharyngeal arches of E11.5 mutant embryos confirmed a dramatic reduction of Dicer expression (Supplemental Figure IC, ID). We could still detect residual Dicer expression level, primarily because pharyngeal arch tissues also contain non-NCC cell lineage. These data demonstrate that Dicer is indispensable for the development of NCCs.
Neural-crest-cell-specific Dicer mutant mice display severe craniofacial defects
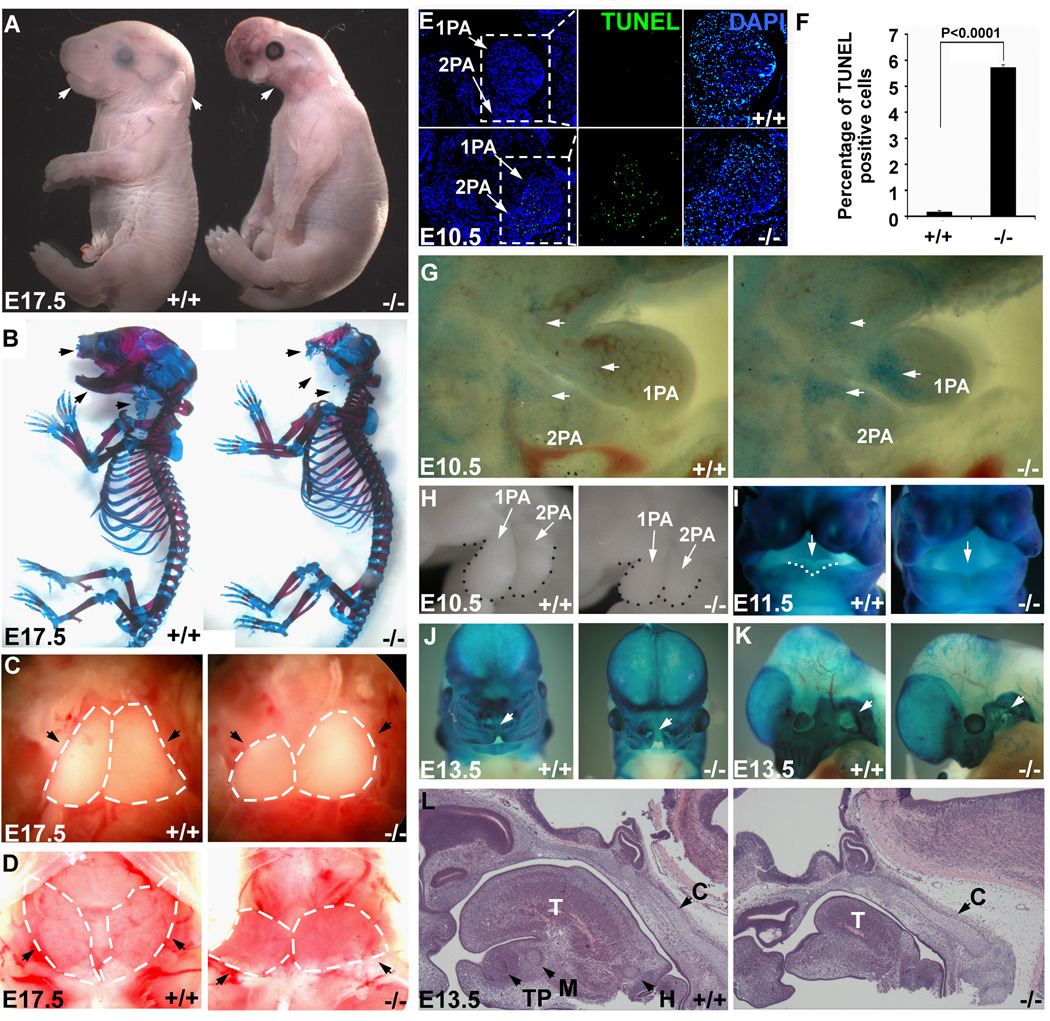
All neural-crest-cell-specific-Dicer (herein referred to as NCC-Dicer) mutants exhibit severe craniofacial defects with 100% penetrance. These defects include the complete absence of maxilla, nose, mandible, external ear and eyelid at E17.5 (Figure 1A, arrows). Skeletal preparation of E17.5 embryos clearly demonstrated developmental defects of craniofacial skeleton and cartilage in NCC-Dicer mutants. As shown in Figure 1B (arrows), the maxilla, mandible and nasal bones are completely absent in mutant embryos. Moreover, most neural-crest-derived cartilages, including Meckel’s cartilage, hyoid, thyroid cartilage and tracheal cartilages, did not form in NCC-Dicer mutants (Figure 1B). In addition to contributing to the development of craniofacial structures, NCCs are also known to contribute to the development of thymus and parathyroid glands25. Accordingly, we observed that NCC-Dicer mutant embryos displayed hypoplastic thymus and the parathyroid glands were underdeveloped (Figures 1C, D, arrows). Our data therefore established Dicer as an essential factor for the development of NCC-derived structures and are consistent with recent reports 21, 22.
Figure 1. Neural-crest-specific ablation of Dicer resulted in severe craniofacial defects.
In all images, wild type (+/+) control and NCC-Dicer mutant (−/−) embryos are shown. (A) Gross morphology of E17.5 embryos. Neural-crest-specific Dicer mutant embryos displayed severe craniofacial defects (arrows). (B) Skeletal preparation of E17.5 embryos. Most of the neural-crest-derived cartilages and bones, including maxilla, mandible and nasal bones (arrows), were absent in Dicer mutants. (C) Thoracotomies showed the hypoplasia of thymus (arrows) in mutant embryos. (D) Dicer mutant mice exhibited underdeveloped thyroid glands (arrows). (E) TUNEL assay showed an abnormal apoptosis in the 1st (1PA) and 2nd (2PA) pharyngeal arches in mutant embryos. (F) Statistics of TUNEL-positive cells. Three discontinuous sections containing 1st and 2nd pharyngeal arches were stained and counted. (G) Whole mount NBS staining consistently revealed the abnormal apoptosis in mutants. (H) The morphology of 1st and 2nd pharyngeal arches of E10.5 embryos showed the reduced size of pharyngeal arches in Dicer mutants. (I–K) Whole mount β-gal staining of E11.5 and E13.5 embryos containing Rosa-LacZ indicator. E11.5 Dicer mutant embryos failed to form the lateral lingual swellings (I, arrows); E13.5 Dicer mutant embryos displayed a cleft lip (J, arrows) and a hypoplastic pinna (K, arrows). (L) Histological examination of E13.5 embryonic heads. The neural-crest-derived Meckel’s cartilage, hyoid cartilage and tooth primordium failed to form in mutants, while the development of non-neural-crest-derived cartilage primordium of clivus is unaffected. C: cartilage primordium of clivus; H: hyoid cartilage; M: Meckel’s cartilage; T: tongue; TP: tooth primordium.
