Abstract
Jasmonates (JAs) are a family of plant hormones that regulate plant growth, development, and responses to stress. The F-box protein CORONATINE-INSENSITIVE 1 (COI1) mediates JA signaling by promoting hormone-dependent ubiquitination and degradation of transcriptional repressor JAZ proteins. Despite its importance, the mechanism of JA perception remains unclear. Here we present structural and pharmacological data to show that the true JA receptor is a complex of both COI1 and JAZ. COI1 contains an open pocket that recognizes the bioactive hormone, (3R,7S)-jasmonoyl-L-isoleucine (JA-Ile), with high specificity. High-affinity hormone binding requires a bipartite JAZ degron sequence consisting of a conserved α-helix for COI1 docking and a loop region to trap the hormone in its binding pocket. In addition, we identify a third critical component of the JA co-receptor complex, inositol pentakisphosphate, which interacts with both COI1 and JAZ adjacent to the ligand. Our results unravel the mechanism of JA perception and highlight the ability of F-box proteins to evolve as multi-component signaling hubs.
INTRODUCTION
The phytohormone jasmonate (JA) and its metabolites regulate a wide spectrum of plant physiology, participating in normal development and growth processes, as well as defense responses to environmental and pathogenic stressors1. JA is activated upon specific conjugation to the amino acid L-isoleucine (Ile), which produces the highly bioactive hormonal signal, JA-Ile that is functionally and structurally mimicked by the Pseudomonas syringae phytotoxin coronatine2–4. The discovery of coronatine-insensitive mutants enabled the identification of CORONATINE INSENSITIVE 1 (COI1) as a key player in the JA pathway, with further implications of regulated proteolysis in JA perception and signal transduction5.
COI1 is an F-box protein that functions as the substrate-recruiting module of the Skp1-Cul1-F-box protein (SCF) ubiquitin E3 ligase complex. Recent studies have identified the JASMONATE ZIM-domain (JAZ) family of transcriptional repressors as the SCFCOI1 substrate targets, which associate with COI1 in a hormone-dependent manner6–8. In the absence of hormone signal, JAZ proteins actively repress the transcription factor MYC2, which binds to cis-acting elements of JA-response genes. In response to cues that up-regulate JA-Ile synthesis, the hormone stimulates the specific binding of JAZ proteins to COI1, leading to poly-ubiquitination and subsequent degradation of JAZ by the 26S proteasome. JAZ degradation relieves repression of MYC2 and likely other transcription factors, permitting the expression of JA-responsive genes6,9. The role of COI1-mediated JAZ degradation in JA signaling is analogous to auxin signaling through the receptor F-box protein TIR1, which promotes hormone-dependent turnover of the AUX/IAA transcriptional repressors10,11. Supported by its sequence homology and functional similarity to TIR1, COI1 has been assigned a critical role in the direct perception of the JA signal12,13.
Despite the importance of JA signaling in plant physiology, the molecular mechanism of JA perception remains elusive. Here we present crystal structures of COI1 bound to JA-Ile or coronatine, as well as peptides of a bi-partite JAZ1 degron. Our structural and pharmacological studies reveal that the true JA receptor is in fact a co-receptor complex, consisting of the F-box protein COI1, the JAZ degron, and a newly discovered third component, inositol pentakisphosphate.
COI1-JAZ complex as a JA co-receptor
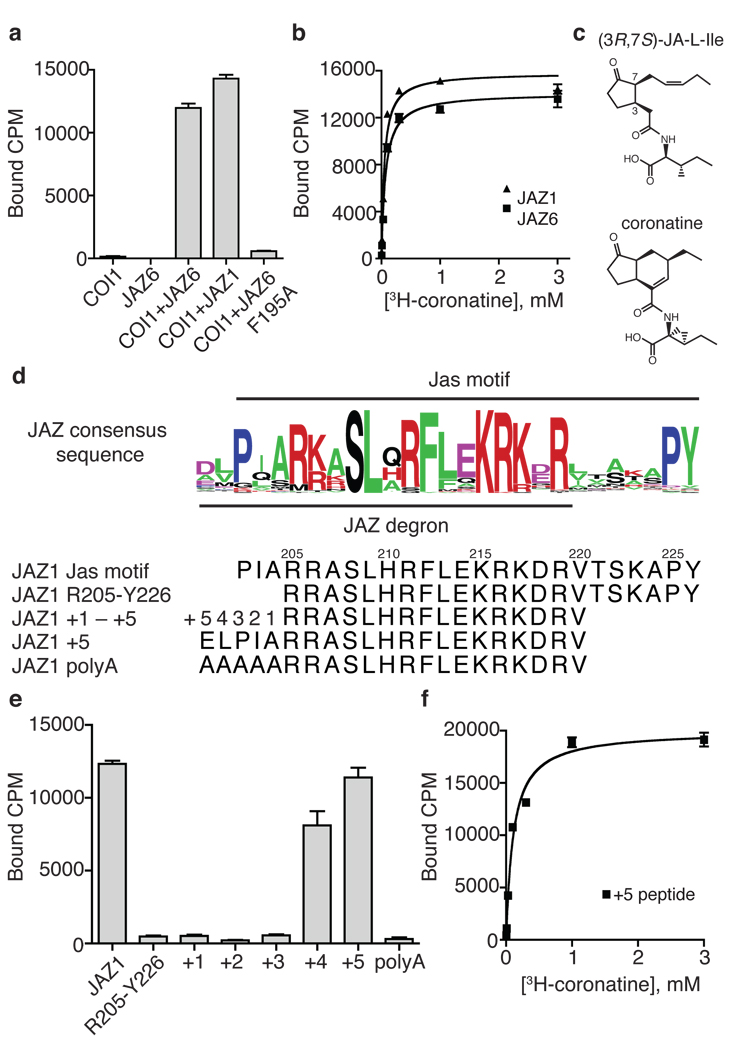
To better characterize the pharmacology of JA perception, we used recombinant proteins and 3H-coronatine to quantitatively define the functional components of the receptor system with an in vitro radioligand binding assay. From saturation binding experiments, we detected high affinity specific binding of 3H-coronatine to COI1 in the presence of two different full-length JAZ proteins, JAZ6 and JAZ1 at a Kd of 68 nM and 48 nM, respectively (Fig. 1a,b). The highly active (3R,7S) isomer of JA-Ile (Fig. 1c), and the less active (3R,7R) isomer, compete with 3H-coronatine for binding to the COI1-JAZ6 complex with Ki values of 1.8 µM and 18 µM, respectively (Supplementary Fig. 1a). In contrast, 3H-coronatine displayed no affinity to the JAZ proteins and exhibited only marginal binding to the F-box protein alone. Hormone binding to COI1 alone elicited < 2% binding signal relative to that of COI1-JAZ at a concentration that saturates the complex (300 nM) (Fig. 1a, Supplementary Fig. 1b). This result, together with the observation that endogenous JA-Ile activates COI1-dependent gene expression in the nM range1,14,15, indicates that the COI1-JAZ complex, rather than COI1 alone, functions as the genuine high affinity JA receptor in a co-receptor form.
