Abstract
Peptidylglycine α-amidating monooxygenase (PAM) is a bifunctional enzyme which catalyzes the post-translational modification of inactive C-terminal glycine-extended peptide precursors to the corresponding bioactive α-amidated peptide hormone. This conversion involves two sequential reactions both of which are catalyzed by the separate catalytic domains of PAM. The first step, the copper-, ascorbate-, and O2-dependent stereospecific hydroxylation at the α-carbon of the C-terminal glycine, is catalyzed by peptidylglycine α-hydroxylating monooxygenase (PHM). The second step, the zinc-dependent dealkylation of the carbinolamide intermediate, is catalyzed by peptidylglycine amidoglycolate lyase. Quantum mechanical tunneling dominates PHM–dependant Cα-H bond activation. This study probes the substrate structure dependence of this chemistry using a set of N-acylglycine substrates of varying hydrophobicity. Primary deuterium kinetic isotope effects (KIEs), molecular mechanical docking, alchemical free energy perturbation, and equilibrium molecular dynamics were used to study the role played by ground-state substrate structure on PHM catalysis. Our data show that all N-acylglycines bind sequentially to PHM in an equilibrium-ordered fashion. The primary deuterium KIE displays a linear decrease with respect acyl chain length for straight-chain N-acylglycine substrates. Docking orientation of these substrates displayed increased dissociation energy proportional to hydrophobic pocket interaction. The decrease in KIE with hydrophobicity was attributed to a pre-organization event which decreased reorganization energy by decreasing the conformational sampling associated with ground state substrate binding. This is the first example of pre-organization in the family of non-coupled copper monooxygenases.
Introduction
Peptidylglycine α-amidating monooxygenase (PAM) is a bifunctional metallo-oxygenase of considerable interest across many fields, including neurobiology, mechanistic enzymology, metabolomics, and process biochemistry.1 Industrially, PAM has been used for the in vitro production of salmon calcitonin.2 In vivo, PAM catalyzes the oxidative cleavage of the glycyl Cα-N bond yielding an amide and glyoxylate (Scheme 1).3 This chemistry is essential to biosynthesis of α-amidated peptide hormones in mammals,1a insects,4 and cnidarians5 and also may have a role in the biosynthesis of mammalian lipid amides like oleamide.6
Scheme 1.
The reactions catalyzed by bifunctional PAM. PHM (peptidylglycine α-hydroxylating monooxygenase) domain is dependent on copper reduction by two ascorbate (ASC) molecules to activate the Cu/O2 species for Cα-Hs cleavage and hydroxylation. Deakylation of the stable α-hydroxylated intermediate is catalyzed by the PAL (peptidylamidoglycolate lyase) domain to yield glyoxylate and the corresponding amide. Oxidation of ascorbate yields semidehydroascorbate (SDA).
The amidation reaction, as catalyzed by PAM, occurs in two steps.7 The first step is the copper-dependent hydroxylation of the glycyl α-carbon in a reaction that requires O2 and a reductant.3b,7b,8 The second step is the dealkylation of carbinolamide in a reaction that requires zinc.9 Metals other than zinc have been reported to be important for the dealkylation reaction,10 but the most recent data suggest a direct catalytic role only for zinc.11
PHM shares mechanistic, sequence, and structural homology with dopamine β-monooxygenase (DβM).1e Both enzymes contain two bound copper atoms that are required for catalytic activity.8,12 The crystal structure of PHM shows that the two copper atoms are separated by a distance of 10.6 Å across a solvent filled active site,13 results consistent with EXAFS data for DβM indicating that two DβM-bound copper atoms were > 4 Å apart.14 EPR and EXAFS studies of PHM and DβM indicate that the active site copper atoms behave completely as non-blue type II, uncoupled mononuclear metal centers.15 Consistent with the EPR and EXAFS data are the finding that the two PHM-bound copper atoms have different functions in catalysis and amino acid ligands.15,16 One copper atom, CuH, has three Nδ-histidine ligands (His107, His108, and His172) and is involved in electron transfer. The other copper atom, CuM, has two Nε-histidines ligands (His240 and His242) and a methionine sulfur ligand (Met314) and is involved in O2 activation and substrate oxidation.13
The reactions catalyzed by PHM and DβM involve hydrogen atom transfer from substrate to an activated Cu/O species, resulting either in the stereospecific hydroxylation of a glycyl α-carbon (PHM) or benzylic carbon (DβM).7a,17 The first step in the catalytic cycle for both enzymes is reduction of the enzyme-bound Cu(II) atoms with an exogenous reductant, most likely ascorbic acid in vivo. Reduction is ping-pong with release of oxidized reductant from E•2Cu(I) followed by binding O2 or oxidizable substrate to the reduced enzyme.18,19 The dependence of 18O-kinetic isotope effect on deuteration of the oxidizable substrate for both enzymes indicates that C-H bond cleavage precedes cleavage of the activated O2 complex, yielding a Cu(II)-hydroperoxo and a substrate-based radical.20 The nucleophilic species responsible for hydrogen abstraction being a Cu(II)-superoxo.21 The oxidative species responsible for hydroxylation of the aliphatic radical is the subject of much debate.1e,22 Recent data from our laboratory suggest that an end-on/η1 Cu(II)-oxyl is the oxidant in PHM.19
Klinman and co-workers have demonstrated that Cα-H bond cleavage as catalyzed by PHM is dominated quantum mechanical tunneling.20b,23 This result was surprising because of the high degree of solvent accessibility of the PHM active site.13 Detailed studies of enzymes catalyzing H-transfer reactions have demonstrated that semi-classical transition state theory, including the Bell tunneling correction, cannot adequately account for the wealth of kinetic isotope effect obtained on these systems.24–26 Over the past few years, a full tunneling model has emerged that can account for the kinetic isotope effect data obtained for H-transfer enzymes, including their pressure-dependence.24–27 The hallmark of this model is the coupling of environmental reorganization to enzyme-catalyzed hydrogen tunneling (for the transfer of H+, H−, or H•) through the reaction barrier. Environmental reorganization involves two types of motion which are coupled to H-tunneling, (a) Marcus-like, thermally-equilibrated, relatively slow (ns - ms) motions of the enzyme and substrate which increase the possibility of attaining an E•S configuration optimized for tunneling (referred to as pre-organization) and (b) non-equilibrium, relatively rapid (fs - ps) “gating” motions optimize the H-donor and acceptor distance for wave function overlap (referred to as reorganization or protein promoting vibrations). The parameters most important in relating environmental reorganization to tunneling probability is the reorganization energy, λ, and the gating frequency, ωg.24,25 Work on soybean lipoxygenase (SLO) and PHM has shown that magnitude of the λ is approximately the same for both, 19.5 kcal/mol for SLO25 and 20 kcal/mol for PHM.23 However, the gating frequencies are different for the two enzymes, 400 cm−1 for SLO and 45 cm−1 for PHM.23 This difference in ωg between the two enzymes likely results from the increased flexibility of PHM relative to more rigid SLO with a greater participation of the gating motions in PHM catalysis. The SLO active site is non-polar to accommodate the binding of it fatty acid substrates rendering the gating motions energetically difficult. Thus, H• tunneling in SLO is dominated by active site pre-organization with only a small contribution from gating motions.
Francisco et al.23 provided the first evidence of tunneling in the hydrogen atom abstraction reaction catalyzed by PHM. The hallmark features of this study, using N-benzoylglycine as a substrate, were a temperature-independent intrinsic primary deuterium KIE of 10.6 ± 0.8, an Arrhenius pre-factor deuterium isotope effect of 5.9 ± 3.2, and an Ea(D) - Ea(H) value of 0.37 ± 0.33 kcal/mol. All of these experimentally determined values are well outside the semi-classical limits for C-H bond cleavage pointing toward a tunneling mechanism. Fits of these data to the model incorporating environmentally-coupled motions pointed toward an important participation of gating motions in PHM catalysis. This result seems consistent with the large, solvent accessible PHM active site (required to accommodate peptide substrates that can be in excess of 100 amino acids long) that is not likely optimized for tunneling in the ground state. Evidence that more global conformational dynamics are also important in PHM catalysis comes from pH-dependent changes in the Debye-Waller factor for the Cu-SMet314 component of the X-ray absorption spectrum of reduced enzyme.28 Bauman et al.28 report that reduced PHM exists in two forms: an inactive “met-on” form with a strong Cu-SMet314 interaction and an active “met-off” form with a relative weak Cu-SMet314 interaction. These authors found that the pH dependence of “met-off” → “met-on” transition and the PHM reaction was similar (pKa ~ 5.9). The transition between reduced Cu-SMet314 sub-states resulted from a global change in PHM structure with conformational mobility associated with the “met-off” state serving as an indirect probe into the essential role protein dynamics contributes to the reaction coordinate.
