Abstract
Fibroblast growth factor 21 (FGF21) is a novel master regulator of metabolic profile. The biological actions of FGF21 are elicited upon its klotho beta (KLB)-facilitated binding to FGF receptor 1 (FGFR1), FGFR2 and FGFR3. We hypothesised that common polymorphisms in the FGF21 signalling pathway may be associated with metabolic risk. At the screening stage, we examined associations between 63 common single-nucleotide polymorphisms (SNPs) in five genes of this pathway (FGF21, KLB, FGFR1, FGFR2, FGFR3) and four metabolic phenotypes (LDL cholesterol – LDL-C, HDL-cholesterol – HDL-C, triglycerides and body mass index) in 629 individuals from Silesian Hypertension Study (SHS). Replication analyses were performed in 5478 unrelated individuals of the Swiss CoLaus cohort (imputed genotypes) and in 3030 directly genotyped individuals of the German Myocardial Infarction Family Study (GerMIFS). Of 54 SNPs that met quality control criteria after genotyping in SHS, 4 (rs4733946 and rs7012413 in FGFR1; rs2071616 in FGFR2 and rs7670903 in KLB) showed suggestive association with LDL-C (P=0.0006, P=0.0013, P=0.0055, P=0.011, respectively) and 1 (rs2608819 in KLB) was associated with body mass index (P=0.011); all with false discovery rate q<0.5. Of these, only one FGFR2 polymorphism (rs2071616) showed replicated association with LDL-C in both CoLaus (P=0.009) and men from GerMIFS (P=0.017). The direction of allelic effect of rs2071616 upon LDL-C was consistent in all examined populations. These data show that common genetic variations in FGFR2 may be associated with LDL-C in subjects of white European ancestry.
Keywords: fibroblast growth factor 21, fibroblast growth factor receptor 2, cholesterol, single-nucleotide polymorphism, genome-wide association studies
Introduction
Genetic epidemiology has made significant progress over the last few years. With the availability of genotyping platforms that can analyse hundreds of thousands of single-nucleotide polymorphisms (SNPs) it has become feasible to perform high resolution scans over the whole human genome (genome-wide association studies – GWAS), thereby identifying genetic variants associated with phenotypes of interest. The major advantage of GWAS is that they do not require a priori assumptions about a potential role of alleles, genes and pathways in regulation of the examined trait. However, because of the scale of such analysis the problem of substantial correction for multiple testing arises and, therefore, representative cohorts and replication are needed to reduce the probability of false-positive reports. Furthermore, despite the extremely large number of SNPs on the platforms currently available, many single loci are poorly covered on a majority of platforms and, therefore, a significant proportion of potentially relevant variants may not be directly genotyped. Thus, GWAS are a tool of choice to discover disease genes that have not been suspected before, but may have limited value in analysis of candidate genes or pathways with strong functional potential to affect the phenotype of interest. For this type of investigation, systematic exploration of all common allelic variants in genes that encode molecules within the selected pathway is the method of choice. We used this approach to examine associations of common genetic variation in the fibroblast growth factor 21 (FGF21) signalling pathway with metabolic phenotypes in subjects of white European ancestry.
The FGF21 pathway is a novel metabolic signalling cascade recently identified as a master hormonal regulator of glucose, lipids and overall energy balance.1 Specifically, endogenous FGF21 was shown to facilitate insulin-independent uptake of glucose in adipocytes2 and to promote lipolysis3 contributing to reduction of fat storage.4 Exogenous FGF21 administration in experimental models was demonstrated to lower both glucose and triglyceride (TG) plasma levels and promote weight loss.2 These effects may be mediated via three types of FGF receptor (termed FGFR1, FGFR2 and FGFR3, respectively).5, 6, 7 Activation of these FGFRs by FGF21 is conditional upon coexpression of klotho beta (KLB),5, 6, 7 a transmembrane protein that has been discovered through its structural similarity to klotho (which mediates receptor binding of FGF23).
We have hypothesised that common polymorphisms in five genes of FGF21 signalling pathway may be associated with determinants of metabolic risk in subjects of European origin.