Next, we sought to determine the developmental timing of craniofacial defects found in NCC-Dicer mutants. Cranial NCCs migrate through the 1st and 2nd pharyngeal arches (PAs) to give rise to many of the craniofacial structures26. We examined the proliferation, apoptosis and migration of cranial NCCs in NCC-Dicer mutant embryos. There was no difference in cell proliferation of cranial NCCs in E10.5 NCC-Dicer mutant embryos, when compared with controls (Supplemental Figure II). However, massive apoptosis occurred in the 1st and 2nd PAs as early as E10.5 in NCC-Dicer mutant embryos, evidenced by a significant increase in TUNEL staining (Figure 1E). Increased apoptosis in 1st and 2nd PAs was further confirmed by the increased signal of cleaved-caspase 3 (Supplemental Figure III). In addition, whole-mount Nile blue sulfate (NBS) was applied in E10.5 embryos to further demonstrate an increased apoptosis in NCC-Dicer mutant embryos (Figure 1G). As a consequence, the 1st and 2nd PAs of NCC-Dicer mutant embryos were substantially smaller than that of the controls. (Figure 1H). NCC-Dicer mutant embryos failed to form the lateral lingual swellings in mandibular portion of the 1st pharyngeal arch (Figure 1I, arrows), and there was a dramatic decrease in the formation of tongue (Figure 1L). Despite the increased apoptosis in cranial NCCs, the overall morphogenesis of the 1st and 2nd PAs was not significantly affected in E10.5 NCC-Dicer mutant embryos, evidenced by a normal PECAM staining pattern, which marks in the pharyngeal endothelium (Supplemental Figure IV). At E13.5, NCC-Dicer mutant embryos also displayed a cleft lip (Figure 1J, arrows) and hypoplastic pinna (Figure 1K, arrows). Interestingly, the development of cartilage primordium of clivus, which is not derived from cranial NCCs, was unaffected in NCC-Dicer mutant embryos (Figure 1L, arrows), demonstrating the specificity of defects. Finally, we examined the development of neural-crest-derived cranial ganglia in DCC-Dicer mutant embryos. We found that the development of ganglion of cranial nerve V, revealed by whole mount immunostaining using an anti-neurofilament antibody, was slightly affected at E10.5, however, such defects become more profound in E13.5 NCC-Dicer mutant embryos (Supplemental Figure V). These data demonstrate that Dicer is indispensable for the survival of cranial NCC progenitor cells and NCC-specific deletion of Dicer severely perturbed the proper development of neural-crest-derived craniofacial structures in a cell-autonomous manner.
NCC-specific Dicer mutation leads to multiple cardiovascular defects
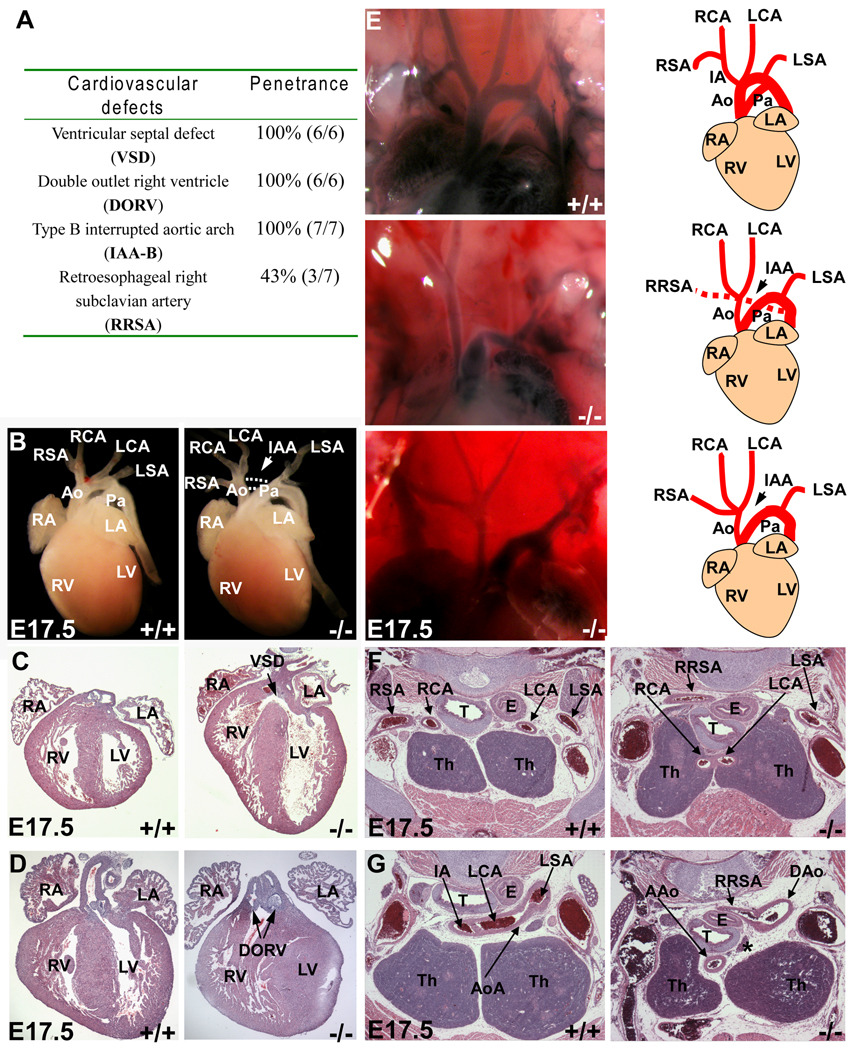
Cardiac NCCs migrate into the embryonic heart field and contribute to the development of aortic arch arteries and outflow tract. It is therefore not surprising that NCC-Dicer mutant embryos exhibited multiple cardiovascular defects (Figure 2A). All NCC-Dicer mutant embryos examined displayed interrupted aortic arch (IAA) at E17.5 (n=7), in which the aortic arch is not connected to the descending aorta (Figure 2B, arrows). Histological examination also revealed ventricular septal defect (VSD) in the heart of all NCC-Dicer mutant embryos (Figure 2C). We found that all NCC-Dicer mutant embryonic hearts displayed double outlet right ventricle (DORV), in which the right ventricle was connected to both the pulmonary artery and aorta (Figure 2D). In order to view aortic artery defects in more detail, we injected ink into the ventricle of E17.5 embryonic hearts. This allowed us to clearly define a Type B interrupted aortic arch (IAA-B), in which the aortic arch is interrupted between the common carotid artery and the left subclavian artery, in NCC-Dicer mutant embryos (Figure 2E), In addition, about 50% of NCC-Dicer mutant embryos exhibited retroesophageal right subclavian artery (RRSA), in which a right subclavian artery ectopically originated from descending aorta instead of innominate artery and extended rightward behind esophagus (Figure 2E). Histological sections further confirmed RRSA in the NCC-Dicer mutant embryos (Figures 2F, G). Furthermore, NCC-Dicer expression is required for the correct position and patterning of the carotid arteries and thymus. As shown in Figure 2, both carotid arteries, the left carotid artery (LCA) and right carotid artery (RCA), were misplaced in NCC-Dicer mutant embryos: instead of lying behind the thymus as in the normal control embryos, both carotid arteries were found passing through the thymus in NCC-Dicer mutant embryos (Figure 2F). Together, our data established that Dicer played a crucial role in cardiac NCCs and is required for the proper development of NCC-derived cardiovascular system.