Figure 1. COI1-ASK1 and JAZ proteins form a high-affinity JA co-receptor.
a. Binding of tritium-labeled coronatine (300 nM) to recombinant COI1-ASK1 and JAZ proteins. b. Saturation binding of 3H-coronatine to the complex of COI1-ASK1 in the presence of JAZ6 or JAZ1, with a Kd of 68±15 nM and 48±13 nM, respectively. c. Chemical structures of (3R,7S)-JA-Ile and coronatine. d. The consensus sequence of the Jas motif from 61 JAZ proteins from two monocot and three dicot plant species. Corresponding peptide sequences from JAZ1 in (e) are listed below. e. 3H-coronatine binding at 300 nM to COI1 in the presence of a series of synthetic JAZ1 peptides with the N-terminus of R205-Y226 systematically extended as described in (d). f. Saturation binding of COI1-ASK1 and the JAZ1 +5 degron peptide, with a Kd of 108±29 nM. All results are the mean ± S.E. of two to three experiments performed in duplicate.
We have previously mapped the COI1-binding region of the JAZ proteins to the C-terminal Jas motif, which is characterized by the SLX2FX2KRX2RX5PY consensus sequence preceded by two consecutive basic residues16,17 (Fig. 1d). A single Ala mutation of the central strictly conserved phenylalanine residue in the Jas motif is sufficient to abolish the formation of the high affinity JA co-receptor (Fig. 1a). Previous studies showed that the highly conserved PY sequence at the C-terminus of the Jas motif plays a role in JAZ localization and stability in vivo, but is not required for ligand-dependent COI1-JAZ interaction16,18,19. Consistent with these findings, truncation of the PY motif in JAZ1 has little effect on the in vitro ligand binding activity (Supplementary Fig. 1c).
To further map the minimal region of the Jas motif required for high affinity ligand binding with COI1, we replaced the recombinant protein with synthetic peptides of JAZ1 in the ligand binding assay (Fig. 1d, e). A 22-amino-acid JAZ1 peptide (Arg205–Tyr226) spanning the central conserved Jas motif plus the two N-terminal basic residues was not sufficient to form the high affinity JA co-receptor with COI1, suggesting that amino acids N-terminal to Arg205 also participate in the COI1-JA interaction. Because several JAZ proteins exhibit sequence homology in this region (Fig. 1d), we tested a series of JAZ1 peptides in which the N-terminus was systematically extended by one amino acid. Strikingly, inclusion of four but not three amino acids N-terminal to Arg205 allows ligand-dependent co-receptor formation, whereas addition of the fifth residue (Glu200) to the JAZ1 peptide permits 3H-coronatine binding with a Kd comparable to that of the full-length JAZ1 protein (Fig. 1e, f). Despite the sequence variation among different JAZ members in this region, only select amino acids are functional in this five amino acid extension, as a penta-alanine sequence fails to elicit the same effect (Fig. 1e). Together, these results indicate that the JAZ1 protein uses a minimal sequence (Glu200–Val220) within the Jas motif, which consists of a highly conserved central and C-terminal region and a more variable N-terminal region, to interact with COI1 and perceive the JA signal. Consistent with our in vitro ligand binding data, the minimal sequence in JAZ1 is sufficient for coronatine-induced COI1-JAZ1 interaction (Supplementary Fig. 1d). Therefore, we conclude that the interactions among COI1, coronatine, and the JAZ1 peptide are highly cooperative and that the short Glu200–Val220 sequence functions as the JAZ1 degron.
JA-binding pocket on COI1
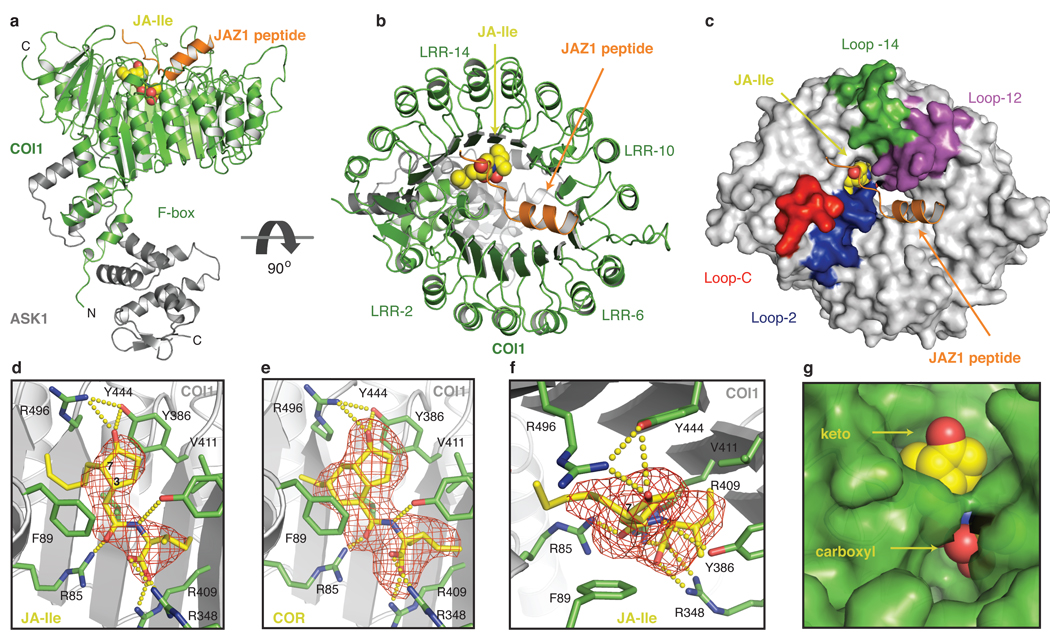
To elucidate the structural mechanism by which the COI1-JAZ1 co-receptor senses JA, we crystallized and determined the structures of the COI1-ASK1-JAZ1 degron peptide complex together with either (3R,7S)-JA-Ile or coronatine (Supplemental Table 1). The crystal structure of COI1 reveals a TIR1-like overall architecture20, with an N-terminal tri-helical F-box motif bound to ASK1 and a C-terminal horseshoe-shaped solenoid domain formed by 18 tandem leucine-rich repeats (LRRs; Fig. 2a, b). Similar to TIR1, the top surface of the COI1 LRR domain has three long intra-repeat loops (Loop-2, 12, and 14) that are involved in hormone and polypeptide substrate binding. Unlike TIR1, however, a fourth long loop (Loop-C) in the C-terminal capping sequence of the COI1 LRR domain folds over Loop-2, partially covering it from above (Fig 2b, c).
Figure 2. Crystal structure of the COI1-ASK1 complex with JA-Ile and the JAZ degron peptide.
a, b. COI1-ASK1 (green and grey ribbon, respectively), with JAZ degron peptide (orange ribbon) and (3R,7S)-JA-Ile in yellow spacefill. c. Surface representation of COI1 (grey) with Loops-2 (blue), -12 (purple) and -14 (green) forming the JA-Ile binding pocket. d, e. Side view of (3R,7S)-JA-Ile (JA-Ile) and coronatine (COR) binding. Hormones are shown as stick models, along with positive Fo-Fc electron density, calculated before they were built into the model (red mesh). Hydrogen bond and salt bridge networks are shown with yellow dashes. f. Top view of JA-Ile pocket showing the Fo-Fc electron density, calculated before JA-Ile was built into the model (red mesh). The electron density of the pentenyl side chain of (3R,7S)-JA-Ile cannot accommodate the (3R,7R)-JA-Ile side chain, which is constrained by the chiral configuration at the C7 position. g. When bound to COI1, JA-Ile (yellow spacefill) is solvent accessible at both the keto group (top) and carboxyl group (bottom).