The present study deepens our understanding of the underlying relationship between PHM dynamics and substrate structure and is built upon the earlier work of Wilcox et al.29 and Merkler et al.30 Herein, we define how substrate hydrophobicity affects equilibrium binding to reduced PHM in the ground state and Cα-H cleavage, as observed in the steady-state. Using a series of unbranched, non-conjugated, N-acyglycines (from N-acetylglycine to N-decanoylglycine), we determined the acyl chain-length effect on both the primary kinetic isotope effects (KIEs) for Cα-H bond cleavage and the viscosity-dependence of apparent VMAX and VMAX/KM terms. Alchemical free energy perturbation calculations (AFEP) and equilibrium molecular dynamics simulations were performed in conjunction with the biochemical experiments to define the relationship between acyl chain length of N-acylglycine substrates and the relative dissociation energies of the reduced PHM•acylglycine and the reduced PHM•acylglycine•O2 complexes. Overall, our data suggest that N-acylglycine binding utilizes hydrophobic interactions coupled to salt-bridge formation with Arg240 in the enzyme•acylglycine•O2 central complex as the likely mechanism by which PHM pre-organization regulates catalysis. Our work provides the first evidence of pre-organization in the family of non-coupled di-copper monooxygenases and yields a novel perspective on the role ground state substrate structure has on conformational dynamics in the PHM tunneling reaction.
Materials and Methods
Materials
Morpholino-ethane-sulfonic acid (MES), sodium ascorbate, acetyl chloride, propionic anhydride, butyric anhydride, hexanoic anhydride, octanoic anhydride, decanoic anhydride, and cuprous nitrate were obtained from Sigma, N-acetylglycine (TCI, America), [α-2H2]-glycine (98%) was purchased from CDN Isotopes, Triton-X-100 was purchase from Fisher, and bovine liver catalase was supplied by Worthington. Recombinant type A rat medullary thyroid carcinoma bifunctional PAM was produced and purified as described31 and was a gift from Unigene Laboratories, Inc. (Fairfield, NJ, see www.unigene.com). All other experimental reagents were purchased from commercial sources at highest purity grade available and used without additional modification.
N-Acylglycine and [α-2H2]-N-Acylglycine Synthesis
The N-acylglycines and the [α-2H2]-N-acylglycines were synthesized according to literature procedures.29,32 To a cooled solution of glycine or [α-2H2]-glycine at 0 °C containing 1.2 equivalents of NaOH, one equivalent of the desired acyl anhydride was added drop wise with stirring. The reaction was allowed to stir for an additional 3 hours with the temperature slowly rising to room temperature and the pH maintained at 7–8 by the manual addition of NaOH, as necessary. An acid extraction of the aqueous phase into EtOAc (3X) was washed with brine and was subsequently dried over anhydrous MgSO4. Solvent was removed in vacuo, the product first recrystallized from a minimum volume of hot ethyl acetate, and then precipitated with hexane to yield white crystals. The NMR spectra (1H and 13C) for the N-acylglycines we synthesized are provided in the supplementary information.
[2H2]-N-Acetylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 1.80 (singlet, 3H, CH3)~δ 8.10 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 172.1 (C=O, carboxylic acid), δ 170. 3 (C=O, amide), δ 22.9 (CH3, methyl) mp. 205–207 °C (lit. 208 °C).32a
N-Propionylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.91 (triplet, J = 7.6 Hz, 3H, CH3), δ 2.07 (quartet, J = 7.6 Hz, 2H, CH2), δ 3.67 (singlet, 2H, CH2), and δ 7.96 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 174.5 (C=O, carboxylic acid), δ 172.1 (C=O, amide), δ 41.2 (CH2, α-glycine), δ 28.8 (CH2, n-alkyl methylene linker), δ 10.2 (CH3, n-alkyl terminal methyl). mp. 122–124 °C (lit. 125.5–7 °C).32b
[2H2]-N-Propionylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.93 (triplet, J = 7.6 Hz, 3H, CH3), δ 2.07 (quartet, J = 7.2 Hz, 2H, CH2), and δ 8.01 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 173.9 (C=O, carboxylic acid), δ 172.1 (C=O, amide), δ 28.8 (CH2, n-alkyl methylene linker), δ 10.4 (CH3, n-alkyl terminal methyl). mp. 122–124 °C.
N-Butyrylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.80 (triplet, J = 7.4 Hz, 3H, CH3), δ 1.45 (multiplet, 2H, CH2), δ 2.04 (triplet, J = 7.2 Hz, 2H, CH2), δ 3.68 (singlet, 2H, CH2), and δ 8.05 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 172.5 (C=O, carboxylic acid), δ 171.5 (C=O, amide), δ 40.5 (CH2, α-glycine), δ 37.0 (CH2, n-alkyl methylene linker), δ 18.6 (CH2, n-alkyl methylene linker), δ 13.5 (CH3, n-alkyl terminal methyl). mp. 69–70 °C (lit. 68.5–70 °C).32b
[2H2]-N-Butyrylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.72 (triplet, J = 7.4 Hz, 3H, CH3), δ 1.38 (multiplet, 2H, CH2), δ 1.95 (triplet, J = 7.2 Hz, 2H, CH2), and δ 7.95 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 172.6 (C=O, carboxylic acid), δ 171.6 (C=O, amide), δ 37.1 (CH2, n-alkyl methylene linker), δ 18.7 (CH2, n-alkyl methylene linker), δ 13.7 (CH3, n-alkyl terminal methyl). mp. 69–71 °C.
N-Hexanoylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.79 (triplet, J = 7.6 Hz, 3H, CH3), δ 1.24 (multiplet, 4H, (CH2)2), δ 1.51 (multiplet, 2H, CH2), δ 2.09 (triplet, J = 6.8 Hz, 2H, CH2), δ 3.74 (singlet, 2H, CH2), and δ 8.05 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 173.3 (C=O, carboxylic acid), δ 172.1 (C=O, amide), δ 41.2 (CH2, α-glycine), δ 39.7 (CH2, n-alkyl methylene linker), δ 35.7 (CH2, n-alkyl methylene linker), δ 25.6 (CH2, n-alkyl methylene linker), δ 22.6 (CH2, n-alkyl methylene linker), δ 14.6 (CH3, n-alkyl terminal methyl). mp. 88–89 °C (lit. 92 °C).32b
[2H2]-N-Hexanoylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.77 (triplet, J = 6.7 Hz, 3H, CH3), δ 1.17 (multiplet, 4H, (CH2)2), δ 1.41 (multiplet, 2H, CH2), δ 2.02 (triplet, J = 7.5 Hz, 2H, CH2), and δ 7.99 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 172.7 (C=O, carboxylic acid), δ 171.5 (C=O, amide), δ 35.1 (CH2, n-alkyl methylene linker), δ 30.9 (CH2, n-alkyl methylene linker), δ 24.9 (CH2, n-alkyl methylene linker), δ 21.9 (CH2, n-alkyl methylene linker), δ 13.9 (CH3, n-alkyl terminal methyl). mp. 88–89 °C.