Figure 2. Neural-crest-specific ablation of Dicer led to multiple cardiovascular defects.
(A) The penetrance of cardiovascular defects displayed in Dicer mutant mice. (B) Gross morphology of heart and great vessels from E17.5 embryos. The Dicer mutant mice exhibited a type B interrupted aortic arch (arrow). (C–D) Histological examination of E17.5 embryonic hearts. The mutant hearts possessed ventricular septal defect (C) accompanied with a double outlet right ventricle (D). (E) Great vessels shown by ink injection. The type B interrupted aortic arch was consistently found in mutant embryos. (F–G) Histological examination of defects in great vessels. A right subclavian artery was ectopically originated from the descending aorta (G) and extended rightward behind the esophagus (F, G). The asterisk in (G) indicates the interruption of aortic arch. AAo: ascending aorta; Ao: aorta; AoA: aortic arch; DAo: descending aorta; DOSV: double outlet right ventricle; E: esophagus; H: heart; IA: innominate artery; IAA: interrupted aortic arch; LA: left atrium; LCA: left carotid artery; LSA: left subclavian artery; LV: left ventricle; Pa: pulmonary artery; RA: right atrium; RCA: right carotid artery; RSA: right subclavian artery; RRSA: retroesophageal right carotid artery; RV: right ventricle; T: trachea; Th: thymus; VSD: ventricular septal defect.
Distinct requirement for Dicer and miRNAs in cranial and cardiac NCC development
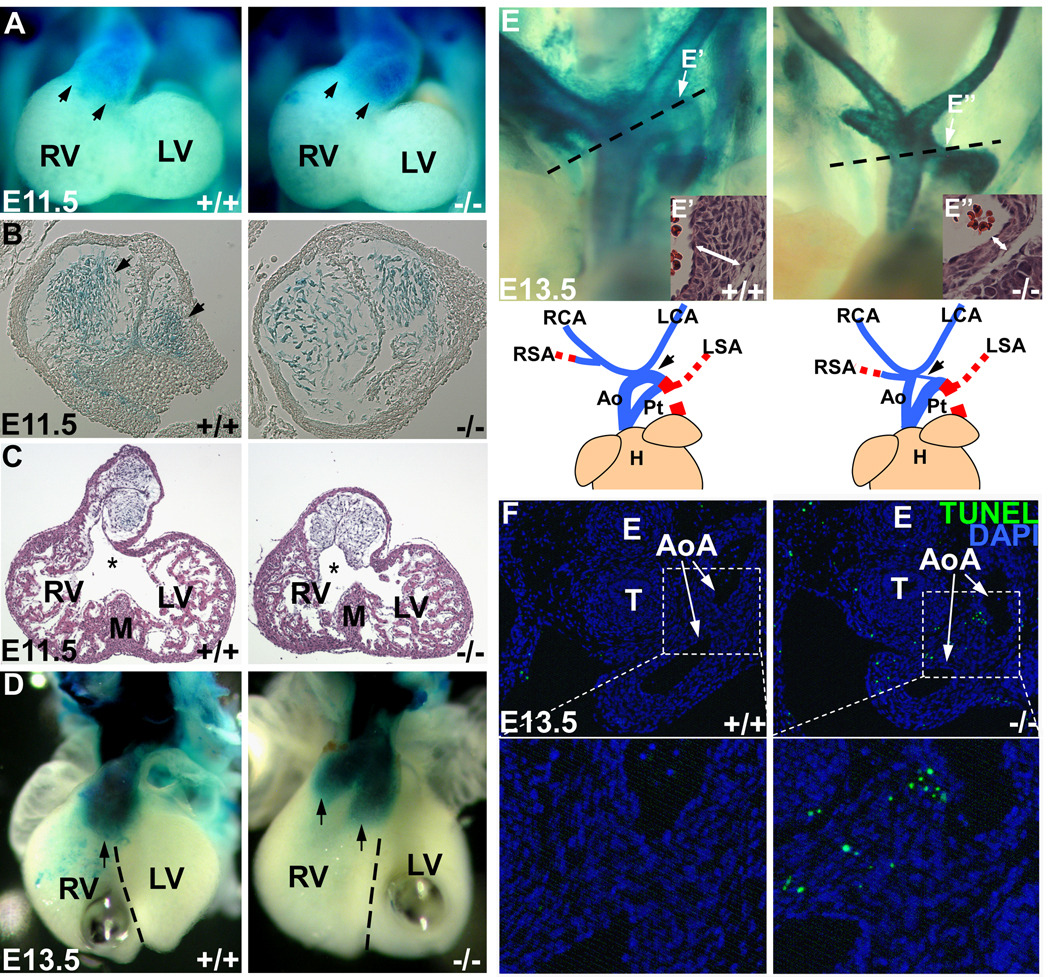
NCC-Dicer mutant mice displayed defects in craniofacial and cardiovascular development. However, the severity of defects was quite distinct, loss-of-Dicer in NCCs led to almost complete absence of craniofacial structures, whereas the defects in cardiovascular system were less severe and were primarily associated with patterning disorders. We asked whether there was a distinct requirement for Dicer and miRNAs in cranial versus cardiac NCCs during development. Cranial NCCs primarily migrate through the 1st and 2nd PAs and Dicer is required for their survival (Figures 1E, F, G). In contrast, cardiac NCCs migrate into the outflow tract, primarily through the 3rd, 4th and 6th PAs, to form the aorticopulmonary septum, and two prongs of cardiac NCCs will further extend to anterior heart field and contribute to the closure of ventricular septum27. To track the migration of NCCs, the Rosa-LacZ indicator was introduced into NCC-Dicer mutant mice. Whole mount β-gal staining revealed that NCC-specific deletion of Dicer did not prevent NCCs from migrating into all pharyngeal arches and the outflow tract at E11.5 (Figure 3A and Supplemental Figures VI A, B). Next, we examined whether NCC-Dicer mutant embryos displayed defects in their proliferation or apoptosis in cardiac NCC-derived structures. Interestingly, no aberrant cellular proliferation or apoptosis was found in the 3rd, 4th and 6th PAs or in the outflow tract of E10.5 NCC-Dicer mutant embryos (Supplemental Figures VI C–F). This is in sharp contrast to the dramatic increase of apoptosis detected in the 1st and 2nd PAs (Figures 1E–G). However, the migration pattern of cardiac NCCs in the outflow tract was altered in NCC-Dicer mutant embryos (Figures 3B, D). At E11.5, Dicer-deficient cardiac NCCs failed to form two columns of condensed conotruncal cushion in the outflow tract, when compared to wildtype controls (Figure 4B, arrows). In addition, a malalignment between the outflow tract entrance for future aorta and the muscular interventricular septum occurred in NCC-Dicer mutant mice (Figure 3C). At E13.5, two prongs of cardiac NCCs, which are normally aligned with ventricular septum in a fore-and-aft position, rotated improperly and were located in right ventricle in a side-by-side manner, leading to a malalignment with ventricular septum in mutant mice (Figure 3D). The aberrant rotation and malalignment of conus cushion between E11.0 to E12.5 have been reported to result in VSD and DORV28. Our data suggested that abnormal migration and patterning of cardiac NCCs in outflow tract led to the rotation and alignment defects of conus cushion, which eventually contribute to the VSD and DORV defects observed in NCC-Dicer mutant mice.