Despite their similar overall fold, COI1 has evolved a hormone-binding site that is distinct from TIR1. Configured in between Loop-2 and the inner wall of the LRR solenoid, the ligand-binding pocket of COI1 is exclusively encircled by amino acid side chains (Fig. 2d–f). Many of the pocket-forming residues on COI1 are large in size and carry a polar head group (Supplementary Fig. 2). These properties allow them to mold a binding pocket into a specific shape while forming close interactions with each chemical moiety of the ligand. These close interactions are critical to proper hormone-sensing of the complex — in yeast two-hybrid assays, mutation of any of these large side-chain amino acids on COI1 is sufficient to disrupt the interaction of COI1 with JAZ1 in the presence of coronatine (Supplementary Fig. 3).
In the binding pocket, both JA-Ile and coronatine sit in an “upright” position with the keto group of their common cyclopentanone ring pointing up and forming a triangular hydrogen bond network with Arg496 and Try444 of COI1 at the pocket entrance (Fig. 2d–f). Without the JAZ degron peptide bound, the keto group of the ligand is accessible to solvent (Fig. 2g). The rest of the cyclopentanone ring of both JA-Ile and coronatine is sandwiched between the aromatic groups of Phe89 and Tyr444 of COI1, stabilized by hydrophobic packing. The cyclohexene ring of coronatine provides a rigid surface area for close packing with Phe89, whereas the more flexible and extended pentenyl side chain of JA-Ile is more loosely accommodated by a hydrophobic pocket formed by Ala86, Phe89, and Leu91 from Loop-2 as well as Leu469 and Trp519 from the LRRs (Supplementary Fig. 4a). Differences at this interface likely explain the approximately 10-fold higher affinity of coronatine over (3R,7S)-JA-Ile, as detected in our binding assays.
Deeper in the ligand binding pocket, the common amide and carboxyl groups of JA-Ile and coronatine bind to the bottom of the binding site by forming a salt bridge and hydrogen bond network with three basic residues of COI1, Arg85, Arg348, and Arg409 (Fig. 2d, e). Together, these arginine residues constitute the charged floor of the ligand pocket. Tyr386 reinforces the interactions from above by making a hydrogen bond with the amine group of the ligand. In doing so, Tyr386 approaches the cyclopentanone ring of the ligand, narrowing the pocket entrance, and creating a hydrophobic cave below. The rest of the basin is carved out by Val411, Ala384, and the aliphatic side chain of Arg409 (Supplementary Fig. 4b). The ethyl-cyclopropane group of coronatine and the isoleucine side chain of JA-Ile can both comfortably fit in this space due to their similar size and hydrophobicity. The nature of the cave explains the preference of COI1 for JA conjugates containing a moderately-sized hydrophobic amino acid13. Although most of the ligand is buried inside the binding site, the keto group at the top and the carboxyl group at the bottom remain exposed, available for additional interactions with the JAZ portion of the co-receptor (Fig. 2g).
Structural roles of the bipartite JAZ degron
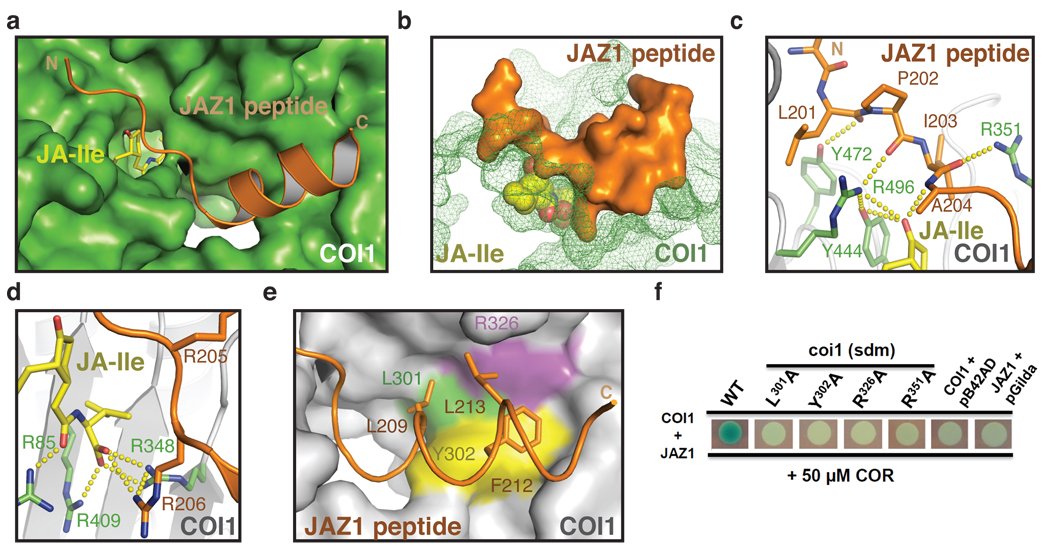
The JAZ1 degron peptide adopts a bipartite structure with a loop region followed by an α-helix to assemble with the COI1-JA complex. The hallmark of the JAZ1 degron is the N-terminal five amino acids identified in the radioligand binding assay. In a largely extended conformation, this short sequence lies on top of the hormone-binding pocket and simultaneously interacts with both COI1 and the ligand, effectively trapping the ligand in the pocket (Fig. 3a, b). At the N-terminal end, Leu201 of the JAZ1 peptide is embedded in a hydrophobic cavity presented by surface loops on top of COI1 (Fig. 3c). At the C-terminal end, Ala204 of JAZ1 uses its short side chain to pack against the keto group of the ligand and Phe89 of COI1 (Fig. 3c, Supplementary Fig. 4a). The same alanine residue of JAZ1 also donates a hydrogen bond through its backbone amide group to the keto moiety of the ligand emerging from the pocket (Fig. 3c). The middle region of the five amino acid sequence is secured to the COI1-JA complex through a hydrogen bond formed between the backbone carbonyl of Pro202 in JAZ1 and the ligand-interacting COI1 residue Arg496, which is critical for hormone-dependent COI1-JAZ interaction (Supplementary Fig. 3). In agreement with its important role in forming the JA-Ile co-receptor, this short N-terminal region of the JAZ1 degron completely covers the opening of the ligand-binding pocket, conferring high affinity binding to the hormone. The close interaction between the hormone and the co-receptor complex provides a plausible structural explanation for the favorable binding of (3R,7S)-JA-Ile isomer, as the stereochemistry at the 7 position of (3R,7R)-JA-Ile may place the aliphatic chain unfavorably close to nearby JAZ1 and COI1 residues (Supplementary Fig. 4a).
Figure 3. The bi-partite JAZ degron peptide.
a. Top view of the complete JAZ1 degron peptide (orange) bound to COI1 (green) and JA-Ile (yellow). b. Side view and surface representation of the JAZ peptide, which acts as a clamp to lock JA-Ile in the pocket. c. Interactions of the N-terminal region of the JAZ1 degron with COI1 and JA-Ile. Hydrogen bonds are shown with yellow dashes. d. Structural role of the Arg206 residue from the JAZ1 degron in coordinating the carboxyl group of JA-Ile with three basic residues of the COI1 ligand pocket floor. e. Top view of the amphiphathic JAZ1 degron helix bound to COI1 with three hydrophobic residues of JAZ1 shown in orange stick, and COI1 residues in colored surface representation. f. Coronatine-induced interactions of wild type and mutant COI1 with JAZ1 detected by yeast two-hybrid assay (sdm: site-directed mutants). Blue color indicates interaction.