Methods
Subjects
Primary association analyses were carried out in the Silesian Hypertension Study (SHS) that has been previously described.8 In brief, SHS is a collection of 207 Polish Caucasian families (629 individuals, 49.8% men) recruited through essentially hypertensive offspring. Each individual was phenotyped according to a standard protocol (collection of clinical information by coded questionnaire, weight, height and blood pressure measurements, basic anthropometry, biochemical phenotyping). Blood samples were taken under fasting conditions. Total cholesterol, HDL-C and TG were determined using standard enzymatic methods on a Cobas Bio-Autoanalyzer (Roche Products, Basel, Switzerland), and LDL cholesterol (LDL-C) was calculated using the Friedewald formula. None of the subjects was treated pharmacologically with lipid-lowering medication.
Replication analyses were carried out in the Swiss CoLaus cohort and the German Myocardial Infarction Family Study (GerMIFS).
CoLaus is a random sample of 6188 unrelated individuals (2937 men) from the general population of Lausanne, Switzerland, and has been previously described in detail.9 In short, randomly selected Caucasian Lausanne inhabitants aged 35–75 years were invited to join the study. Eligible subjects were phenotyped in the outpatient clinic of the Centre Hospitailier Universitaire Vaudois (CHUV). Phenotyping included taking clinical information, anthropometry and blood pressure measurements. Blood samples were taken after overnight fasting. LDL-C was calculated using the Friedewald formula (in subjects with TG <4.6 mmol/l) and was adjusted for lipid-lowering treatment by adding 1.5 mmol/l to the measured values of subjects who were managed pharmacologically for hyperlipidaemia.10
Analyses in GerMIFS were based upon 1261 families (3030 patients; 60.6% men) of white, European ancestry. The details of this study have been previously described.11 In brief, families were recruited through index patients with coronary artery disease at 15 cardiac rehabilitation centres distributed throughout Germany. All index patients had MI before 60 years of age. If at least one sibling presented with MI or severe coronary artery disease before 70 years of age, the entire nuclear family (index patient, available parents, and all affected and unaffected siblings) was contacted and invited to participate in the study. Blood was drawn from non-fasting individuals. Total cholesterol, LDL-C, HDL-C and TG were determined directly using standard enzymatic methods. For the purpose of replication study of LDL-C, we excluded all patients on lipid-lowering treatment at the time of recruitment, leaving 3115 patients available for analysis. After excluding patients with missing phenotypes, we were able to perform association analyses with LDL-C in 3030 patients (1261 families, 1835 men).
The local Ethics Committees approved the study protocols, and all participants gave written, informed consent. All protocols conform to the principles outlined in the Declaration of Helsinki.
SNP selection
Tagging SNPs in FGF21, FGFR1, FGFR2, FGFR3 and KLB were selected from the HapMap database to reach a gene coverage of >90% with r2>0.8 for all SNPs with a minor allele frequency of ≥0.1 in the CEU population. In addition, each locus was saturated with additional common potentially functional polymorphisms (CpG islands, DNAse I hypersensitive sites, microRNA target sites).12 Altogether, 63 SNPs were selected for genotyping in SHS. Of these, after genotyping, quality filtering and association testing, five SNPs with high probability of a true association in SHS were selected for further replication.
Genotyping
Genotyping in SHS was carried out using iPLEX assays on the Sequenom MassArray system (Sequenom Inc., San Diego, CA, USA). SNPs in CoLaus were retrieved from a genome-wide scan conducted using the Affymetrix Genome-Wide Human SNP Array 5.0 (Affymetrix, Santa Clara, CA, USA). Specifically SNP rs2608819 was directly genotyped and the other four SNPs were imputed using the Quicktest imputation software (Computational Biology Group, Department of Medical Genetics, University of Lausanne; freely available online at: http://www2.unil.ch/cbg/index.php?title=Genome _Wide_Association_Studies) (the quality metrics were rs7012413: 0.999, rs4733946: 0.824, rs2071616: 0.302 and rs7670903: 0.914). Replication genotyping of rs2071616 in GerMIFS was performed using the TaqMan assays (Applied Biosystems, Foster City, CA, USA) on ABI PRISM 7900HT Sequence Detection System (Applied Biosystems).
Statistical and bioinformatic analysis
The following quality control filters were applied to SNPs before the statistical analyses: call rate ≥0.9, no deviation from Hardy–Weinberg equilibrium in the parental generation (Hardy–Weinberg equilibrium P>0.01) and number of Mendelian errors <3.