Figure 3. Dicer and miRNAs are required for proper development of neural crest cells.
(A) Whole mount β-gal staining of E11.5 embryos containing Rosa-LacZ indicator showed the migration of cardiac neural crest to the caudal limit of outflow tract (arrows). (B) Histological examination of the stained outflow tract. The Dicer-deficient cardiac NCCs failed to form two columns of condensed mesenchyme in outflow tract. (C) Histological examination of E11.5 heart. The entrance of outflow tract for future aorta (asterisks) had a rightward malalignment with muscular interventricular septum. (D) Whole mount β-gal staining of E13.5 embryonic hearts indicated the rotation defect and malalignment of conus cushion. Two prongs of Dicer-deficient cardiac NCCs (arrows) located in right ventricle in a side-by-side manner. The dash lines indicate the ventricular septum. (E) Neural-crest-derived great vessels in E13.5 embryos were shown by β-gal staining. The blue lines in cartoon indicate the neural-crest-derived vessels, while the red dash lines indicate the vessels not contributed by cardiac NCCs, which cannot be shown in β-gal staining. Mutant embryos displayed a hypoplastic aortic arch artery. Histological examination of aortic arch was performed in the orientation indicated by dashed lines. The regions (E’, E’’) indicated by arrows were shown at bottom right and Supplemental Figure VII. The aortic arch in mutant embryos had a thinner vessel wall with fewer smooth muscle cells. (F) TUNEL assay with E13.5 embryo sections containing aortic arch showed an abnormal apoptosis of neural-crest-derived smooth muscle cells in the vessels in Dicer mutant embryos. (G) Statistics of TUNEL-positive cells. Three discontinuous sections containing aortic arches were stained and counted. Ao: aorta; AoA: aortic arch; E: esophagus; H: heart; LA: left atrium; LCA: left carotid artery; LSA: left subclavian artery; LV: left ventricle; M: muscular interventricular septum; Pt: pulmonary trunk; RA: right atrium; RCA: right carotid artery; RSA: right subclavian artery; RV: right ventricle; T: trachea.
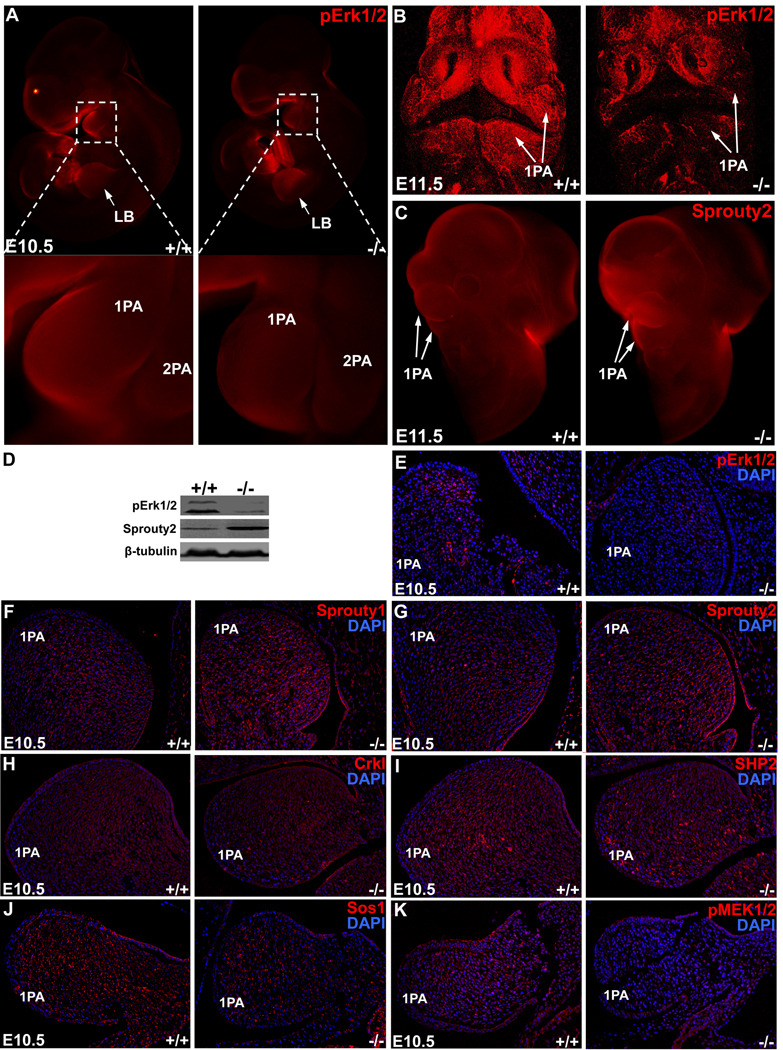
Figure 4. Dysregulation of gene expression of components in MEK/ERK signaling cascade in NCC-Dicer mutants.
(A–B) Whole mount immunochemistry detecting phospho-Erk1/2 (pErk1/2) in E10.5 (A, side view) and E11.5 (B, frontal view) embryos. pErk1/2 was downregulated in neural-crest-derived pharyngeal arches but not in limb buds of Dicer mutant embryos. (C) Whole mount immunochemistry detecting Sprouty2 in E11.5 embryos. Sprouty2 was upregulated in neural-crest-derived pharyngeal arches of Dicer mutant embryos. (D) The dysregulation of pErk1/2 and Sprouty2 was shown by Western blot with pharyngeal arch tissue samples. (E–K) Examination of gene expression by immunohistochemistry detecting pErk1/2 (E), Sprouty1 (F), Sprouty2 (G), Crkl (H), SHP-2 (I), Sos1 (J) and pMEK1/2 (K) expression in first pharyngeal arches of E10.5 Dicer mutant embryos. 1PA: 1st pharyngeal arch; 2PA: second pharyngeal arch; LB: limb bud.