Within the JAZ1 degron, two conserved basic residues, Arg205 and Arg206, were previously shown to play an important role in hormone-induced COI1 binding17. In the structure, Arg205 contributes to COI1 binding by directly interacting with Loop-12, whereas Arg206 points in the opposite direction and inserts deeply into the central tunnel of the COI1 solenoid. Approaching the bottom of the ligand-binding pocket, the guanidinium group of the Arg206 side chain joins the three basic COI1 residues that form the pocket floor and interacts directly with the carboxyl group of the ligand (Fig. 3d). Thus, the N-terminal seven amino acids (ELPIARR) of the JAZ1 degron peptide acts as a clamp that wraps the ligand-binding pocket from top to bottom, closing it completely (Fig. 3b).
The highly conserved C-terminal half of the JAZ1 degron forms an amphiphathic α-helix that strengthens JAZ1-COI1 interaction by binding to the top surface of the COI1 LRR domain, adjacent to the ligand-binding site (Fig. 3a). With its N-terminal end directly packing against Loop-2 of COI1, the Jas motif helix blocks the central tunnel of the COI1 LRR solenoid like a plug. The N-terminal half of the Jas motif helix is characterized by three hydrophobic residues, Leu209, Phe212, and Leu213, which are aligned on the same side of the helix and form a hydrophobic interface with COI1 (Fig. 3e). By soaking the COI1-ASK1 crystals with coronatine and a sufficiently high concentration of JAZ1 degron peptide lacking the N-terminal ELPIA sequence, we were able to trap a complex formed by COI1, coronatine, and the isolated Jas motif helix in the crystal (Supplementary Table 1). This suggests that the α-helix may provide a low affinity anchor for docking the JAZ protein on COI1. In support of this idea, single amino acid mutations at the complementary surface on COI1 readily disrupt hormone-induced COI1-JAZ1 interaction (Fig. 3f).
InsP5 as a cofactor of COI1
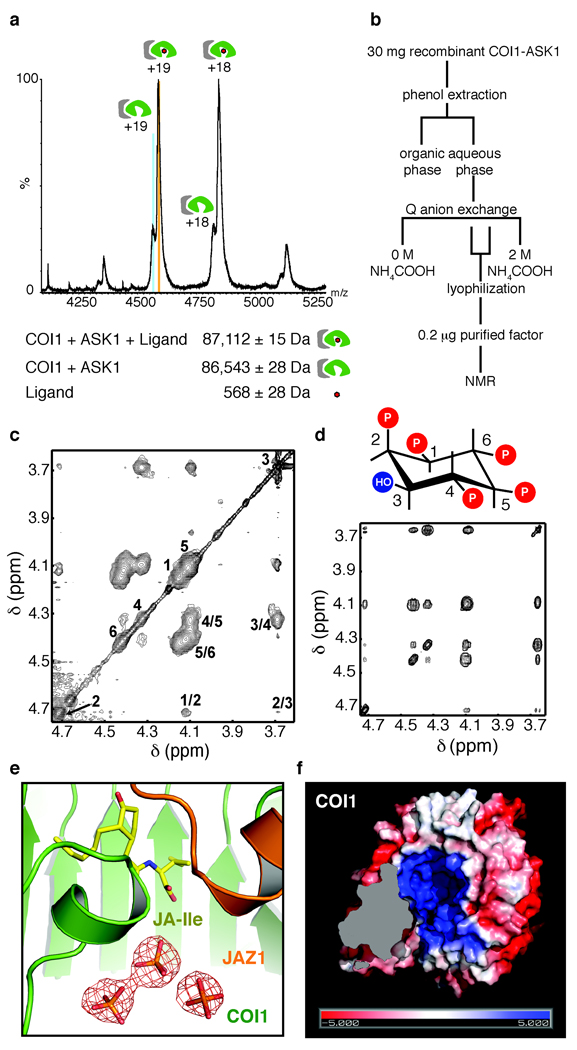
The crystal structure of TIR1 revealed an unexpected inositol hexakisphosphate (InsP6) molecule bound in the center of the protein underneath the auxin-binding pocket20. The sequence homology between COI1 and TIR1 suggests that COI1 might also contain a similar small molecule. Prior to crystallization, we analyzed the recombinant COI1-ASK1 complex by structural mass spectrometry (MS). Nano-electrospray mass spectra of the intact COI1-ASK1 complex revealed two populations differing by a mass of ~568 Da, indicating that a small molecule was indeed co-purified with the proteins (Fig. 4a, Supplementary Fig. 5). The MS-derived molecular weight of the unknown compound is different from the mass of InsP6 (651 Da) but matches that of an inositol pentakisphosphate (InsP5) molecule. Unfortunately, MS analyses of either the native COI1-ASK1 complex or the denatured proteins were unable to achieve direct mass analysis of the small molecule.
Figure 4. Identification of an inositol pentakisphosphate cofactor in COI1.
a. Nano-electrospray MS of the intact COI1-ASK1 complex. Low intensity charge series corresponds in mass to the cofactor-free COI1-ASK1 complex. High intensity charge series corresponds to cofactor-bound COI1-ASK1 complex. b. Optimized cofactor purification scheme. c. Proton TOCSY spectrum of the purified cofactor. Numbers along the diagonal indicate the positions of the six protons of Ins(1,2,4,5,6)P5. The cross-peaks corresponding to direct couplings are labeled. Other cross-peaks correspond to relayed connectivities. d. TOCSY spectrum of a synthetic Ins(1,2,4,5,6)P5 as a standard. e. Islands of positive Fo-Fc electron density (red mesh) below the hormone-binding pockets, which likely belong to inorganic phosphate molecules from the crystallization solutions that displace InsP5 from the InsP5-binding site. f. Bottom view of a surface electrostatic potential representation of COI1 from positive (blue) to negative (red).
To investigate the identity of the unknown compound, we first estimated that the molecule contains four or five phosphate groups by 31P nuclear magnetic resonance (NMR) of trypsin-digested COI1-ASK1 complex (data not shown). To unequivocally identify the unknown molecule, we set out to purify it away from the COI1-ASK1 complex in a quantity sufficient for 1H NMR analysis. The high phosphate content of the molecule allowed us to trace it through a multi-step purification procedure (Fig. 4b). Following isolation of 150 nanomoles of the purified small molecule, we acquired a series of 1D and 2D NMR data, including a highly informative homonuclear total correlation (TOCSY) spectrum. The observed chemical shifts and TOCSY cross-peak patterns are clearly characteristic of inositol phosphates (Fig. 4c). A comparison with previously reported NMR spectra of various inositol phosphates established that the unknown compound is either D- or L-inositol (1,2,4,5,6) pentakisphosphate, or Ins(1,2,4,5,6)P5 (Fig. 4c)21. This conclusion was further supported by the TOCSY spectrum of synthetic Ins(1,2,4,5,6)P5 (Fig. 4d) and subsequently acquired negative-ion electrospray ionization-MS spectrum of the compound (Supplementary Fig. 6).