N-Octanoylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.73 (triplet, J = 6.4 Hz, 3H, CH3), δ 1.14 (multiplet, 8H, (CH2)4), δ 1.38 (multiplet, 2H, CH2), δ 1.98 (triplet, J = 7.2 Hz, 2H, CH2), δ 3.60 (singlet, 2H, CH2), and δ 7.98 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 173.3 (C=O, carboxylic acid), δ 172.1 (C=O, amide), δ 41.2 (CH2, α-glycine), δ 39.7 (CH2, n-alkyl methylene linker), δ 35.8 (CH2, n-alkyl methylene linker), δ 31.9 (CH 2, n-alkyl methylene linker), δ 29.3 ((CH2)2, n-alkyl methylene linker), δ 22.8 (CH2, n-alkyl methylene linker), δ 14.6 (CH3, n-alkyl terminal methyl). mp. 103–105 °C. (lit. 105 °C).32c
[2H2]-N-Octanoylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.79 (triplet, J = 6.4 Hz, 3H, CH3), δ 1.20 (multiplet, 8H, (CH2)4), δ 1.47 (multiplet, 2H, CH2), δ 2.07 (triplet, J = 7.2 Hz, 2H, CH2), and δ 8.01 (singlet, 1H, NH). 13C NMR (100 MHz, Me2SO-d6) δ 173.0 (C=O, carboxylic acid), δ 171.7 (C=O, amide), δ 39.9 (CH2, n-alkyl methylene linker), δ 35.4 (CH2, n-alkyl methylene linker), δ 31.5 (CH2, n-alkyl methylene linker), δ 28.9 ((CH2)2, n-alkyl methylene linker), δ 25.5 (CH2, n-alkyl methylene linker), δ 22.4 (CH2, n-alkyl methylene linker), δ 14.6 (CH3, n-alkyl terminal methyl). mp. 102–104 °C.
N-Decanoylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.84 (triplet, J = 6.6 Hz, 3H, CH3), δ 1.24 (multiplet, 12H, (CH2)6), δ 1.48 (multiplet, 2H, CH2), δ 2.08 (triplet, J = 7.4 Hz, 2H, CH2), δ 3.70 (singlet, 2H), and δ 8.08 (singlet, 1H). 13C NMR (100 MHz, Me2SO-d6) δ 173.2 (C=O, carboxylic acid), δ 172.1 (C=O, amide), δ 41.7 (CH2, α-glycine), δ 35.7 (CH2, n-alkyl methylene linker), δ 31.9 (CH2, n-alkyl methylene linker), δ 29.5 ((CH2)3, n-alkyl methylene linker), δ 25.8 ((CH2)2, n-alkyl methylene linker), δ 22.7 (CH2, n-alkyl methylene linker), δ 14.5 (CH3, n-alkyl terminal methyl). mp. 113–114 °C (lit. 114 °C).32c
[2H2]-N-Decanoylglycine
1H-NMR (400 MHz, Me2SO-d6) δ 0.67 (triplet, J = 6.0 Hz, 3H, CH3), δ 1.06 (multiplet, 12H, (CH2)6), δ 1.31 (multiplet, 2H, CH2), δ 1.92 (triplet, J = 7.2 Hz, 2H, CH2), and δ 7.89 (singlet, 1H). 13C NMR (100 MHz, Me2SO-d6) δ 173.3 (C=O, carboxylic acid), δ 172.1 (C=O, amide), δ 35.7 (CH2, n-alkyl methylene linker), δ 31.0 (CH2, n-alkyl methylene linker), δ 30.0 ((CH2)3, n-alkyl methylene linker), δ 25.8 ((CH2)2, n-alkyl methylene linker), δ 22.8 (CH2, n-alkyl methylene linker), δ 14.5 (CH3, n-alkyl terminal methyl). mp. 113–114 °C.
Steady State Kinetics
Reactions at 37.0 ± 0.1 °C were initiated by the addition of 0.12–0.18 μM PAM (4–5 μL) into 2.0 mL of 100 mM MES/NaOH pH 6.0, 30 mM NaCl, 1.0% (v/v) ethanol, 0.001% (v/v) Triton X-100, 1.0 μM Cu(NO3)2, 5.0 mM sodium ascorbate, with N-acylglycine or [α-2H2]-N-acylglycine at concentrations of 0.2 to 10-fold KM. The concentration of dissolved O2 under these conditions was 217 μM.33 Initial rates were measured by following the PAM-dependent consumption of O2 using a Yellow Springs Instrument Model 53 oxygen monitor interfaced with a personal computer using a Dataq Instruments analogue/digital converter (model DI-154RS). VMAX,app values were normalized to controls performed at 11.0 mM N-acetylglycine to account for differences in specific activity between different lots of enzyme. Background O2 consumption rates were first determined without enzyme and were subtracted from the rate obtained upon PAM addition. Ethanol was added to protect the catalase against ascorbate-mediated inactivation34 and Triton X-100 was included to prevent nonspecific absorption of PAM to the sides of the oxygen monitor chambers.
O2-Dependence of the Primary Deuterium Kinetic Isotope Effects
The O2-dependence of the KIEs expressed by N-acetylglycine was determined by the addition of 0.18 μM PAM into 2.0 mL of 100 mM MES/NaOH pH 6.0, 30 mM NaCl, 1.0% (v/v) ethanol, 0.001% (v/v) Triton X-100, 1.0 μM Cu(NO3)2, 5.0 mM sodium ascorbate, 2–50 mM N-acetylglycine (or [α-2H2]-N-acetylglycine) and 25–830 μM O2. The [O2] was varied by mixing different proportions of N2:O2 gas into the headspace of the electrode chamber above the stirring reaction for 4 min. The resulting [O2] was determined from percent saturation observed with the O2 electrode compared to the ambient [O2] as a reference.
Viscosity Dependence of the PAM-Catalyzed Oxidation of N-Acylglycines
An Ubbelholde viscometer (Industrial Research Glassware Ltd, Union, NJ, size 1B) was used to determine the relative microviscosity (ηrel) of the control solution containing 100 mM MES/NaOH pH 6.0, 30 mM NaCl, 1.0% (v/v) ethanol, and 0.001% (v/v) Triton X-100. Ten trials were performed in a temperature-controlled water bath, with viscometer and buffer equilibrated for 10 minutes at 37.0 ± 0.1 °C and the values (centistokes) averaged with corresponding standard deviation. The relative microviscosity was determined by comparing buffer solution supplemented with sucrose (at the desired concentrations) to the control solution. The relative microviscosities used were 1.0 (no microviscogen), 2.08, 3.69, and 5.33. Macroviscosity was measured in a similar fashion using a Ficoll-400 solution (pH 6.0) to alter the macroviscosity of the reaction environment. The relative macroviscosities were 1.0 (no macroviscogen), 3.27, and 5.17. The dependence of the steady-state kinetic parameters was determined by measuring the consumption of O2 for N-acetylglycine or N-decanoylglycine in solutions fixed at each relative microviscosity and macroviscosity. Reactions were initiated by the addition of 0.12 μM PAM into 100 mM MES/NaOH pH 6.0, 30 mM NaCl, 1.0% (v/v) ethanol, 0.001% (v/v) Triton X-100, 1.0 μM Cu(NO3)2, 5.0 mM sodium ascorbate, N-acetylglycine or N-decanoylglycine, and the desired concentration of viscogen (sucrose or Ficoll-type 400). The VMAX,app values were normalized to 11.0 mM N-acetylglycine (no viscogen).
Analysis of Steady-State Kinetic Data
Steady-state kinetic parameters (± standard error) were obtained by a Kaleida-Graph™ fit of the initial velocity (rate) vs. initial substrate concentration ([S]) to the Michaelis-Menten equation (eq. 1) where KM,app is the apparent Michaelis constant for the N-acylglycine at fixed [ascorbate] and [O2] and VMAX,app is the apparent maximum velocity at saturating N-acylglycine and fixed [ascorbate] and [O2].
| (1) |
Values for the D(VMAX/KM)app and DVMAX,app were obtained from the quotient of appropriate constants for the light/heavy N-acylglycine substrate.35 Initial rate data generated to determine the minimal kinetic mechanism were fit to either a steady-state (eq. 2) or equilibrium preferred (eq. 3) minimal kinetic mechanism using the ENZKIN programs, where [AG] is the concentration of the N-acetylglycine, [O2] is the concentration of O2, VMAX represents the maximal velocity for the reaction at infinite concentrations of both O2 and N-acetylglycine, KM values for either substrate are the Michaelis constant at saturating concentrations of the second substrate, and KI,AG is the steady-state dissociation constant for N-acetylglycine at a zero [O2].