The defective development of 4th pharyngeal arch (PA) is often associated with the type B IAA in congenital heart disease29. To investigate the etiology of type B IAA found in NCC-Dicer mutant mice, we examined the morphogenesis of the 4th PA during the outflow tract remodeling in wildtype controls and NCC-Dicer mutant embryos. No obvious defect of 4th PAA was found in E10.5 NCC-Dicer mutant embryos, when compared with that of wildtype controls (Supplemental Figure IV). However, a hypoplastic aortic arch artery became evident in E13.5 NCC-Dicer mutant embryos (Figure 3E, arrows). Histology analyses clearly demonstrated much thinner vessel wall in the aortic arch artery of NCC-Dicer mutant embryos, when compared with controls (Figures 3E’, E’’ and Supplemental Figure VII). TUNEL assays further revealed an increased apoptosis in the aortic arch artery of NCC-Dicer mutant embryos (Figures 3F, G). We noticed that apoptosis only occurred at a restricted region of the aortic arch artery and no increase in apoptosis was observed in the ascending aorta or carotid arteries of mutant embryos. These observations suggested that apoptosis occurred in the developing aortic arch artery likely contributed to IAA-B of NCC-Dicer mutant embryos. These data further suggest that there is a distinct requirement for Dicer and miRNAs during the development of the 4th versus 3rd/6th PAs during outflow tract remodeling.
Next, we examined whether loss of Dicer in NCCs affected the specification of vascular smooth muscle cell (SMC) fate and the morphogenesis of aortic arch arteries. Immunohistochemistry using smooth muscle markers, including smooth muscle actin (SM-actin) and SM-α-22, demonstrated that SMC differentiation was not affected in NCC-Dicer mutant aortic arteries (Supplemental Figure VIII). However, deletion of Dicer from NCCs appeared to affect the assembly of smooth muscles in aortic arch arteries (Supplemental Figure IX). The ascending aorta, but not the descending aorta (which is not derived from cardiac NCCs), displayed fewer layers of smooth muscle cells in E17.5 NCC-Dicer mutant embryos (Supplemental Figure IX).
Dicer and miRNAs participate the MEK/ERK signaling cascade to regulate neural crest cell and pharyngeal arch artery development
Signal transduction pathways, in particular the MEK/ERK cascade, play a central role in cell proliferation, differentiation, migration and survival30. Recent studies established that components of this signaling cascade are important for the development of NCCs10. Dysregulation of their expression or genetic mutations of members of this signaling pathway have been associated with human congenital birth defects31. Targeted deletion of MEK or ERK1/2 in mouse NCCs led to severe craniofacial and cardiovascular defects, reminiscent that of NCC-Dicer mutant mice10. We examined the expression level of ERK proteins in NCC-Dicer mutant embryos. Whole mount immunostaining, examined by confocal microscopy, showed that the expression of phosphorylated Erk1/2 (pErk1/2) was significantly decreased in the pharyngeal arches (PAs) of E10.5 and E11.5 NCC-Dicer mutant embryos (Figures 4A, B). Western blot analyses further demonstrated a decreased expression level of pErk1/2 proteins in the PAs of E11.5 NCC-Dicer mutant embryos (Figure 4D). To confirm the above observation, additional immunohistochemistry was performed on E10.5 embryonic sections. Indeed, the expression of pErk1/2 was dramatically decreased in the 1st PA (Figure 4E). We also examined the expression level of total Erk1/2 proteins and found that it was not significantly altered in mutant embryos (data not shown). Notably, the expression level of pErk1/2 proteins did not change in the limb buds of NCC-Dicer mutant embryos (Figure 4A), suggesting that the decreased pErk1/2 expression in the NCC-derived structure is due to the loss of Dicer and miRNAs.
miRNAs are known to repress the expression of their target genes, primarily by degrading the target mRNAs and/or inhibiting their translation18. The observation that pErk1/2 expression level was significantly decreased in the PAs of NCC-Dicer mutant embryos suggested that Erk1/2 is unlikely a direct target of miRNAs in this setting. We speculated that increased expression of a negative regulator/inhibitor of the ERK signaling could explain the observation. Interestingly, sprouty proteins have been characterized as intrinsic inhibitors of the MEK/ERK pathway32. We tested the expression level of sprouty proteins and found that the expression of sprouty2 was significantly increased in NCC-Dicer mutant pharyngeal arches (Figure 4C). Immunostaining using tissue sections confirmed the increase of sprouty1/2 protein level in the 1st PA of NCC-Dicer mutant embryos (Figures 4F, G). Increased sprouty2 expression in NCC-Dicer mutant embryonic PAs was further quantified by Western blot analyses (Figure 4D). We examined the expression of additional components of the MEK/ERK signaling pathway in PAs of NCC-Dicer mutant embryos. Whereas the expression levels of Crkl and SHP2 appeared not affected (Figures 4H, I), the protein levels for SOS1 and pMEK1/2 were decreased in NCC-Dicer mutant embryos (Figures 4J, K).