Consistent with the binding of a small molecule cofactor, the crystal structure of COI1 showed strong unexplained electron densities clustered in the middle of the COI1 LRR domain. Like InsP6 in TIR1, these extra densities in COI1 are located directly adjacent to the bottom of the ligand-binding pocket of the JA co-receptor, interacting with multiple positively charged COI1 residues (Fig. 4e). Unexpectedly, these islands of electron density cannot be explained by an Ins(1,2,4,5,6)P5 molecule. Instead, their intensity, overall symmetry, and poor connectivity strongly suggest that they belong to multiple free phosphate molecules. Because a high concentration of ammonium phosphate was used as the major precipitant for crystallizing the JA co-receptor, we postulate that the InsP5 molecule co-purified with COI1 was later displaced by phosphate molecules in the crystallization drops. In support of this scenario, the concave surface of the COI1 solenoid fold surrounding the phosphates is highly basic and decorated with residues conserved in plant COI1 orthologues, indicating a functionally important surface area (Fig. 4f, Supplementary Fig. 2,7).
InsP5 potentiates JA perception by COI1-JAZ1
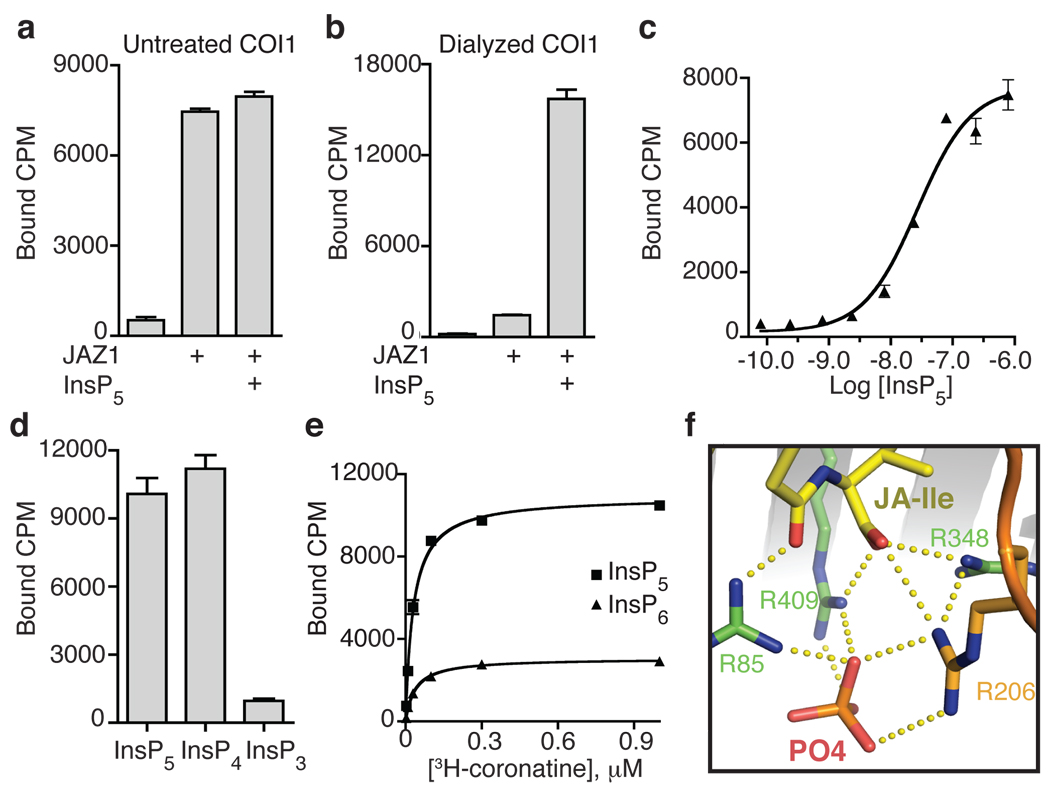
The highly selective co-purification of two different inositol phosphates, InsP5 and InsP6, with two homologous plant hormone receptors, COI1 and TIR1, implies that the proper function of the two F-box proteins might require the binding of specific inositol phosphates. To assess the functional role of Ins(1,2,4,5,6)P5 in the COI1-JAZ1 co-receptor, we took advantage of our crystallographic observation and developed a protocol to strip the co-purified InsP5 from COI1 without denaturing the protein. The resulting COI1-ASK1 complex was then tested in a ligand binding-based reconstitution assay. As shown in Fig. 5a, untreated COI1 formed a high affinity JA co-receptor with JAZ1. Addition of exogenous Ins(1,2,4,5,6)P5 did not significantly change its activity. In contrast, the dialyzed COI1 sample completely lacked ligand binding by itself and showed only trace activity in the presence of JAZ1. Supplementation with either synthetic Ins(1,2,4,5,6)P5 (Fig. 5b) or the purified and NMR-analyzed InsP5 sample (data not shown) rescued the interaction in a dose-dependent manner and with an EC50 of 27 nM (Fig. 5c). From this reconstitution result, we conclude that Ins(1,2,4,5,6)P5 binding is crucial for the JA co-receptor to perceive the hormone with high sensitivity.
Figure 5. Inositol phosphate is an essential component of the COI1-JAZ co-receptor.
a. Binding of 3H-coronatine at 100 nM to a complex of COI1 and JAZ1, with addition of 1 µM synthetic Ins(1,2,4,5,6)P5 (InsP5). b. With extensive dialysis to remove the co-purified InsP5 cofactor, 100 nM 3H-coronatine no longer binds dialyzed COI1 in the presence of JAZ1. Synthetic Ins(1,2,4,5,6)P5 rescues binding. c. Ins(1,2,4,5,6)P5 rescues the binding of 100 nM 3H-coronatine to dialyzed COI1-ASK1 in the presence of JAZ1 with an EC50 of 27±12 nM. d. Binding assays performed with 100 nM 3H-coronatine, dialyzed COI1, and 1 µM synthetic Ins(1,2,4,5,6)P5 (InsP5), Ins(1,4,5,6)P4 (InsP4), or Ins(1,4,5)P3 (InsP3). e. Saturation binding of 3H-coronatine to dialyzed COI1 in the presence of 1 µM of Ins(1,2,4,5,6)P5 (InsP5) and Ins(1,2,3,4,5,6)P6 (InsP6) at a Kd of 30±5 nM and 37±8 nM, respectively. All results are the mean ± S.E. of up to three experiments performed in duplicate. f. A phosphate-binding site in the complex structure reveals an interwoven hydrogen bond network that may explain the mechanism by which the InsP cofactor potentiates the JA co-receptor.
Although further effort is needed to reveal how InsP5 binds to COI1, a close examination of the phosphate molecules in the available COI1 structure suggests a mechanism by which the inositol phosphate molecule may modulate the activity of the JA co-receptor. Among four COI1-bound phosphates, one stands out by binding at a critical position in the JA co-receptor. This phosphate molecule interacts simultaneously with four basic residues at the bottom of the ligand-binding pocket, namely Arg206 in the JAZ1 degron and the three COI1 arginine residues that form the floor of the pocket. As a result, a tetragonal bipyramidal interaction network is formed among four molecules at the core of the JA co-receptor assembly. The four arginines from COI1 and JAZ1 sit at the four corners of the central plane, interacting with the hormone above and the phosphate below (Fig. 5f). As the free phosphate molecule likely mimics the action of a phosphate group on InsP5, this four-molecule junction, together with additional phosphate-COI1 interactions seen in the crystal, conceivably represents the structural basis for InsP5 potentiation of the JA co-receptor. Consistent with this interpretation, coronatine-induced formation of a COI1-JAZ1 complex was readily abolished by mutations of select COI1 residues adjacent to the phosphates, but not in contact with the hormone (Supplementary Fig. 8).