The primary association analyses in SHS were performed using family-based association testing (FBAT version 2.0.2)13 under the null hypothesis of no linkage and no association. Replication analyses in CoLaus were based on linear regression models constructed in SAS v.9.1 (SAS Institute Inc., Cary, NC, USA). In GerMIFS, association analysis was performed using generalised estimating equation regression with an exchangeable familial correlation structure, as implemented in the geepack library in R.14
We used FBAT for our exploratory primary analysis in SHS because this tends to produce less false-positive results than generalised estimating equation15 and performs well in samples consisting of nuclear families with available parents (such as SHS). Generally, FBAT and generalised estimating equation use different approaches to handle family structure: FBAT is based on Mendelian laws in families and therefore requires certain minimum information content of the pedigree, which depends on family structure, genotypes and phenotypes. The GerMIFS cohort had initially been recruited for microsatellite linkage analyses and consists, to a large extent, of sib-pairs. Given that about 50% of the pedigrees would not contribute to the FBAT statistics, we did not use FBAT in this cohort. Generalised estimating equation regression, on the other hand, is based on classical population-based association and therefore is more susceptible to bias owing to stratification and admixture effects. However, in the GerMIFS study, the vast majority of index cases has been included in GWAS,16, 17 showing no signs of population substructure.
All association tests were performed assuming an additive model of inheritance. All models were adjusted for age, age2 and gender (where applicable). All P-values are given for two-tailed tests.
To correct for multiple testing in the primary family-based analysis, we calculated false discovery rates (q-values). False discovery rates describe the expected number of false-positives among all positive results. The q-values were calculated using the R-based tool q-value (smoother algorithm, λ range 0.0–0.9),18 including all 216 performed association tests on the four traits analysed in SHS. At the replication stage, we used Bonferroni correction.
Haplotypes and linkage disequilibrium (LD) in FGFR2 were constructed using the Haploview program19 and visualised using the LocusView software. LocusView is a Java-based application that visualizes chromosomal regions annotated with genomic features and experimental data (such as results from association analysis (http://www.broadinstitute.org/science/programs/medical -and-population-genetics/locusview20)). The software was developed by Petryshen, Kirby, and Ainscow and is freely available.
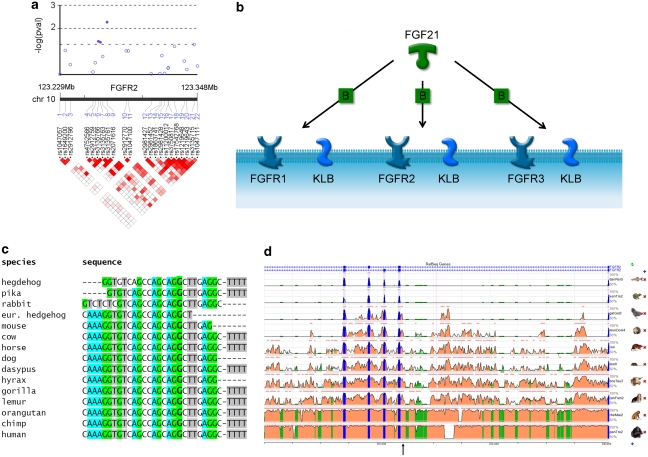
The graphical representation of FGF21 signalling cascade (Figure 1b) was constructed using the GeneGo software (http://www.genego.com). Sequence alignment (Figure 1c) was derived from the ClustalW2 portal (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Graphical presentation of evolutionary conservation (Figure 1d) was derived from the Evolutionary Conserved Regions (ECR) browser website (http://ecrbrowser.dcode.org). Information on functional potential of SNPs was obtained in silico from SNP Function Annotation Portal (http://brainarray.mbni.med.umich.edu/ Brainarray/Database/SearchSNP/snpfunc.aspx).
Figure 1.
Fibroblast growth factor receptor 2 gene (FGFR2)-association and evolutionary analysis. (a) Upper: −log transformed probability values from the analysis of association between FGFR2 and LDL-C in Silesian Hypertension Study; lower: linkage disequilibrium (LD) map (r2 coefficient-based) of FGFR2 SNPs. Note: dark-red squares indicate high r2 and therefore high LD, white squares indicate r2=0 and no LD. (b) FGFR2 binds FGF21 in the presence of KLB (a transmembrane cofactor); B-binding. (c) Sequence alignment of region containing SNP rs2071616 in FGFR2 across 15 mammalian species, position of rs2071616 is marked in red. (d) Evolutionary conservation of region in FGFR2 surrounding SNP rs2071616, the position of which is marked by the arrow.