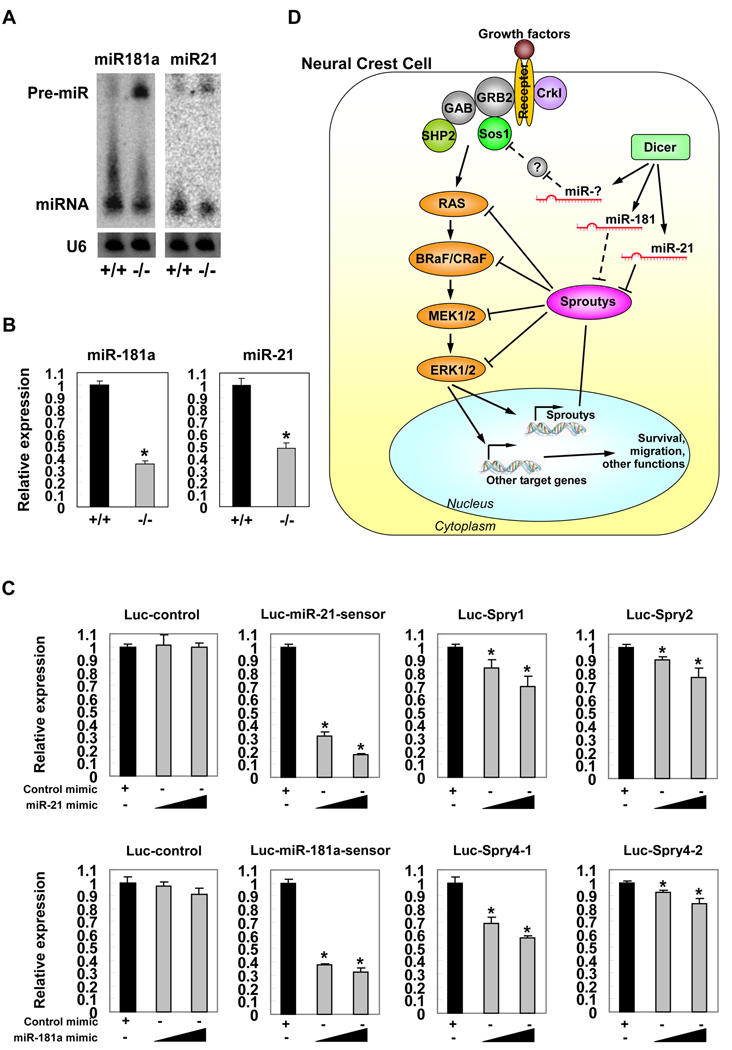
Searching for miRNAs that target sprouty proteins, we found that miR-21 and miR-181a are putative repressors. Interestingly, a recent report showed that miR-21 inhibited the expression level of sprouty1 in cardiac fibroblasts, leading to cardiac hypertrophy and fibrosis33. We examined the expression of miR-21 and miR-181a and found that both miRNAs were expressed in the PAs of E11.5 mouse embryos (Figure 5A). Most importantly, the expression level of mature miRNAs was significantly decreased, accompanying with an accumulation of their precursor miRNAs in the PAs of NCC-Dicer mutants (Figure 5A). Quantitative real-time PCR assays further confirm the reduction of mature miR-21 and miR-181a level (Figure 5B). Such a decreased miR-21 and miR-181a expression inversely correlated with an increased protein level of their target gene, sprouty2. In order to directly test whether miR-21 and miR-181a could repress the expression of sprouty genes, we constructed two repeats of the predicted miR-21 or miR-181a target sequences from the 3’UTRs of indicated sprouty genes downstream of a luciferase gene (Luc-Spry) and co-transfected with miRNA mimics. We have also included Luc-miR-21-sensor and Luc-miR-181a–sensor to serve as controls for miRNA-mediated repression. Transfection of either miR-21 mimic or miR-181a mimic repressed luc-Spry luciferase activity (Figure 5C), suggesting that miR-21 and miR-181a specifically target sprouty genes at 3’ UTRs.
Figure 5. Repression of sprouty expression by miRNAs.
(A) Detection of the haploinsufficiency of mature miRNAs and the accumulation of miRNA precursors in mutant pharyngeal arch tissue samples by Northern blot. (B) Quantitative real-time PCR assays to measure the expression level of mature miR-181a and miR-21 in mutant pharyngeal arch tissues. *: P < 0.01. (C) Hela cells were transfected with indicated luciferase reporters, along with either miR-21 or miR-181a duplex mimic. A Renilla luciferase vector was cotransfected to serve as an internal control for normalization. Cells were harvested and luciferase activity measured 24 hours after transfection. Values are presented as mean luciferase activity ± SD relative to the luciferase activity of reporters with control duplex mimic. *: P < 0.05. (D) Dicer and miRNAs are suggested to participate in a regulatory cascade to modulate the expression levels and activities of MEK/ERK signaling pathway in neural crest cells.
Discussion
Neural crest cells are one of the most important embryonic progenitor cells which can give rise to multiple cell and tissue types, including that of the craniofacial structures and the great arteries, during vertebrate development12. In this study, we demonstrated that Dicer and miRNAs are essential for the development of neural crest cells. Targeted deletion of Dicer in neural crest cells led to severe craniofacial and cardiovascular defects, which are reminiscent to features of human congenital neuro-craniofacial-cardiac defects. Our studies also suggested that perturbation of MEK/ERK signaling pathway by miRNAs contributed to the developmental defects.
The MEK/ERK signaling is one of the most important signal transduction pathways in mammalian cells. Activated Erk1/2 can participate a broad array of cellular functions including proliferation, survival, apoptosis, motility, transcription, metabolism and differentiation34–36. Deregulation of its expression or inhibition of its activity was reported in many developmental defects and pathophysiological conditions37–39. Furthermore, mutation in the components of the MEK/ERK signaling cascade is associated with human disease31. Neural crest cell-specific Erk1/2 double knockout mice displayed craniofacial and cardiovascular defects10, similar to what we observed in NCC-specific Dicer mutant mice described in this study. These observations suggest that inhibition of the ERK signaling pathway is likely responsible for many of the developmental defects in NCC-Dicer mutant mice. Our studies therefore implied that ERK signaling is regulated by miRNAs during neural crest development. Indeed, recent reports showed that ERK activity was positively regulated by miR-2133, 40, 41.
The fact that most miRNAs negatively regulate the expression level of their target genes, together with our observations in which deletion of Dicer (therefore the depletion of miRNAs) resulted in a down-regulation of pErk level, strongly suggested to us that miRNAs in NCCs likely directly targeted an inhibitor of the Erk signaling pathway, not the Erk itself. The Erk signaling antagonist, Sprouty family members (Sprouty1, Sprouty2 and Sprouty4), are strong inhibitors of the MEK/ERK signaling pathway, which can interfere with the MEK/ERK signaling at many levels32. Indeed, our data showed that NCC-Dicer mutation resulted in an increased level of Sprouty proteins and decreased expression of MEK1/2, ERK1/2. In this sense, our results uncovered miRNAs as critical components of the feedback loop to modulate the activities of RTK signaling in neural crest development (Figure 5D). Our data are consistent with recent reports, for example, Sprouty133 and Sprouty241 have been reported to be regulated by miR-21, whereas the Sprouy4 3’ UTR possesses two predicted miR-181a binding sites42. Importantly, both miR-181a and miR-21 were highly expressed in NCCs, suggesting their potential role there. Interestingly, targeted deletion of Dicer in limb mesenchyme led to hypoplastic forelimb and hindlimb development43. Furthermore, Dicer-deficient limbs also displayed a decrease in the expression level of pErk and an increase in sprouty2 expression43. These data further support the view that the relation between Dicer, miRNAs, Sprouty2 and MEK/ERK signaling is common in the development of multiple organs.
miRNAs have emerged as a novel class of key regulators for gene expression in many biological processes. Multiple studies have demonstrated distinct perturbations in miRNA expression under various cardiac remodeling conditions and disease models44–50. However, there were only very limited genetic studies to define the in vivo function of miRNA(s) in cardiovascular system. Our studies provided genetic evidence to support the causal role of miRNAs in neural crest cells for the development of craniofacial and cardiovascular structures. Interestingly, several recent studies reported that deletion of Dicer and depletion of miRNAs from vascular smooth muscle cells or endothelial cells resulted in a variety of vascular development defects51–53, further highlighting the central role of miRNAs in cardiovascular system. It will be important for the future to determine the involvement of individual miRNAs in NCCs and the development of aortic arch arteries. Given the profound defects revealed in this study, we speculate that dysregulation and/or mutation in miRNA genes will be identified in human patients with neural-cranial-facial-cardiac defects.