We used the reconstitution assay to further investigate the specificity of JA co-receptor regulation by inositol phosphates (Fig. 5d). Intriguingly, inositol (1,4,5,6) tetrakisphosphate supports the activity of the COI1-JAZ1 co-receptor, whereas the second messenger signaling molecule inositol (1,4,5) triphosphate does not. Addition of a phosphate to InsP5, which gives rise to InsP6, is also not favorable for activity. Although saturation binding of 3H-coronatine is stimulated by both Ins(1,2,4,5,6)P5 and InsP6 with similar Kd values (30 nM and 37 nM, respectively), the two inositol phosphates yield drastically different Bmax values for coronatine binding, indicating that InsP6 is significantly less efficacious in activating the co-receptor despite having equal affinity as Ins(1,2,4,5,6)P5 (Fig. 5e). Functional selectivity of COI1 for the inositol phosphate cofactor is consistent with the conservation of the putative inositol phosphate-binding site, which is distinct in amino-acid sequence from the InsP6-binding site in TIR120 (Supplementary Fig. 2).
Discussion
Our structural and pharmacological analyses reveal not only the essential components of the receptor system, but also the detailed mechanism by which these components cooperatively assemble and recognize the hormonal signal through a network of interactions. Our data identify the true JA receptor as a three-molecule co-receptor complex, consisting of COI1, JAZ degron, and inositol pentakisphosphate, all of which are indispensable for high affinity hormone binding. Our analyses also define the JAZ degron boundaries as a unique bi-partite sequence that binds COI1 and directly participates in hormone recognition. Unexpectedly, the N-terminal clamp region of the JAZ1 degron that is critical for hormone binding is diverse amongst JAZ proteins. This variable sequence might create a family of COI1-JAZ co-receptors that respond differentially to the hormone.
The crystal structure of the COI1-JAZ1 co-receptor in complex with JA-Ile revealed a drastically different binding mode of the hormone as predicted by computational modeling12. Although COI1 shares high sequence homology with TIR1, subtle structural differences and the integration of two additional factors critical for ligand binding give rise to a hormone-binding pocket in COI1 that is challenging to model. For the same reason, the structural nature of the ligand-free form of the F-box protein cannot be modeled with accuracy. The direct interactions of the hormone with both COI1 and the JAZ protein as observed in the crystal, nonetheless, support a molecular glue mechanism previously proposed for the auxin system20.
Discovery of the inositol pentakisphosphate cofactor of COI1 offers profound implications for the role of inositol phosphates in plant hormone signaling. COI1 co-purifies with a single isoform of InsP5, Ins(1,2,4,5,6)P5, indicating selectivity at the receptor level. However, both inositol (1,2,4,5,6) pentakisphosphate and inositol (1,4,5,6) tetrakisphosphate support high affinity hormone binding in our reconstitution assays, leaving the identity of the physiologically relevant form of inositol phosphate an open question.
Lastly, our study is the latest in a series of receptor structures for plant hormones, including auxin20, gibberellin22,23, and abscisic acid24–28. Despite different structural mechanisms, a common theme of hormone-mediated protein interactions emerges as a unique strategy favored by plant systems throughout evolution.
METHODS SUMMARY
The full Methods provides detailed information about all experimental procedures, including: (1) description of protein preparation, purification, and mutagenesis; (2) description of protein crystallization, data collection, and structure determination; (3) details for conducting in vitro radioligand binding assay; (4) details for conducting yeast-two-hybrid assay; (5) description of inositol phosphate purification scheme; (6) details for conducting in vitro inositol phosphate reconstitution assays; (7) description of structural mass spectrometry analysis of the intact protein complex; (8) description of nuclear magnetic resonance NMR analysis of the inositol phosphate; and (9) description of mass spectrometry analysis of the inositol phosphate.
METHODS
Protein preparation
The full-length Arabidopsis thaliana COI1 and ASK1 were co-expressed as a glutathione S-transferase (GST)-fusion protein and an untagged protein, respectively, in Hi5 suspension insect cells. The COI1-ASK1 complex was isolated from the soluble cell lysate by glutathione affinity chromatography. After on-column tag cleavage by tobacco etch virus protease, the complex was further purified by anion exchange and gel filtration chromatography and concentrated by ultrafiltration to 12–18 mg mL−1. Full-length JAZ substrate proteins were expressed as 6xHIS-fusion proteins in Escherichia coli and purified on Ni-NTA resin with subsequent dialysis into 20 mM Tris-HCl, pH=8.0, 200 mM NaCl, and 10% glycerol. For truncation mutants, a stop codon was introduced in JAZ1 proteins using the Quick-Change II site-directed mutagenesis kit (Stratagene). Synthetic JAZ degron peptides were prepared by United Biochemical Research, Inc. JAZ degron fusion peptides were prepared with N-terminal 6x-HIS tag and C-terminal glutathione S-transferase (GST)-fusion tag and expressed in Escherichia coli. The protein was isolated by glutathione affinity resin for pull-down assay with untagged COI1-ASK1 complex.
Site-directed mutagenesis
Individual amino acid residues in the LRR domain of COI1 proteins were mutated to alanine using the Quick-Change II site-directed mutagenesis kit (Stratagene). Mutant proteins were co-expressed with JAZ1 (JAZ1:pB42AD) in yeast to detect protein-protein interactions.
Crystallization, data collection, and structure determination
The crystals of the COI1-ASK1-JAZ1 peptide complexes bound to either coronatine or JA-Ile were grown at 4°C by the hanging-drop vapor diffusion method with 1.5 µl protein complex samples containing COI1-ASK1, JAZ1 peptide, and hormone compound at 1:1:1 molar ratio mixed with an equal volume of reservoir solution containing 100 mM BTP, 1.7–1.9 M ammonium phosphate, 100 mM NaCl, pH=7.0. Diffraction quality crystals were obtained by the micro-seeding method at 4°C. The crystals all contain eight copies of the complex in the asymmetric unit. The data sets were collected at the BL8.2.1 beamline at the Advanced Light Source in Lawrence Berkeley National Laboratory as well as the GM/CA-CAT 23 ID-B beamline at the Advanced Photon Source in Argonne National Laboratory using crystals flash-frozen in the crystallization buffers supplemented with 15%–20% ethylene glycol at −170°C. Reflection data were indexed, integrated, and scaled with the HKL2000 package29. All crystal structures were solved by molecular replacement using the program Phaser30 and the TIR1-ASK1 structure as search model. The structural models were manually built in the program O31 and refined using CNS32 and PHENIX30. All final models have 96–98% of residues in the favoured region and 0% in disallowed region of the Ramachandran plot.