Results
The brief clinical and anthropometric characteristics of the three study populations are listed in Table 1. Additional characteristics of the cohorts have been described elsewhere.9, 11, 20
Table 1. General clinical characteristics of the study cohorts.
| Phenotype | SHS | CoLaus | GerMIFS |
|---|---|---|---|
| No. of subjects | 629 | 5478 | 3030 |
| No. of families | 207 | 0 | 1261 |
| No. of men (%) | 313 (49.8) | 2,612 (47.7) | 1,835 (60.6) |
| Age, years | 45.8±15.7 | 53.2±10.8 | 59.1±10.7 |
| BMI, kg/m2 | 26.8±4.7 | 25.8±4.6 | 27.2±3.9 |
| LDL-cholesterol, mmol/l | 4.0±1.10 | 3.47±1.01 | 4.17±1.14 |
| HDL-cholesterol, mmol/l | 0.98±0.29 | 1.63±0.44 | 1.36±0.38 |
| Triglycerides, mmol/l | 1.76±1.10 | 1.40±1.21 | 2.19±1.42a |
| Lipid-lowering medication (%) | 0 | 472 (8.6) | Excluded |
Abbreviations: CoLaus: population-based cohort of Lausanne; GerMIFS: German Myocardial Infarction Family Study; SHS: Silesian Hypertension Study.
Data are means±SD or counts and percentages.
Non-fasting.
Primary association analysis – SHS
Of the 63 selected and genotyped polymorphisms, nine SNPs were excluded before the association analysis owing to poor genotyping quality. The mean call rate of the remaining 54 SNPs (Supplementary Table 1) was 97%. At r2=0.8, we reached a good tagging coverage of all HapMap SNPs in our candidate genes (100% for FGF21, 96% for FGFR1, 90% for FGFR2, 100% for FGFR3 and 78% for KLB). Under additive model of inheritance, primary family-based analysis revealed 14 nominal genetic associations with metabolic phenotypes in SHS – 6 with body mass index (rs1982738, rs7674434, rs12152703, rs17431867, rs2608819, rs3135859), 6 with LDL-C (rs4733946, rs7012413, rs3135766, rs3135763, rs2071616, rs7670903), 1 with HDL-C (rs2608819) and 1 with TG (rs3135761); as illustrated in Table 2. Of these, only five (rs7012413, rs4733946, rs2071616, rs7670903 with LDL-C, rs2608819 with body mass index) showed q-values <0.5 (suggestive associations; Table 2). The details of this analysis are given in Supplementary Table 2.
Table 2. Nominally significant associations between SNPs of FGF21 signalling pathway and metabolic phenotypes in primary age-, age2- and gender-adjusted family-based analysis (SHS).
| Trait | Gene | SNP | Alleles | MA | MAF | Risk allele | Z-value | P-value | q-value |
|---|---|---|---|---|---|---|---|---|---|
| BMI | FGFR3 | rs3135859 | G/T | T | 0.22 | G | 2.13 | 0.033 | 0.68 |
| KLB | rs1982738 | A/G | T | 0.21 | C | 2.04 | 0.041 | 0.68 | |
| rs7674434 | G/T | G | 0.29 | G | 2.30 | 0.021 | 0.59 | ||
| rs12152703 | G/T | T | 0.23 | T | 2.11 | 0.035 | 0.68 | ||
| rs17431867 | C/T | T | 0.18 | T | 1.98 | 0.048 | 0.68 | ||
| rs2608819* | C/T | T | 0.11 | C | 2.55 | 0.011 | 0.42 | ||
| LDL-C | FGFR1 | rs7012413* | C/T | T | 0.30 | T | 3.21 | 0.0013 | 0.14 |
| rs4733946* | G/T | T | 0.09 | T | 3.42 | 0.0006 | 0.13 | ||
| FGFR2 | rs3135766 | C/T | G | 0.45 | G | 2.09 | 0.037 | 0.68 | |
| rs3135763 | G/T | A | 0.18 | A | 2.05 | 0.041 | 0.68 | ||
| rs2071616* | C/T | T | 0.35 | T | 2.78 | 0.0055 | 0.39 | ||
| KLB | rs7670903* | A/G | A | 0.38 | A | 2.57 | 0.011 | 0.47 | |
| HDL-C | KLB | rs2608819 | C/T | T | 0.11 | C | −2.24 | 0.025 | 0.68 |
| Triglycerides | FGFR2 | rs3135761 | C/T | A | 0.23 | G | 2.39 | 0.026 | 0.68 |
Abbreviations: BMI, body mass index; HDL-C, HDL-cholesterol; LDL-C, LDL-cholesterol; MA, minor allele; MAF, minor allele frequency in the parental generation of Silesian Hypertension Study; SHS, Silesian Hypertension Study; SNP, single-nucleotide polymorphism.