Materials and Methods
Generation of Dicer conditional null mice
Dicer floxed (Dicerflox/flox) mice23 and Wnt1-Cre transgenic mice24 have been described. Dicerflox/flox/Wnt1-Cre mice were generated by breeding Dicerflox/flox mice with Wnt1-Cre mice. To track the neural crest-derived cells, Rosa-lacZ reporter was introduced by breeding Dicerflox/flox mice with Rosa-lacZ reporter mice54. Dicerflox/flox/Rosa-lacZflox/flox double homozygous offsprings were used in the further mating with Dicerflox/flox/Wnt1-Cre mice. The genotype of all the offsprings were determined by PCR using the following primers: Cre1: 5'-GTTCGCAAGAACCTGATGGACA-3'; Cre2: 5'-CTAGAGCCTGTTTTGCACGTTC-3'; Dicer forward: 5'-ATTGTTACCAGCGCTTAGAATTCC-3'; Dicer reverse: 5'-GTACGTCTACAATTGTCTATG-3'; R26R1: 5'-GCGAAGAGTTTGTCCTCAACC-3'; R26R2: 5'-GGAGCGGGAGAAATGGATATG-3'; R26R3: 5'-AAAGTCGCTCTGAGTTGTTAT-3'.
Northern blot analyses, RT-PCR, TaqMan qPCR and Western blot analyses
Total RNAs were isolated using Trizol Reagent (Invitrogen) from wildtype or Dicer mutant embryonic samples as indicated in figure legends. Northern blot detecting miRNAs was done as described55. For RT-PCR, 2.0 µg RNA samples were reverse-transcribed to cDNA by using random hexamers and MMLV reverse transcriptase (Invitrogen) in 20µl reaction system. 0.25µl cDNA pool was used per qPCR reaction. The sequences of the PCR primers are: DicerRTF: 5’-AGCCGTTGAAGAGGATGACT-3’; DicerRTR: 5’-CACACAGTCCGCTATGCTCT-3’. TaqMan-based quantitative real-time PCR was used to measure the expression levels of mature miR-181a, miR-21 and was performed as recommended by manufacturer (Applied Biosystem). Western blot analyses were performed as described55 using protein extracts from embryonic samples as indicated in figure legends.
Histological examination, immunohistochemistry, Verhoeff's elastic staining and TUNEL assay
Whole embryos and dissected tissues were fixed in 4% paraformaldehyde (pH 8.0) overnight. After dehydration through a series of ethanol baths, samples were embedded in paraffin wax according to standard laboratory procedures. Sections of 5 µm were stained with haematoxylin and eosin (H&E) for routine histological examination with light microscope. For immunohistochemistry, paraffin sections containing the structures of interest were selected. After rehydration for paraffin sections, slides were heated in the 89°C in retrievagen A solution (BD Pharmingen) for 10 min and washed in PBS for 5 min at room temperature. The remaining steps for immunostaining were performed as previously described55. Sections were examined and photographed using regular upright fluorescent microscope. Verhoeff's Elastic Staining, showing the elastic fibre (in black), was performed as described56. TUNEL assay was performed according to the protocol provided in ApopTag Fluorescein In Situ Apoptosis Detection Kit (CHEMICON). At least three pairs of embryos (wildtype controls and mutants) at the indicated developmental stages were used in each experiment.
Whole mount immunostaining, β-gal staining and NBS staining
Embryos or tissues were carefully dissected out and rinsed for several times in ice-cold PBS. Samples were fixed in fresh 4% paraformaldehyde (pH 8.0) at 4°C for 2 hours and washes with PBS for more than 30 minutes. Whole mount immunostaining was performed as previously described57. Samples were examined and photographed using confocal fluorescent microscope. For β-gal staining, samples were stained with a solution containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 1 mg/ml X-gal substrate at 37°C for 2 to 4 hours before photographing. For whole mount Nile blue sulfate (NBS) staining, E10.5 embryos were dissected out and washed in PBS, and incubated for 1 hours at 37°C in filtered NBS solution (10 mg/ml NBS (Sigma N-5632) in PBS containing 0.1% Tween 20). Embryos were washed multiple times in PBS at room temperature and photographed immediately. At least three pairs of embryos (wildtype controls and mutants) at the indicated developmental stages were used in each experiment.
Skeleton preparation
Embryos were dissected out, fixed in 100% ethanol, skinned, and eviscerated, then stained with alizarin red and alcian blue overnight at 37°C. Samples were cleared in 1% KOH for 10 hr, and then further incubated in 50:50 1% KOH/glycerol for several days before photography.
Antibodies
The primary antibodies used in this study include: Dicer (Abcam), β-tubulin(Sigma), Sprouty1 (Santa Cruz Biotechnology), Sprouty2 (Santa Cruz Biotechnology), phospho-Erk1/2 (Cell Signaling), phosphor-MEK1/2 (Cell Signaling), Crkl (Santa Cruz Biotechnology), SHP-2 (Santa Cruz Biotechnology), Sos1 (Santa Cruz Biotechnology), smooth muscle 〈 22 (Abcam), smooth muscle actin (Sigma), phospho-histone H3 (Upstate Biotechnology), cleaved-caspase 3 (Cell Signaling) and PECAM-1 (1:100, BD Pharmingen). Alexa Fluor secondary antibodies were purchased from Invitrogen.
Luciferase reporter assays
A modified pGL3-control vector (pGL3cm) for 3′ UTR–luciferase reporter assays was described previously55. The luc-Spry1, luc-Spry2, luc-Spry4-1 and luc-Spry4-2 luciferase reporters were generated by annealing two tandem 40 bp repeat oligonucleotides from the 3’ UTR sequences of indicated sprouty genes which contain the predicted binding site for either miR-21 or miR-181a. Since two miR-181a binding sites are presented in the 3’UTR of the sprouty4 gene, two reporters, luc-Spry4-1 and luc-Spry4-2, were generated. We also generated miRNA sensor constructs in which perfect complementary sequences against either miR-21 or miR-181a were cloned downstream of the luciferase reporter gene. Luciferase reporter assays were performed using 24-well plates in Hela cells. All experiments were repeated at least 3 times in triplicate. Unless otherwise indicated, transfection was performed using 25 ng reporter plasmid and 150 or 450 ng miRNA duplex mimics. A Renilla luciferase vector was used to serve as an internal control for normalization.