Hormone and IP reagents
3H-coronatine was synthesized by Amersham13. Coronatine was purchased from Sigma; JA-Ile conjugates were chemically synthesized as previously described33. Synthetic inositol phosphates were purchased as sodium salts from Cayman Chemicals. InsP6 was purchased from Sigma.
Radioligand binding assay
Radioligand binding was assayed on purified proteins, with 2 µg COI1-ASK1 complex and JAZ proteins at 1:3 molar ratio, and/or 10 µM synthetic peptides. Reactions were prepared in 100 µL final volume and in a binding buffer containing 20 mM Tris-HCl, 200 mM NaCl, and 10% glycerol. Saturation binding experiments were conducted with serial dilutions of 3H-coronatine in binding buffer. Non-specific binding was determined in the presence of 300 µM coronatine. Competition binding experiments were conducted with serial dilutions of JA-Ile in the presence of 100 nM 3H-coronatine with nonspecific binding determined in the presence of 300 µM coronatine. Total binding was determined in the presence of vehicle only. Two-point binding experiments were performed in the presence of 100 nM or 300 nM 3H-coronatine with nonspecific binding determined in the presence of 300 µM coronatine. Following incubation with mixing at 4°C, all samples were collected with a cell harvester (Brandel, Gaithersburg, MD) on polyethyleneimine (Sigma)-treated filters. Samples were incubated in liquid scintillation fluid for >1 hour before counting with a Packard Tri-Carb 2200 CA liquid scintillation analyzer (Packard Instrument Co. Inc., Rockville, MD). Saturation binding experiments were analyzed by non-linear regression, competition binding experiments by non-linear regression with Ki calculation as per the method of Cheng and Prousoff34, and concentration-response data by sigmoidal dose-response curve fitting, all using GraphPad Prism version 4.00 for MacOSX.
Yeast two-hybrid (Y2H) assay
The coding sequences (CDS) of the Arabidopsis thaliana gene COI1 (At2g39940) and coi1 site-directed mutants were cloned into the Y2H bait vector pGILDA (Clontech) using XmaI and XhoI restriction enzyme recognition sequences previously added to the 5’ and 3’ end of the COI1 CDS, respectively, creating DNA-binding domain (LexA:COI1 and LexA:coi1) protein fusions. The CDS of Arabidopsis thaliana JAZ1 gene (At1g19180) was cloned into the Y2H prey vector pB42AD (Clontech) creating a transcriptional activation domain (AD:JAZ1) fusion protein. Individual wild-type and mutant COI1 constructs were co-transformed with JAZ1 constructs into Saccharomyces cerevisae strain EGY48 (p8opLacZ) using the frozen-EZ yeast transformation II kit (Zymo Research). Transformants were selected on SD-glucose medium (BD Biosciences) supplemented with –Ura/-Trp/–His drop-out solution (BD Biosciences). To detect the interaction between COI1 and JAZ1, transformants that had been selected in SD-Glu medium were resuspended in sterile water. Ten µl of each suspension was spotted onto inducing media (SD-Galactose/Raffinose –UWH; BD Biosciences) supplemented with 80 µg mL−1 X-Gal and 50 µM coronatine (Sigma). Y2H assays plates were incubated in the dark at 20°C and photographed 7 days later. Induced yeast cells were analyzed for COI1 and JAZ1 expression levels by western blotting using epitope-specific antibodies (data not shown).
Inositol phosphate purification
Phenol was melted at 68°C and equilibrated with equal parts 0.5 M Tris-HCl, pH=8.0 until a pH of 7.8 was reached. The equilibrated phenol was then topped with 0.1 volume 100 mM Tris-HCl, pH=8.0 and stored at 4°C. For extraction, 30–40 mg of 1 mg mL−1 COI1-ASK1 protein was mixed in small batches with equal parts equilibrated phenol at room temperature. The samples were inverted and incubated for 30 minutes until phase separation occurred. With 30 seconds vortexing, the samples were incubated at room temperature for 30 minutes and spun at 15,000 RPM for 5 minutes. The aqueous phase was removed as a primary extraction. Equal parts of a solution containing 25 mM Tris-HCl, pH=8.0 was added to the phenol and collected as above as a secondary extraction. The primary and secondary extractions were then combined and diluted 10× in 25 mM Tris-HCl, pH=8.0, then further purified by gravity flow on Q sepharose high performance anion exchange resin (GE Healthcare). Following column wash with 10× column volumes 0.1N formic acid, stepwise elution was performed with 2× column volumes of 0.1N formic acid (Thermo Scientific) with increasing concentrations of ammonium formate (Sigma), from 0 to 2M.
Fractions were analyzed for phosphate content by wet-ashing method with perchloric acid in Pyrex culture tubes (13× 100mm). Typically samples of 50–100 µl were ashed with 100–200 µl 70% perchloric acid (purified by redistillation, Sigma). Ashing was performed by heating the sample over a Bunsen-type burner with continuous shaking to prevent bumping. When the sample stopped emitting white smoke, the reaction was considered complete and then heated to dryness. 500 µl of distilled water was added to the room temperature tubes and vortexed. 100 µl samples containing up to 10 nmole inorganic phosphate were assayed for phosphate by a modification of a published procedure35. 125 µl of acid molybdate color reagent was added and the samples were incubated, covered, at room temperature for 12–14 hr (overnight) for full color development (total volume 225µl). Plates were read at 650 nm and unknowns were determined from the linear regression of the standard curve (0–10 nmole NaH2PO4 per well). All assays were done in triplicate. Final fractions containing phosphate were combined and lyophilized repeatedly to remove residual ammonium formate.
Inositol phosphate reconstitution assays
COI1-ASK1 complex was separated from pre-bound inositol phosphate by dialysis. Briefly, proteins were mixed with 10% glycerol and incubated in 2 M ammonium phosphate, 100 mM Bis-Tris propane pH=7.0, 200 mM NaCl, 10% glycerol, at 4°C for >24hrs with a minimum of 3x buffer changes at 100× sample volume. Samples were then transferred to 20 mM Tris-HCl, pH=8.0, 200 mM NaCl, 10% glycerol, at 4°C for >24hrs with a minimum of 3 buffer changes at 100x sample volume. Inositol phosphate rescue experiments were conducted according to the radioligand binding assays described above in the presence of 300 nM 3H-coronatine with nonspecific binding determined in the presence of 300 µM coronatine.
Structural mass spectrometry analysis of the intact protein complex
Nano-electrospray ionization mass spectrometry (MS) and tandem MS (MS/MS) experiments were performed on a Synapt HDMS instrument. Prior to MS analysis, 50 µl of a 16 mg mL−1 solution of COI1-ASK1 in 20 mM Tris-HCl pH=8, 0.2 M NaCl and 5 mM DTT, was buffer-exchanged twice into 0.5 M ammonium acetate solution by using Bio-Rad Biospin columns. To improve desolvation during ionization, samples were diluted 1:4 in 0.5 M ammonium acetate and isopropanol was added to a final concentration of 5%. Typically an aliquot of 2 µl solution was loaded for sampling via nano-ESI capillaries which were prepared in house from borosilicate glass tubes as described previously36. The conditions within the mass spectrometer were adjusted to preserve non-covalent interactions. The following experimental parameters were used: capillary voltage up to 1.26 kV, sampling cone voltage 150 V and extraction cone voltage 6 V, MCP 1590. For tandem MS experiments peaks centered at m/z 4,564 and 4,588 were selected in the quadrupole and collision energy up to 65 V was employed. Argon was used as a collision gas at maximum pressure. All spectra were calibrated externally by using a solution of cesium iodide (100 mg mL−1). Spectra are shown with minimal smoothing and without background subtraction.