| (2) |
| (3) |
Predicted Active-Site Docking Conformations of N-Acylglycines
The initial coordinates of the reduced PHM pre-catalytic complex at 1.85 Ǻ resolution were obtained from the Protein Data Bank (http://www.rcsb.org/pdb/, 1SDW).13c Poses were predicted using quantum polarized ligand docking (QPLD) to generate high accuracy substrate binding modes utilizing molecular mechanics (MM)36 and ab initio programs of the Schrödinger First Discovery suites, Glide37 and Q-site.38
Molecular Dynamics (MD) Simulations
i.) Parameterization and Protein Equilibration
A protein starting configuration for MD simulations was taken from the above referenced crystal structure for reduced PHM pre-catalytic complex that consists of residues 43–356 of rat PAM that includes the PHM catalytic core. MD was performed with NAMD 2.6 using the CHARMM22 force field parameters where available, including treatments for (non-catalytic site) PHM, N-acylglycines, and N-benzoylglycine (hippurate).39 Additional required parameters for the reactive center were calculated using the GAMESS electronic structure code on a representative molecular fragment shown in Fig. 4. Density functional theory40 calculations were performed with the B3LYP41 hybrid exchange-correlation functional and the SBKJC42 basis set using effective core potentials on the copper. The intramolecular force field parameters for both CuH and CuM were determined by normal mode analysis. Specifically, the force constant matrix was calculated and diagonalized for each Cu-containing fragment separately. Partial charges for a subset of the reactive center atoms in Fig. 4 (i.e. copper and the oxygen atoms of O2) were fit to the electrostatic potential surface using the Connelly algorithm implemented in GAMESS. The van der Waals parameters for the Cu and O2 atoms are taken from the UFF43 force field.
Figure 4.
The active-site analogue that was used in the electronic structure calculations. The histidine and methionine residues were methyl-capped with the methyl cap carbons frozen. This geometry was optimized and then used in the normal mode analysis and ESP calculations.
To obtain an initial protein structure consistent with the force field, energy minimizations were performed using the conjugate gradient algorithm in NAMD for a system solvated using the relevant VMD44 functionality. Note, the crystal structure includes a different peptide substrate and these coordinates were used to place the substrates-of-interest in approximately the correct position in the active site using a minimum least squares overlap of peptide coordinates. Explicit solvent was included in a cubic box with 24490 water molecules using the TIP3P model;45 the equilibrium density was obtained from NPT simulations at 1.0 atmosphere and 310 K (with Langevin therm/baro stating as implemented in NAMD). Long range electrostatic interactions were included via the Particle Mesh Ewald method.46 Equilibration of the solvated PHM system was determined by the convergence of root mean square deviation values of the system volume to less than one percent.
ii.) Alchemical Free Energy Perturbation (AFEP) Calculations
AFEP is a dual topology hybrid molecule approach used to calculate the relative free energy differences and was employed as implemented in NAMD to determine the Helmholtz free energy difference, ΔA, between two N-acylglycine substrates bound to PHM.
The relative free energy was calculated using eq. 4 (below), where ΔAa→b is the Hemholtz free energy determined for each “window” in the AFEP calculation. In Eq. 4, kB is Boltzmann’s constant, T is the absolute temperature, Hb(r,p) and Ha(r,p) are the Hamiltonians characteristic of states a and b, and <…>a denotes an ensemble average over configurations representative of the initial state, a.
| (4) |
In this system, convergence was found to be optimal with the windows divided into (dimensionless) intervals of 0.1 with the endpoints being sampled every 0.05 to promote convergence. For each window, 10,000 molecular dynamics equilibration steps equilibration and 50,000 ensemble averaging steps (both of 1.0 femtosecond duration) were performed. The alchemically permutated N-acylglycine substrates were N-decanoyl-, N-octanoyl-, N-hexanoyl-, N-butyryl-, N-propionyl- and N-acetyl-. Specifically, N-decanoylglycine was chosen as the initial state for each trial varying the final permutated ligand state to a shorter N-acylglycine derivative. The variance over each ensemble window was calculated in order to estimate the error of the derived values.
Equilibrium dynamic NVT (310 K) simulations were performed for all experimentally tested N-acylglycine substrates, as well as an N-benzoylglycine control to assess the dynamic structure of the protein substrate system.
Results
Steady-State Kinetic Data and Substrate Hydrophobicity
The (VMAX/KAG)app for the PAM-catalyzed oxidation of the N-acylglycines at ambient O2 exhibited a parabolic relationship with the increase in chain length (R = number of carbon atoms in the linear acyl chain) (Supplementary Information, Fig. S1). The (VMAX/KAG)app increased over 250-fold as the N-acyl chain lengthened from R = 1 (N-acetylglycine) to 9 (N-decanoylglycine), with the value from 510 ± 50 M−1s−1 at R = 1 to (1.3 ± 0.08) × 105 M−1s−1 at R = 9, respectively (Table 1). The increase in the VMAX,app with chain length is only ~1.4-fold; thus, the increase in (VMAX/KAG)app results in a decrease in KAG,app as a function of R (~165-fold effect) (Table 1). These data agree nicely with those reported by Wilcox et al.29 The primary deuterium kinetic isotope effect for the Cα-H bond cleavage for the set of N-acylglycines employed here decreased linearly as the N-acyl chain length increased, with the D(VMAX/KAG)app decreasing from 3.21 ± 0.31 at R = 1 to 1.24 ± 0.12 at R = 9, respectively (Fig. 1 and Table 1).
Table 1.
Steady-State Kinetic Constants and Deuterium Kinetic Isotope Effects for the PHM-Mediated Oxidation on N-Acylglycinesa,b
| N-Acylglycine Substrate (AG) | R | (VMAX,AG)app (s−1) | (KM,AG)app (mM) | (VMAX/KAG)app (mM−1 s−1) | D(VMAX/KAG)app |
|---|---|---|---|---|---|
| N-acetylglycine | 1 | 9.2 ± 0.3 | 18 ± 1.6 | 0.51 ± 0.05 | 3.2 ± 0.31 |
| N-propionylglycine | 2 | 9.8 ± 0.3 | 2.4 ± 0.3 | 2.6 ± 0.3 | 2.9 ± 0.39 |
| N-butyrylglycine | 3 | 11 ± 0.4 | 2.3 ± 0.3 | 4.6 ± 0.6 | 2.3 ± 0.33 |
| N-hexanoylglycine | 5 | 11 ± 0.2 | 0.58 ± 0.05 | 18 ± 1.5 | 2.1 ± 0.22 |
| N-octanoylglycine | 7 | 13 ± 0.3 | 0.20 ± 0.02 | 66 ± 6.1 | 1.6 ± 0.21 |
| N-decanoylglycine | 9 | 13 ± 0.2 | 0.11 ± 0.01 | 130 ± 8.0 | 1.2 ± 0.12 |
All experiments were carried out at ambient O2 meaning that the intial O2 concentration was 217 μM.
The steady-state parameters are reported as the experimental value ± the standard error.
Figure 1.
The decrease in the D(VMAX/KAG)app at ambient O2 (217 μM) as the length of the N-acyl chain increases for PAM catalysis. The dashed line is drawn solely to emphasize the linearity of these data and is not model-based.
The values for (VMAX/KO2)app increased linearly as the length of the acyl chain increased from N-acetylglycine (R = 1) to N-decanoylglycine (R = 9) (Fig. 2). As shown in Table 2, the (VMAX/KO2)app value increases from 37 ± 1.9 mM-1s-1 for N-acetylglycine to 204 ± 23 mM−1s−1 for N-decanoylglycine, respectively. Interestingly, the VMAX is relatively insensitive to the acyl chain length for N-acylglycine substrates, with an average value of 20 ± 0.7 s−1 for all the N-acylglycine substrates included here (R = 1 to 9)
Figure 2.
Values for the (VMAX/KO2)app measured at a saturating concentration of the indicated N-acylglycine and 217 μM O2.