Z-value – family-based association test that illustrates the magnitude of the genetic association and the direction of the allelic association (‘−' refers to a decrease in the quantity of the trait); P-value – level of statistical significance of family-based association test; q-value – false discovery rate. *denotes suggestive associations, as described in the text.
Replication
A total of five SNPs (rs7012413, rs4733946, rs2071616, rs7670903, rs2608819) with q-values <0.5 was taken forward for in silico replication in CoLaus. Of these, FGFR2 rs2071616 polymorphism showed a significant association with LDL-C (P=0.009; Table 3), even after correcting for multiple testing (Bonferroni correction threshold: 0.01; 0.05/5). The direction of the allelic effect on LDL-C was consistent with the findings from SHS – in both cohorts the minor allele was associated with higher LDL-C. The quantitative analysis in CoLaus showed that each minor allele copy of rs2071616 increased LDL-C by ∼0.092 mmol/l (β=0.092, SE=0.004).
Table 3. Replication of suggestive associations identified in Silesian Hypertension Study in CoLaus cohort.
| Trait | Gene | SNP | MA | MAF | Risk allele | β-coefficient | SE | P-value |
|---|---|---|---|---|---|---|---|---|
| LDL-C | FGFR1 | rs7012413 | T | 0.30 | T | 0.018 | 0.021 | 0.37 |
| rs4733946 | T | 0.08 | T | 0.027 | 0.037 | 0.48 | ||
| FGFR2 | rs2071616 | T | 0.31 | T | 0.092 | 0.035 | 0.009 | |
| KLB | rs7670309 | A | 0.42 | G | 0.024 | 0.019 | 0.21 | |
| BMI | KLB | rs2608819 | T | 0.13 | T | 0.014 | 0.128 | 0.91 |
Abbreviations: BMI, body mass index; LDL-C, LDL-cholesterol; MA, minor allele; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
β-coefficient, estimated quantitative effect of each SNP minor allele copy on a metabolic phenotype (adjusted for age, age2 and sex).
We genotyped rs2071616 in the third independent cohort – the GerMIFS study – and found that rs2071616 was associated with LDL-C amongst men only (P=0.017), but not in the entire cohort of subjects (P=0.32). However, both the direction of allelic association and the magnitude of minor allele effect of rs2071616 on LDL-C in GerMIFS men were strikingly similar to those for CoLaus subjects (β=0.094, SE=0.039).
Discussion
The most important finding from our analysis is a suggestive association between a genetic variant of FGFR2 (SNP rs2071616) and LDL-C in subjects of white European ancestry. This association was identified through a systematic screening of all common variants in five genes of the FGF21 signalling cascade in Polish families and was replicated in subjects from the general Swiss population and German men from families with clustering of coronary artery disease. To the best of our knowledge, no associations between FGFR2 and lipids have been reported by GWAS or candidate gene studies.
The selection of FGF21 signalling cascade as a novel candidate pathway of metabolic regulation is supported by multiple lines of evidence from experimental models. The central molecule of this pathway – FGF21 – promotes glucose uptake into adipocytes and improves glucose tolerance in diabetic rodents without inducing hypoglycemia.2 Furthermore, mice overexpressing FGF21 have lower TG plasma levels and are less prone to diet-induced obesity.2 When administered to diabetic monkeys, FGF21 improves glucose tolerance (without inducing hypoglycemia), lowers LDL-C and elevates HDL-C.21 Knockdown of FGF21 in mice leads to rapid hepatic fat accumulation and prominent lipid disturbances.22 Acting downstream in the PPARα signalling cascade, FGF21 regulates metabolic adaptation to the fasting state. Under a ketogenic diet or in the fasting state, FGF21 stimulates lipolysis in white adipose tissue and hepatic ketogenesis.3, 4, 22 These metabolic effects of FGF21 become apparent upon activation of at least three types of FGF receptor (FGFR1, FGFR2 and FGFR3) along with transmembrane mediator protein KLB.5, 6, 7 The further signalling cascade has been shown to involve PPARγ-coactivator protein 1α.23 Interestingly, both the FGF21-induced tyrosine phosphorylation activity of FGFR2 and circulating FGF21 levels are augmented by the PPARγ-ligand rosiglitazone,24, 25 suggesting a feedback mechanism in metabolic regulation.