Nuclear magnetic resonance (NMR) analysis
NMR spectra were acquired on a Varian INOVA600 spectrometer equipped with a cold probe using 200 µM samples of purified compound X or synthetic inositol (1,2,4,5,6) pentakisphosphate (Cayman Chemical) dissolved in D2O. TOCSY spectra were acquired with mixing times of 35 or 50 ms, processed with NMRPipe37 and visualized with NMRView38.
Mass spectrometry analysis of inositol phosphate purified from COI1-ASK1
MS experiments were conducted on a Finnigan (San Jose, CA) LTQ linear ion-trap mass spectrometer (ITMS) with Xcalibur operating system. Methanol was continuously infused (10 µL min−1) to the ESI source, where the skimmer was set at ground potential, the electrospray needle was set at 4.5 kV, and the temperature of the heated capillary was 275°C. The sample was diluted with equal volume of 2% ammonia in methanol and 10 µl was flow injected. The automatic gain control of the ion trap was set at 2×104, with a maximum injection time of 50 ms. Helium was used as the buffer and collision gas at a pressure of 1×10−3 mbar (0.75 mTorr). The MSn (n=2,3,4,5) experiments were carried out with an optimized relative collision energy ranging from 12–16% with an activation q value at 0.25. The activation time was set at 30–60 ms. The mass spectra were acquired in the profile mode and were accumulated for 3–5 min for MSn-spectra. The mass resolution of the instrument was tuned to 0.6 Da at half peak height.
Supplementary Material
Acknowledgements
We thank the beamline staff of the Advanced Light Source at the University of California at Berkeley and the Advanced Photon Source at Argonne National Laboratory for help with data collection. We also thank Ponni Rajagopal and Rachel Klevit for 31P NMR analysis, Martin Sadilek for MS analysis, Leron Katsir, Chris Hague, and John Lyssand for radioligand binding assay assistance, and Christy Mecey and Maeli Melotto for initial cloning of coi1(sdm) mutants. We also thank members of the Zheng laboratory and Wenqing Xu laboratory, and Rich Gardner for discussion and help. This work is supported by Howard Hughes Medical Institute and grants from the National Institutes of Health (R01 CA107134 to N.Z., T32 GM07270 to L.B.S., R01GM57795 to G.A.H., R01AI068718 to S.Y.H.), National Science Foundation (0929100 to N.Z.), U.S. Department of Energy (DE-FG02-99ER20323 to J.B. and DE-FG02-91ER20021 to G.A.H. and S.Y.H.), Michigan State University Plant Science Fellowship (J.W.), the Welch Foundation (I-1304 to J.R.), and the European Research Council (ERC) under the European Community’s Seventh Framework Program (FP7/2007-2013)/ ERC Grant agreement n° 239679 (G.B.N. and M.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. L.B.S., G.H., and N.Z. conceived and L.B.S. conducted radioligand binding and additional functional experiments. X.T., H.M., and L.B.S. purified COI1-ASK1 complex and conducted crystallographic experiments. X.T. crystallized and determined the structures of the COI1-ASK1-JAZ1-hormone complexes. L.B.S., X.T., and N.Z. analyzed crystallographic data. J.W. and S.Y.H. conceived and J.W. conducted Y2H experiments. G.B.N. and M.S. conducted and interpreted the structural mass spectrometry experiments. L.B.S., H.M., T.H., F.H., J.R., and N.Z. conceived and conducted experiments for inositol phosphate purification and identification. Y.K. synthesized JA stereoisomers. L.B.S. and N.Z. wrote the manuscript with comments from all authors.
Author Information. Structural coordinates and structural factors have been deposited in the Protein Data Bank under accession numbers 3OGK, 3OGL, and 3OGM. Authors declare no financial interest.
REFERENCES
- 1.Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. doi:10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 2.Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. doi:10.1105/tpc.6.5.751 6/5/751 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. doi:10.1105/tpc.104.023549 tpc.104.023549 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. doi:nchembio.161 [pii] 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 5.Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 6.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. doi:nature06006 [pii] 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 7.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. doi:nature05960 [pii] 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 8.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. doi:tpc.107.050708 [pii] 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. doi:10.1105/tpc.022319 tpc.022319 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. doi:nature03542 [pii] 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 11.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. doi:nature03543 [pii] 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 12.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. doi:tpc.109.065730 [pii] 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. doi:0802332105 [pii] 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suza WP, Staswick PE. The role of JAR1 in Jasmonoyl-L: -isoleucine production during Arabidopsis wound response. Planta. 2008;227:1221–1232. doi: 10.1007/s00425-008-0694-4. doi:10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
- 15.Koo AJ, Gao X, Jones AD, Howe GA. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009;59:974–986. doi: 10.1111/j.1365-313X.2009.03924.x. doi:TPJ3924 [pii] 10.1111/j.1365-313X.2009.03924.x. [DOI] [PubMed] [Google Scholar]
- 16.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–145. doi: 10.1105/tpc.108.064097. doi:tpc.108.064097 [pii] 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melotto M, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. doi:TPJ3566 [pii] 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grunewald W, et al. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009;10:923–928. doi: 10.1038/embor.2009.103. doi:embor2009103 [pii] 10.1038/embor.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung HS, et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 2010 doi: 10.1111/j.1365-313X.2010.04265.x. doi:TPJ4265 [pii] 10.1111/j.1365-313X.2010.04265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. doi:nature05731 [pii] 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 21.Stephens LR, et al. myo-inositol pentakisphosphates. Structure, biological occurrence and phosphorylation to myo-inositol hexakisphosphate. Biochem J. 1991;275(Pt 2):485–499. doi: 10.1042/bj2750485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimada A, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. doi:nature07546 [pii] 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 23.Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. doi:nature07519 [pii] 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 24.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. doi:nature08591 [pii] 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 25.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. doi:nature08613 [pii] 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. doi:nature08583 [pii] 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. doi:1181829 [pii] 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. doi:nsmb.1730 [pii] 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW, Sweet RM, editors. Vol. 276. New York: Academic Press; 1997. pp. 307–326. [Google Scholar]
- 30.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. doi:S0907444902016657 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa N, Kobayashi Y. Strategy for synthesis of the isoleucine conjugate of epi-jasmonic acid. Tetrahedron Lett. 2008;49:7124–7127. [Google Scholar]
- 34.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 35.Sadrzadeh SM, Vincenzi FF, Hinds TR. Simultaneous measurement of multiple membrane ATPases in microtiter plates. J Pharmacol Toxicol Methods. 1993;30:103–110. doi: 10.1016/1056-8719(93)90013-5. doi:1056-8719(93)90013-5 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Nettleton EJ, et al. Protein subunit interactions and structural integrity of amyloidogenic transthyretins: evidence from electrospray mass spectrometry. J Mol Biol. 1998;281:553–564. doi: 10.1006/jmbi.1998.1937. doi:S0022-2836(98)91937-4 [pii] 10.1006/jmbi.1998.1937. [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. doi:1-59259-809-9:313 [pii] 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.