Table 2.
| N-Acylglycine Substrate (AG) | R | (VMAX,O2)app (s−1) | (KM,O2)app (μM) | (VMAX/KO2)app (mM−1 s−1) |
|---|---|---|---|---|
| N-acetylglycinec | 1 | 28 ± 3.3 | 550 ± 130 | 51 ± 7.0 |
| N-acetylglycined | 1 | 21 ± 0.9 | 230 ± 30 | 92 ± 12 |
| N-propionylglycine | 2 | 19 ± 0.4 | 210 ± 11 | 90 ± 5.0 |
| N-butyrylglycine | 3 | 17 ± 0.7 | 170 ± 20 | 99 ± 12 |
| N-hexanoylglycine | 5 | 20 ± 0.6 | 150 ± 14 | 140 ± 13 |
| N-octanoylglycine | 7 | 17 ± 0.3 | 110 ± 7.0 | 160 ± 10 |
| N-decanoylglycine | 9 | 21 ± 0.6 | 100 ± 11 | 200 ± 20 |
For these experiments, O2 was the variable substrate with the indicated N-acylglycine fixed at a saturating concentration, 10–15 × (KM,AG)app value.
The steady-state parameters are reported as the experimental value ± the standard error.
This value for the VMAX/KO2 using N-acetylglycine as the oxidizable was calculated from a fit the to the equilibrium–preferred equation (equation 3).
This value for the (VMAX/KO2)app using N-acetylglycine as the oxidizable was measured by varying the intial [O2] at saturating [N-acetylglycine].
Minimal Kinetic Mechanism
The N-acylglycine exhibiting the highest D(VMAX/KAG)app at ambient O2 was N-acetylglycine, 3.2 ± 0.31 (Table 1). The minimal kinetic mechanism for N-acetylglycine was determined by measuring the dependence of the D(VMAX/KAG)app as a function of O2 concentration and D(VMAX/KO2)app as a function of N-acetylglycine concentration. Initial rate data were fit to the bi-substrate kinetic equations representing either the sequential or equilibrium-ordered mechanisms (equation 2 and 3), respectively. The data best fit an equilibrium-ordered kinetic mechanism (equation 3) with σ values of 0.27 (H) and 0.13 (D) and variance values 0.074 (H) and 0.016 (D), respectively. By comparison, data fit to the sequential kinetic mechanism had comparable σ and variance values though many calculated parameters were negative and had high error suggesting a lack of significance for many of the terms.
The magnitude of the D(VMAX/KAG) term was constant, 1.9 ± 0.2 (Supporting Information, Table S1), though lower than the observed D(VMAX/KO2) value of 2.7 ± 0.4 (Table 3). Replot of the (VMAX,AG)app, vs. [O2] to determine the D(VMAX) (Supporting Information, Fig. S2), yielded VMAX values of 21 ± 1.6 s−1 for N-acetylglycine and 24 ± 2.0 s−1 for [α-2H2]-N-acetylglycine giving a D(VMAX) of 0.88 ± 0.10. These values are in reasonable agreement with those obtained by a fit of the kinetic data to the rate equation for an equilibrium-ordered mechanism (equation 3): VMAX values of 28 ± 3 s−1 (H) and 22 ± 1.4 s−1 (D) N-acetylglycine with a D(VMAX) of 1.3 ± 0.2 (Tables 2 and 3).47
Table 3.
Deuterium Kinetic Isotope Effects for N-Acetylglycine Oxidationa
| Oxidizable Substrate | |||
|---|---|---|---|
| Parameter | N-Acetylglycine | [2H2]-N-Acetylglycine | DKIE |
| VMAX (s−1) | 28 ± 3.3 | 22 ± 1.3 | 1.3 ± 0.2 |
| VMAX/KO2 (mM−1 s−1) | 51 ± 7.0 | 19 ± 0.7 | 2.7 ± 0.4 |
| KI,AG (mM) | 17 ± 3.0 | 14 ± 0.7 | 1.2 ± 0.2 |
The deuterium isotope effects (DKIE) for each relevant kinetic parameter (± standard error) are included.
It should also be noted, that the D(VMAX/KO2) calculated by re-plot analysis yielded a value of 2.5 ± 0.6 (Supporting Information, Fig. S2). These data show apparent terms for the D(VMAX/KAG)app kinetic isotope effect to be constant as a function of oxygen concentration. The dissociation constant for the N-acetylglycine substrate, KI,AG, was independent of α-carbon deuterium substitution with DKI,AG values of 1.2 ± 0.2.
Viscosity Effects
Values measured for both VMAX and VMAX/KM as a function of Ficoll-400 concentration (the macroviscogen) show no deviation from controls for the oxidation of either N-acetylglycine or N-decanoylglycine, respectively. Increased microviscosity resulted in a decreased rate of product release (VMAX) for N-acetylglycine and N-decanoylglycine. However, (VMAX/KAG)app showed no significant dependence on microviscosity for both substrates over the relative microviscosity range measured (2.07 → 5.33 ηrel). Alterations in the active site microenvironment were observed to be chain-length independent as the (VMAX/KAG)app term for each substrate was equal and constant over the relative viscosity range studied for both substrates relative to the non-viscogen controls.
As the minimal kinetic mechanism of PHM for substrate addition is sequential with the oxidizable substrate binding first (Scheme 2), viscosity studies are a valid probe for the forward commitment factor, cf = (k5/k4)[1+ (k3[O2]/k2)]. The viscosity independence of VMAX/KM for both N-acetylglycine or N-decanoylglycine (Fig. S5) suggests that binding of both substrates is in true equilibrium and the minimal kinetic mechanism is unaltered as a function of substrate chain length. If k2 significantly decreased as the N-acylglycine chain length increased, the rate constant for the chemical step (k5) would approach the value of k2 resulting in a change in the kinetic mechanism from equilibrium-ordered to sequential. Under these conditions, the isotope effect on the VMAX/KM would decrease as the forward commitment increased and would vary as a function of [O2]. The viscosity dependence of N-acylglycine binding provides a decrease in VMAX for both N-acetylglycine and N-decanoylglycine vs. non-viscogenic controls. The observed decrease in VMAX for both N-acetylglycine and N-decanoylglycine vs. non-viscogenic controls suggests that product release (k7) is rate-limiting for PHM, consistent with the lack of a VMAX isotope effect (Table 3).
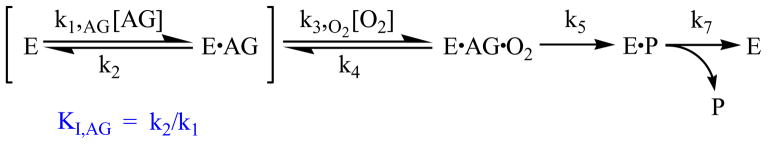
Scheme 2.
Representative minimal kinetic mechanism for an equilibrium-ordered, sequential mechanism for the binding of the N-acylglycine (AG) and O2 binding to PHM (E).
Furthermore, the viscosity studies directly probe the relative ‘stickiness’ of the N-acylglycine to PHM. The lack of a microviscosity effect on the VMAX/KM values for both N-acetylglycine and N-decanoylglycine means that the ratio of the VMAX/KM values for the two substrates are microviscosity independent, despite the differences in the D(VMAX/KAG)app values: 3.2 for N-acetylglycine and 1.2 for N-decanoylglycine. The decrease in the D(VMAX/KAG)app with acyl chain length cannot be attributed to an increased forward commitment factor because the k5/k2 ratio does not change enough to perturb the equilibrium. Using this rationale, it can be concluded that all the N-acylglycines share an equilibrium-ordered mechanism, where k2 is much greater than k5, as well as k3,O2[O2] (Scheme 2). Therefore, the observed KIEs (Fig. 1) cannot be attributed to a change in the minimal kinetic mechanism for this library of substrates. An equilibrium-ordered kinetic mechanism for N-decanoylglycine is supported by the [O2]-independence of the D(VMAX/KAG)app when measured from 30–200 μM O2 (data not shown).