Our most significant association signal maps to FGFR2 (Figure 1) – a molecule known as a critical regulator of cell proliferation. Multiple splice variants of FGFR2 are expressed in many tissues, including vasculature and liver,26 and bind to different FGF ligands, including FGF21. The role of FGFR2 in metabolism is primarily determined by tissue-specific coexpression of KLB:4 FGF receptors bind to the N-terminus of FGF21, whereas KLB interacts with the C-terminus.27 Genetic variation within FGFR2 has been linked to a wide range of phenotypes – from congenital syndromes of complex craniofacial malformations such as Crouzon syndrom (dysostosis craniofacialis), Apert syndrome (acrocephalosyndactyly) and craniosynosthosis to breast cancer and other malignancies such as gastric, lung, ovarian and endometrial cancer.28 The FGFR2 syntenic region in rats is located within a quantitative trait locus for hypertriglyceridemia, type 2 diabetes and body weight (http://www.ucsc.edu/), but, to the best of our knowledge, associations between FGFR2 mutations and LDL-C have not been reported before.
The rs2071616 polymorphism is situated in intron 6 (∼100 bp from the nearest exon–intron boundary) of FGFR2 (isoform 2 precursor). The polymorphism is located within a DNA segment that shows a high level of conservation in mammalian evolution (Figure 1c and d). There are not any known functional polymorphisms that could account for this association through high (r2≥0.8) LD with rs2071616 (Supplementary Table 3, Supplementary Figure 1). The only statistically similar SNP (rs2912762 – in complete LD with rs2071616 in HapMap CEU individuals) maps to another intronic segment of FGFR2 and does not appear to exhibit any obvious biological potential, (Supplementary Table 3) but is conserved (although to a lesser extent than rs2071616) in mammals. Further studies will be needed to clarify whether rs2071616, its proxy (rs2912762) or in fact an unidentified functional polymorphism in LD with rs2071616, is an actual driver of the association.
We should acknowledge several limitations of the study – first, unavailability of plasma glucose levels in SHS did not permit us to include this metabolic phenotype in the primary association analysis. Second, the replication studies in CoLaus cohort were based on data generated using imputation algorithms, and rs2071616 was at the bottom end of what has been accepted as a valid imputation.29 Third, our cohorts were recruited by different strategies: SHS for hypertension, GerMIFS for familial clustering of coronary artery disease, CoLaus from general population. Fourth, we replicated our main association finding (rs2071616 and LDL-C) in German men only and not in women. Whether gender-specific effects may account (at least to some extent) for these findings will require further investigations in other cohorts. Finally, we should also acknowledge the limitations of multiple testing. We analysed over 50 SNPs and four different phenotypes, resulting in over 200 tests in SHS. A conventional adjustment for multiple testing such as Bonferroni correction would have been overly conservative given that neither genotyped SNP nor examined phenotypes were completely independent. Indeed, many SNPs were in LD with each other and lipid fractions showed high levels of correlation. Although calculation of false discovery rate (q-values) does not account for this LD-driven or phenotypic non-independence, we used a non-conservative threshold of 50% (q-value <0.5) to identify suggestive associations. The quantile–quantile plot of −log P-values from all 216 tests performed in SHS revealed deviation in the extreme tail of association statistics distribution from the null distribution, indicating that the findings above the point of the deviation (mainly our suggestive associations) could indeed represent genuine results (Supplementary Figure 2).
Conclusions
In summary, the identified suggestive genetic association between FGFR2 and LDL-C illuminates a novel potential mechanism of cholesterol regulation. We also demonstrate that systematic pathway-oriented genetic analyses may contribute to gene discovery in the era of GWAS. Further replication and functional analyses of FGFR2 in a metabolic context are warranted to validate the association identified through genetic screening of the relevant signalling pathway.