Statistics
Values are reported as means ± SEM unless indicated otherwise. The 2-tailed Mann-Whitney U test was used for comparing 2 means (Prism; GraphPad). Values of P < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgement
We thank members of the Wang and Majesky laboratories for discussion and support, Xiaoyun Hu for excellent technical support. We would like to thank Gregory Hannon (Cold Spring Harbor Laboratory) for reagents. This work was supported by the March of Dimes Birth Defect Foundation (DZW), Muscular Dystrophy Association (DZW) and National Institutes of Health (DZW and MWM). J Regan is a Predoctoral Fellow, Z-P Huang is a Postdoctoral Fellow and DZ Wang is an Established Investigator of the American Heart Association.
Non-standard Abbreviations and Acronyms
- CHD
Congenital Heart Defects
- DORV
Double Outlet Right Ventricle
- IAA
Interrupted Aortic Arch
- LCA
Left Carotid Artery
- NBS
Nile Blue Sulfate
- NCC
Neural Crest Cell
- NCCS
Neuro-Craniofacial-Cardiac Syndromes
- PA
Pharyngeal Arch
- RCA
Right Carotid Artery
- RRSA
Retroesophageal Right Subclavian Artery
- SMC
Smooth Muscle Cell
- VSD
Ventricular Septal Defect
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
References
- 1.Roberts A, Allanson J, Jadico SK, Kavamura MI, Noonan J, Opitz JM, Young T, Neri G. The cardiofaciocutaneous syndrome. J Med Genet. 2006;43:833–842. doi: 10.1136/jmg.2006.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez E, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge and velocardiofacial syndromes) Curr Opin Pediatr. 2002;14:678–683. doi: 10.1097/00008480-200212000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Emanuel BS, Shaikh TH. Segmental duplications: an 'expanding' role in genomic instability and disease. Nat Rev Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 4.McDermid HE, Morrow BE. Genomic disorders on 22q11. Am J Hum Genet. 2002;70:1077–1088. doi: 10.1086/340363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 7.Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 8.Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 9.Bentires-Alj M, Kontaridis MI, Neel BG. Stops along the RAS pathway in human genetic disease. Nat Med. 2006;12:283–285. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- 10.Newbern J, Zhong J, Wickramasinghe RS, Li X, Wu Y, Samuels I, Cherosky N, Karlo JC, O'Loughlin B, Wikenheiser J, Gargesha M, Doughman YQ, Charron J, Ginty DD, Watanabe M, Saitta SC, Snider WD, Landreth GE. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. Proc Natl Acad Sci U S A. 2008;105:17115–17120. doi: 10.1073/pnas.0805239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 12.Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- 13.Trainor PA, Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci. 2000;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- 14.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 15.Deardorff MA, Tan C, Saint-Jeannet JP, Klein PS. A role for frizzled 3 in neural crest development. Development. 2001;128:3655–3663. doi: 10.1242/dev.128.19.3655. [DOI] [PubMed] [Google Scholar]
- 16.Saint-Jeannet JP, He X, Varmus HE, Dawid IB. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc Natl Acad Sci U S A. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 20.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2:152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 22.Zehir A, Hua LL, Maska EL, Morikawa Y, Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Dev Biol. 340:459–467. doi: 10.1016/j.ydbio.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 25.Bockman DE, Kirby ML. Dependence of thymus development on derivatives of the neural crest. Science. 1984;223:498–500. doi: 10.1126/science.6606851. [DOI] [PubMed] [Google Scholar]
- 26.Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- 27.Creazzo TL, Godt RE, Leatherbury L, Conway SJ, Kirby ML. Role of cardiac neural crest cells in cardiovascular development. Annu Rev Physiol. 1998;60:267–286. doi: 10.1146/annurev.physiol.60.1.267. [DOI] [PubMed] [Google Scholar]
- 28.Yasui H, Nakazawa M, Morishima M, Ando M, Takao A, Aikawa E. Cardiac outflow tract septation process in the mouse model of transposition of the great arteries. Teratology. 1997;55:353–363. doi: 10.1002/(SICI)1096-9926(199706)55:6<353::AID-TERA1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 29.Lindsay EA. Chromosomal microdeletions: dissecting del22q11 syndrome. Nat Rev Genet. 2001;2:858–868. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- 30.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 31.Schubbert S, Bollag G, Shannon K. Deregulated Ras signaling in developmental disorders: new tricks for an old dog. Curr Opin Genet Dev. 2007;17:15–22. doi: 10.1016/j.gde.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 33.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 34.Seger R, Krebs EG. The MAPK signaling cascade. Faseb J. 1995;9:726–735. [PubMed] [Google Scholar]
- 35.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 36.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 37.Lunghi P, Tabilio A, Dall'Aglio PP, Ridolo E, Carlo-Stella C, Pelicci PG, Bonati A. Downmodulation of ERK activity inhibits the proliferation and induces the apoptosis of primary acute myelogenous leukemia blasts. Leukemia. 2003;17:1783–1793. doi: 10.1038/sj.leu.2403032. [DOI] [PubMed] [Google Scholar]
- 38.Jiang CC, Chen LH, Gillespie S, Wang YF, Kiejda KA, Zhang XD, Hersey P. Inhibition of MEK sensitizes human melanoma cells to endoplasmic reticulum stress-induced apoptosis. Cancer Res. 2007;67:9750–9761. doi: 10.1158/0008-5472.CAN-07-2047. [DOI] [PubMed] [Google Scholar]
- 39.Jung JY, Yoo CI, Kim HT, Kwon CH, Park JY, Kim YK. Role of mitogen-activated protein kinase (MAPK) in troglitazone-induced osteoblastic cell death. Toxicology. 2007;234:73–82. doi: 10.1016/j.tox.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Loayza-Puch F, Yoshida Y, Matsuzaki T, Takahashi C, Kitayama H, Noda M. Hypoxia and RAS-signaling pathways converge on, and cooperatively downregulate, the RECK tumor-suppressor protein through microRNAs. Oncogene. 2010 doi: 10.1038/onc.2010.23. [DOI] [PubMed] [Google Scholar]
- 41.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 46.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 47.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol. 2008;45:185–192. doi: 10.1016/j.yjmcc.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callis TE, Deng Z, Chen JF, Wang DZ. Muscling through the microRNA world. Exp Biol Med (Maywood) 2008;233:131–138. doi: 10.3181/0709-MR-237. [DOI] [PubMed] [Google Scholar]
- 51.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 54.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 55.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brissie RM, Spicer SS, Thompson NT. The variable fine structure of elastin visualized with Verhoeff's iron hematoxylin. Anat Rec. 1975;181:83–94. doi: 10.1002/ar.1091810107. [DOI] [PubMed] [Google Scholar]
- 57.Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.