Computational Chemistry
i.) Predicted N-Acylglycine Docking Conformations
Our docking results indicate that the N-acylglycine substrates bind into a hydrophobic pocket within the known PHM active site in the ternary AG•O2•enzyme complex. The N-acylglycine substrates are observed to form an ionic bond between the carboxylate of the substrate glycyl residue and the guanidine group of Arg240. This ionic interaction between substrate carboxylate and Arg240 has also been observed in the crystallized PHM structure (1SDW) containing bound N-acetyl-diiodo-tyrosyl-D-threonine.13 Of the N-acylglycines investigated computationally, all of the substrates except N-acetylglycine interacted with an active site, hydrophobic pocket comprised of the residues Leu206, Met208, Ile306, and Met314 (Supporting Information, Fig. S3). The lack of interaction between N-acetylglycine and the hydrophobic pocket can be attributed to the size of the substrate as the Euclidean distance between Arg240, the site of ionic interaction, and the hydrophobic pocket is simply too great. The strength of this interaction increases as a function of increasing substrate N-acyl chain length. These ionic and hydrophobic interactions were observed to be the predominant determinants for substrate binding when forming the ternary reduced PHM•acylglycine•O2 complex in our computational investigation.
ii.) Alchemical Free Energy Perturbation (AFEP) and Equilibrium Dynamics
The AFEP simulations resulted in a thermodynamic plot of ΔG vs. R which illustrates the inverse relationship between relative dissociation energy (ΔGALCHEMICAL) and chain length, increasing and decreasing respectively for N-acylglycine substrates which were observed to interact with the hydrophobic pocket. At R = 1 (N-acetylglycine), the ΔGALCHEMICAL value is ~24 kcal/mole lower than the R = 2 (N-propionylglycine) complex and ~12 kcal/mol lower than R = 9 (N-decanoylglycine) complex, the most stable complex identified from the set of N-acylglycines employed in our work (Fig. 3). The decrease in ΔGALCHEMICAL vs. acyl chain length indicates that increasing the hydrophobicity of the N-acylglycine substrates increases the stability of the AG•O2•enzyme complex. In other words, dissociation of the N-acylglycine from AG•O2•enzyme to form the binary O2•enzyme + AGfree becomes increasingly favorable as the acyl chain decreases.
Figure 3.
A plot of the ΔGALCHEMICAL vs. N-acylglycine chain length. For the above molecular dynamics (MD) simulations, relative free energies were calculated with N-decanoylglycine treated as the initial state (λ≤= 0) with respect to shorter chain N-acylglycine substrates (λ =1). Due to the ‘decanoylglycine-to-acetylglycine’ transformation appearing below the expected trend, ‘butyrylglycine-to-propionylglycine’ and ‘butyrylglycine-to-acetylglycine’ transformations were also performed and plotted (open circles).
The equilibrium dynamics calculations indicate a drastically altered N-acetylglycine pose compared with all other N-acylglycine substrates. The goal of our NPT simulation was to sample each N-acylglycine substrate conformation in the absence of an alchemical permutation (AFEP). Over the 1 nanosecond simulation, N-acetylglycine (R = 1) sampled many more binding conformations than the substrates with longer acyl chains. The N-acetylglycine molecule performed an inversion within the PHM active site during the simulation, consistent with docking orientation poses predicted (Supporting Information, Fig. S3). This phenomenon was partially observed during the AFEP studies, as the Arg240 salt bridge was not observed for the ‘decanoylglycine-to-acetylglycine’ AFEP-transformation as the final state was achieved. For the other substrates, the position of the acyl chain within the active site was modulated by the hydrophobic pocket. Note that equilibrium dynamic (MD) simulation of binding of N-benzoylglycine to PHM displayed no deviation in the salt bridge between Arg240 and the carboxylate of the glycyl moiety. This was similar to the MD simulation for the binding of N-decanoylglycine, although we did observe greater movement of the benzene ring. N-Benzoylglycine (hippurate) has long been known to be a PHM substrate, but with a relative low (VMAX/Khippurate)app value relative to peptide and longer chain N-acylglycine substrates.29,48 The Cα-H ↔ O2 distance minima achieved during the simulations of all the N-acylglycine derivatives approaches the sum of the van der Waals radii for oxygen and hydrogen. The classical force field is not capable of pushing the distances any closer than this without external forces being applied. Note that our intent is to determine the free energy of binding and not to simulate the actual reaction. Other methods are better suited for such a task e.g. QM/MM.
Discussion
Minimal Kinetic Mechanism for the PHM Mediated Oxidation of the N-Acylglycines
Previous work has established that the addition of the oxidizible substrate occurs after the reduction of enzyme-bound Cu(II) atoms.18e,19 Using ascorbate as the exogenous reductant, Merkler et al.49 demonstrated that the two electrons required for the reduction of the PHM-bound Cu(II) atoms are delivered from two one-electron reduction steps, resulting in the oxidation of two ascorbate molecules to two semidehydroascorbate molecules per enzyme turnover. Reduction of PHM-bound Cu(II) is not included in the minimal mechanism as the oxidized reductant dissociates following reduction.18b The initial rate patterns for both N-acetylglycine and [α-2H2]-N-acetylglycine are most consistent with an equilibrium-ordered mechanism (Scheme 2): the convergence of 1/initial rate vs. 1/[N-acetylglycine] at increasing [O2] in the second quadrant while the 1/initial rate vs. 1/[O2] at increasing [N-acetylglycine] intersecting at the abscissa (Supporting Information, Fig. S4).50 An equilibrium-ordered mechanism can be differentiated from equilibrium random as the replot of (KAG/VMAX)app against 1/[O2] passes through the origin.50,51 Therefore, the order of substrate addition to PHM was sequential with the addition of the N-acylglycine followed by O2 to form the central complex, PHM-2Cu(I)•AG•O2. An equilibrium-ordered mechanism is consistent with previous studies of PAM and PHM.18e,19,52 As the rate expression for an equilibrium-ordered kinetic mechanism is unsymmetrical (equation 3),50,51 a signature for this mechanism becomes equivalent magnitudes of the D(VMAX/KM) values for both the N-acylglycine substrates and O2.50,53 Experimentally, this exactly what we found as D(VMAX/KM) for N-acetylglycine and O2 were equivalent within experimental error, 1.9 ± 0.2 for N-acetylglycine and 2.0 ± 0.1 for O2 (Table S1 and 3). The equilibrium-ordered mechanism is rationalized with k2,AG being much faster than k3,O2[O2] at any [O2] (see Scheme 2).18e,51 As shown in equation 5, the D(VMAX/KM) value for each substrate in the equilibrium-ordered mechanism is reduced to the following:
| (5) |
The binding of N-acetylglycine to reduced PHM is in equilibrium, such that the off-rate from the E•acetylglycine complex, k2, is much greater than the rate of catalysis, k5 (Scheme 2).51 The micro-viscosity data show no acyl chain length dependence (Supporting Information, Fig. S5). Therefore, the ~250-fold increase in (VMAX/KAG)app as the acyl chain length increases from N-acetylglycine to N-decanoylglycine (Table 1) catalytic cannot be attributed to either a fully or partially diffusion-limited process.54 This eliminates the possibility of the equilibrium-ordered mechanism found for N-acetylglycine addition to PAM changing to a sequential mechanism for the other N-acylglycine substrates as chain length increases. For an equilibrium-ordered mechanism, the magnitude of D(VMAX/KAG) would differ from the D(VMAX/KO2) as the former term would become dependent on [O2], while the latter would remain constant as a function of [N-acylglycine].51
Acyl Chain Length Dependence of the Binding Mode
The plot of ΔG vs. chain length (R) shows that dissociation energy (ΔGALCHEMICAL) increases as R decreases from 9 to 2. At R = 1, the ΔG values were ~24 kcal/mole lower than that for R = 2 and ~12 kcal/mol lower than that for R = 9, the most stable complex included in our study (Fig. 3). These results are derived from the AFEP calculations by direct comparison with N-decanoylglycine (R = 9). Each N-acylglycine is bound in the PHM active site in the same orientation, ionic bonding between the carboxylate and Arg240 and an interaction between the acyl chain and the hydrophobic pocket defined by Leu206, Met208, Ile306, and Met314. The exception to this was N-acetylglycine (Fig. 3). The anomalous behavior of N-acetylglycine behavior could be attributed to the configurational ensembles not having a large degree of the desired overlap between states; thereby, not providing the requisite accuracy during the AFEP permutations.39a For the substrate range N-propionylglycine through N-decanoylglycine (R = 2 → 9), the increased acyl chain length of the substrate was proportional to an increase in the dissociation energy of substrate from the central complex (Fig. 3). The ΔGALCHEMICAL vs. chain length plot, based on the AFEP calculations, demonstrated that increasing the hydrophobicity of the N-acylglycine substrate results in a proportional increase in the ΔGdissociation for the dissociation of the substrate from the reduced PHM•acylglycine•O2 complex. In other words, as the acyl chain lengthens, the N-acylglycine substrate is less likely to dissociate from the reduced PHM•acylglycine•O2 complex.