Acknowledgments
This project was supported by NIH Fogarty International Research Collaboration Award (R03 TW007165; to MT). BMK's fellowship in Leicester was supported by the EU project Cardiogenics (LSHM-CT-2006-037593). TAB was supported by a British Heart Foundation project Grant (PG/06/097; to MT). NJS holds a British Heart Foundation Chair of Cardiology. This study is a part of the research portfolio supported by the Leicester NIHR Biomedical Research Unit in Cardiovascular Disease. The CoLaus study was supported by research Grants from GlaxoSmithKline and from the Faculty of Biology and Medicine of Lausanne, Switzerland, and is currently supported by the Swiss National Science Foundation (Grant no: 33CSCO-122661). We are grateful to Dr Allan Beveridge for support in bioinformatic analysis.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Moore DD. Physiology. Sister act. Science. 2007;316:1436–1438. doi: 10.1126/science.1144837. [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A, Shiyanova TL, Koester A, et al. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Dutchak P, Zhao G, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Choi M, Ogawa Y, et al. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem. 2007;282:26687–26695. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Kurosu H, Yamamoto M, et al. BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proc Natl Acad Sci USA. 2007;104:7432–7437. doi: 10.1073/pnas.0701600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Uehara Y, Motomura-Matsuzaka K, et al. bet}Klotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol Endocrinol. 2008;22:1006–1014. doi: 10.1210/me.2007-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Dunbar JD, Bina HA, et al. FGF-21/FGF-21 receptor interaction and activation is determined by betaKlotho. J Cell Physiol. 2008;215:1–7. doi: 10.1002/jcp.21357. [DOI] [PubMed] [Google Scholar]
- Tomaszewski M, Charchar FJ, Lynch MD, et al. Fibroblast growth factor 1 gene and hypertension: from the quantitative trait locus to positional analysis. Circulation. 2007;116:1915–1924. doi: 10.1161/CIRCULATIONAHA.107.710293. [DOI] [PubMed] [Google Scholar]
- Firmann M, Mayor V, Vidal PM, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- Broeckel U, Hengstenberg C, Mayer B, et al. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet. 2002;30:210–214. doi: 10.1038/ng827. [DOI] [PubMed] [Google Scholar]
- Tomaszewski M, Charchar FJ, Samani NJ. Association studies in current cardiovascular genetics - functional variants, tags or both. J Hum Hypertens. 2007;21:425–426. doi: 10.1038/sj.jhh.1002197. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Yan J. Yet another package for Generalized Estimating Equations. R-News. 2002;2/3:12–14. [Google Scholar]
- Cupples LA, Arruda HT, Benjamin EJ, et al. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8 (Suppl 1:S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann J, Grosshennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Tomaszewski M, Brain NJ, Charchar FJ, et al. Essential hypertension and beta2-adrenergic receptor gene: linkage and association analysis. Hypertension. 2002;40:286–291. doi: 10.1161/01.hyp.0000029105.21202.fe. [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A, Wroblewski VJ, Koester A, et al. The metabolic state of diabetic monkeys is regulated by fibroblast growth factor-21. Endocrinology. 2007;148:774–781. doi: 10.1210/en.2006-1168. [DOI] [PubMed] [Google Scholar]
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Inagaki T, Satapati S, et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci USA. 2009;106:10853–10858. doi: 10.1073/pnas.0904187106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulides C, Dyson P, Sprecher D, Tsintzas K, Karpe F. Circulating fibroblast growth factor 21 is induced by peroxisome proliferator-activated receptor agonists but not ketosis in man. J Clin Endocrinol Metab. 2009;94:3594–3601. doi: 10.1210/jc.2009-0111. [DOI] [PubMed] [Google Scholar]
- Moyers JS, Shiyanova TL, Mehrbod F, et al. Molecular determinants of FGF-21 activity-synergy and cross-talk with PPARgamma signaling. J Cell Physiol. 2007;210:1–6. doi: 10.1002/jcp.20847. [DOI] [PubMed] [Google Scholar]
- Hughes SE. Differential expression of the fibroblast growth factor receptor (FGFR) multigene family in normal human adult tissues. J Histochem Cytochem. 1997;45:1005–1019. doi: 10.1177/002215549704500710. [DOI] [PubMed] [Google Scholar]
- Yie J, Hecht R, Patel J, et al. FGF21 N- and C-termini play different roles in receptor interaction and activation. FEBS Lett. 2009;583:19–24. doi: 10.1016/j.febslet.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Katoh M. Cancer genomics and genetics of FGFR2 (Review) Int J Oncol. 2008;33:233–237. [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17 (R2:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.