The thermodynamics obtained from the AFEP calculations only provide insight about the relative stability of the optimal conformation for the individual reduced PHM•acylglycine•O2 complexes once these have been achieved. Conversely, the steady-state kinetics and the KIEs describe the probability of reaching the most stable (or optimal) conformation. Increasing the chain length promotes the interaction between the acyl chain and the hydrophobic pocket adjacent to the CuM domain (Supporting Information, Fig. S3) and modulate the fidelity of PHM catalysis. The viscosity effects suggested that the association rate constant for N-acylglycine binding was similar for all the substrates. This coupled to the fact that the magnitude of the dissociation rate constant becomes smaller as the acyl chain length gets longer, suggesting that that the Kdissociation (= koff/kon) would also decrease as the acyl chain lengthens. Therefore, the ΔG°dissociation would become more positive and less favorable as the acyl chain of the substrates becomes longer. The AFEP analysis correlates well with the relative free energy for Cα-H cleavage, providing an internal probe for both the chemistry and binding steps. The dissociation rate constant for the N-acylglycines decreases as the acyl chain lengthens and is perturbed directly into the rate constant for the chemical step (Cα-H cleavage) such that the k5/k2 ratio does not change enough to alter the kinetic mechanism. The lack of a microviscosity effect on the VMAX/KM values for both N-acetylglycine and N-decanoylglycine and [O2]-independence of D(VMAX/KM) for N-decanoylglycine is also consistent an equilibrium-ordered minimal kinetic mechanism for all N-acylglycine substrates included here. Overall, the combination of kinetic and thermodynamic data presented in this study suggests that transient dynamic motions of PHM favor the more hydrophobic N-acylglycines to more efficiently sample conformer distances for optimal wave overlap between Cα-H and Cu/O2 as substrate orientation becomes increasingly constrained proportional to chain length.
Comparison of the α-carbon donor position between both N-benzoylglycine and N-decanoylglycine shows very little difference in the orientation of these atoms over the course of the MD simulation for the glycyl moiety (Supporting Data, movie files). The benzoyl group was observed to display a much greater degree of conformational sampling compared with the decanoyl group. The VMAX/KM and KM values for N-decanoylglycine are 25-fold higher and 13-fold lower than those for N-benzoylglycine.29 The role of Arg240 in protein gating has been addressed with an R240Q PHMcc mutant, with a 2-fold increase in KM and a 200-fold decrease in VMAX.13a Results from this mutation suggest that Arg240 functions to pose a geometrical constraint upon both the oxidizable substrate and protein assisting the Franck-Condon gating dynamics to optimize hydrogen donor-acceptor distances. An advantage for hydrophobic substrates appears to be an extra stabilization provided by interaction with the hydrophobic pocket within the PHM active site which is used to reach the transfer configuration more effectively. Therefore, the increased contact points between enzyme and the N-acylglycine substrates as the acyl chain increases in hydrophobicity most likely assists frequency modulation between the hydrogen donor and Cu(II)-superoxo acceptor to more easily to achieve degenerate states.
Conclusion
The kinetic, viscosity, and KIE data included herein show that N-acylglycine binding to PHM is in equilibrium before catalysis can occur and that the dissociation of the N-acylglycine from the reduced PHM•S becomes less favorable as the length of the acyl chain increases. With the exception of N-acetylglycine, the MD simulations indicate that each N-acylglycine binds to reduced PHM in approximately the same orientation: an ionic interaction between the substrate carboxylate and Arg240 and an interaction between the acyl chain and an active site hydrophobic pocket (Supporting Information, Fig. S3).
The intent of the AFEP poses was to elucidate the contribution of acyl chain length to the relative dissociation energies for each ligand, benchmarked to the most hydrophobic substrate N-decanoylglycine. The equilibrium molecular dynamics simulations showed decreased active site sampling associated with increased chain length. The minima achieved during the simulations of all N-acylglycine derivatives approaches the van der Waals radii for oxygen and the Cα-hydrogen. The classical force field is incapable of creating distances any closer than this without the application of external force(s). The utilization of a hydrophobic pocket in the active site and carboxylate-Arg240 salt bridge for substrate positioning in the PHM ternary complex was the mechanism by which, Cα-H activation became decreasingly rate determining. The decrease in the KIE with hydrophobicity (Fig. 1) suggests that environmentally coupled tunneling phenomena were more efficient as the probability of optimal conformer sampling increased due to the N-acylglycine hydrophobic pocket and Arg240 interaction. The role of the active site hydrophobic pocket and Arg240 appear to pre-organize the bound substrate allowing protein promoting (gating) vibrations to assist the through-barrier tunneling process. Overall, pre-organization of the N-acylglycine substrate facilitated by the hydrophobic pocket and Arg240 provide a greater probability of a transient reduced PHM•S•O2 complex to be productive in the reaction coordinate. The chain length dependence for pre-organization suggests that optimal overlap for acceptor and donor moieties can be regulated by both enzyme dynamics and substrate structure. In this sense, substrate hydrophobicity lends itself to a synergistic event increasing the probability of a hydrogen tunneling event.
Francisco et al.23 suggest that H• tunneling in PHM catalysis was dominated by gating motions. Their results are not inconsistent with the data presented here as Francisco et al.23 did not argue that pre-organization had no role in PHM tunneling. However, there are some other intriguing differences between our work and that of Francisco et al.23 The substrate used by Francisco et al.52 was hippurate and our MD simulations show highly randomized movement of the benzene ring and, thus, pre-organization may be less important for this PHM substrate. Another difference is our use of bifunctional PAM to study PHM while Francisco et al.23 studied monofunctional PHM. It is possible that PAL may modulate the dynamics of the PHM domain such that pre-organization plays a more important role in H• tunneling as catalyzed by the PHM domain of bifunctional PAM. Future studies will address this most interesting possibility.
Given the mechanistic similarities between PHM and DβM, pre-organization is likely to be important for DβM catalysis as well. In fact, an active site glutamine residue in DβM, Gln369, may have a role similar to the active site hydrophobic pocket in PHM as the 3,4-dihydroxbenzene moiety of dopamine bonds weakly with Gln369.55 In addition, the fumarate-dependent activation of DβM could be analogous to the role of substrate carboxylate-Arg240 salt bridge in PHM. It has been proposed that fumarate activation was the result of reduced phenylethylamine motion (pre-organization) due to a weak interaction between anionic fumarate and the primary amine.56 In sum, the observations of this study describe the active site of PHM as extremely well designed to accommodate the spatial requirements for donor and acceptor wave overlap. The decrease in the KIE with hydrophobicity (Fig. 3) suggest that environmentally coupled tunneling phenomena become more efficient as the probability of optimal conformer sampling increased due to interactions between the N-acylglycine and both the hydrophobic pocket and Arg240.
Supplementary Material
Acknowledgments
This work was supported, in part, by grants from the National Institutes of Health - General Medical Sciences (R15-GM067257 and R15-GM073659), the Shirley W. & William L. Griffin Foundation, the Alpha Research Foundation, Inc., the Eppley Foundation for Research, the Gustavus and Louise Pfieffer Research Foundation, the Milheim Foundation for Cancer Research, the Shin Foundation for Medical Research, the Wendy Will Case Cancer Fund, and the University of South Florida - Established Researcher Grant Program, to D.J.M, financial support from Louisiana Cancer Research Consortium (LCRC), Center for Undergraduate Research (CUR) and Research Centers in Minority Institutions (RCMI) to N.R.M, and a predoctoral fellowship to E.W.L. from the American Heart Association (0415259B). This project was supported by an allocation from the TeraGrid Advanced Support Program (TG-MCB070112N) and services of Research Computing at the University of South Florida. The authors dedicate this work to the memory of Dr. Terence C. Owen.
Footnotes
Supporting Information Available: Data referred to in the text, NMR spectra for the N-acylglycines synthesized for this work, and complete lists of authors for references 29, 30, and 39c. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.(a) Eipper BA, Mains RE. Annu Rev Physiol. 1988;50:333–344. doi: 10.1146/annurev.ph.50.030188.002001. [DOI] [PubMed] [Google Scholar]; (b) Bolkenius FN, Ganzhorn AJ. Gen Pharmacol. 1998;31:655–659. doi: 10.1016/s0306-3623(98)00192-x. [DOI] [PubMed] [Google Scholar]; (c) Kulathila R, Merkler KA, Merkler DJ. Nat Prod Rep. 1999;16:145–154. doi: 10.1039/a801346b. [DOI] [PubMed] [Google Scholar]; (d) Prigge ST, Mains RE, Eipper BA, Amzel LM. Cell Mol Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Klinman JP. J Biol Chem. 2006;281:3013–3016. doi: 10.1074/jbc.R500011200. [DOI] [PubMed] [Google Scholar]
- 2.Ray MVL, Van Duyne P, Bertelsen AH, Jackson-Matthews DE, Sturmer AM, Merkler DJ, Consalvo AP, Young SD, Gilligan JP, Shields PP. Biotechnology (N Y) 1993;11:64–70. doi: 10.1038/nbt0193-64. [DOI] [PubMed] [Google Scholar]
- 3.(a) Bradbury AF, Finnie MD, Smyth DG. Nature. 1982;298:686–688. doi: 10.1038/298686a0. [DOI] [PubMed] [Google Scholar]; (b) Eipper BA, Mains RE, Glembotski CC. Proc Natl Acad Sci U S A. 1983;80:5144–5148. doi: 10.1073/pnas.80.16.5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Vanden Broeck J. Peptides. 2001;22:241–254. doi: 10.1016/s0196-9781(00)00376-4. [DOI] [PubMed] [Google Scholar]; (b) Predel R. In: Invertebrate Neuropeptides and Hormones: Basic Knowledge and Recent Advances 2006. Satake H, editor. Transworld Research Network; Kerala, India: 2006. pp. 127–155. [Google Scholar]
- 5.Grimmelikhuijzen CJ, Leviev I, Carstensen K. Int Rev Cytol. 1996;167:37–89. doi: 10.1016/s0074-7696(08)61345-5. [DOI] [PubMed] [Google Scholar]
- 6.(a) Merkler DJ, Chew GH, Gee AJ, Merkler KA, Sorondo JPO, Johnson ME. Biochemistry. 2004;43:12667–12674. doi: 10.1021/bi049529p. [DOI] [PubMed] [Google Scholar]; (b) Farrell EK, Merkler DJ. Drug Discov Today. 2008;13:558–568. doi: 10.1016/j.drudis.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Young SD, Tamburini PP. J Am Chem Soc. 1989;111:1933–1934. [Google Scholar]; (b) Katopodis AG, Ping D, May SW. Biochemistry. 1990;29:6115–6120. doi: 10.1021/bi00478a001. [DOI] [PubMed] [Google Scholar]
- 8.Kulathila R, Consalvo AP, Fitzpatrick PF, Freeman JC, Snyder LM, Villafranca JJ, Merkler DJ. Arch Biochem Biophys. 1994;311:191–195. doi: 10.1006/abbi.1994.1225. [DOI] [PubMed] [Google Scholar]
- 9.Bell J, Ash DE, Snyder LM, Kulathila R, Blackburn NJ, Merkler DJ. Biochemistry. 1997;36:16239–16246. doi: 10.1021/bi970903d. [DOI] [PubMed] [Google Scholar]
- 10.De M, Bell J, Blackburn NJ, Mains RE, Eipper BA. J Biol Chem. 2006;281:20873–20882. doi: 10.1074/jbc.M513886200. [DOI] [PubMed] [Google Scholar]
- 11.(a) Takahashi K, Harada S, Higashimoto Y, Shimokawa C, Sato H, Sugishima M, Kaida Y, Noguchi M. Biochemistry. 2009;48:1654–1662. doi: 10.1021/bi8018866. [DOI] [PubMed] [Google Scholar]; (b) Chufán EE, De M, Eipper BA, Mains RE, Amzel LM. Structure. 2009;17:965–973. doi: 10.1016/j.str.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Ash DE, Papadopoulos NJ, Colombo G, Villafranca JJ. J Biol Chem. 1984;259:3395–3398. [PubMed] [Google Scholar]; (b) Klinman JP, Krueger M, Brenner M, Edmondson DE. J Biol Chem. 1984;259:3399–3402. [PubMed] [Google Scholar]
- 13.(a) Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Science. 1997;278:1300–1305. doi: 10.1126/science.278.5341.1300. [DOI] [PubMed] [Google Scholar]; (b) Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Nat Struct Biol. 1999;6:976–983. doi: 10.1038/13351. [DOI] [PubMed] [Google Scholar]; (c) Prigge ST, Eipper BA, Mains RE, Amzel LM. Science. 2004;304:864–867. doi: 10.1126/science.1094583. [DOI] [PubMed] [Google Scholar]
- 14.Blackburn NJ, Concannon M, Shahiyan SK, Mabbs FE, Collison D. Biochemistry. 1988;27:6001–6008. doi: 10.1021/bi00416a026. [DOI] [PubMed] [Google Scholar]
- 15.(a) Blackburn NJ, Pettingill TM, Seagraves KS, Shigeta RT. J Biol Chem. 1990;265:15383–15386. [PubMed] [Google Scholar]; (b) Eipper BA, Quon AS, Mains RE, Boswell JS, Blackburn NJ. Biochemistry. 1995;34:2857–2865. doi: 10.1021/bi00009a016. [DOI] [PubMed] [Google Scholar]; (c) Boswell JS, Reedy BJ, Kulathila R, Merkler D, Blackburn NJ. Biochemistry. 1996;35:12241–12250. doi: 10.1021/bi960742y. [DOI] [PubMed] [Google Scholar]
- 16.Merkler DJ, Kulathila R, Tamburini PP, Young SD. Arch Biochem Biophys. 1992;294:594–602. doi: 10.1016/0003-9861(92)90730-k. [DOI] [PubMed] [Google Scholar]
- 17.(a) Ramer SE, Cheng H, Palcic MM, Vederas JC. J Am Chem Soc. 1988;110:8526–8532. [Google Scholar]; (b) Ping DS, Katopodis AG, May SW. J Am Chem Soc. 1992;114:3998–4000. [Google Scholar]; (c) Battersby AR, Sheldrake PW, Staunton J, Williams DC. J Chem Soc Perkin 1. 1976:1056–1062. doi: 10.1039/p19760001056. [DOI] [PubMed] [Google Scholar]
- 18.(a) Goldstein M, Joh TH, Garvey TQ., III Biochemistry. 1968;7:2724–2730. doi: 10.1021/bi00848a005. [DOI] [PubMed] [Google Scholar]; (b) Gilligan JP, Lovato SJ, Mehta NM, Bertelsen AH, Jeng AY, Tamburini PP. Endocrinology. 1989;124:2729–2736. doi: 10.1210/endo-124-6-2729. [DOI] [PubMed] [Google Scholar]; (c) Klinman JP, Humphries H, Voet JG. J Biol Chem. 1980;255:11648–11651. [PubMed] [Google Scholar]; (d) Brenner MC, Murray CJ, Klinman JP. Biochemistry. 1989;28:4656–4664. doi: 10.1021/bi00437a022. [DOI] [PubMed] [Google Scholar]; (e) Francisco WA, Merkler DJ, Blackburn NJ, Klinman JP. Biochemistry. 1998;37:8244–8252. doi: 10.1021/bi973004y. [DOI] [PubMed] [Google Scholar]
- 19.McIntyre NR, Lowe EW, Jr, Merkler DJ. J Am Chem Soc. 2009;131:10308–10319. doi: 10.1021/ja902716d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Tian G, Berry JA, Klinman JP. Biochemistry. 1994;33:226–234. doi: 10.1021/bi00167a030. [DOI] [PubMed] [Google Scholar]; (b) Francisco WA, Blackburn NJ, Klinman JP. Biochemistry. 2003;42:1813–1819. doi: 10.1021/bi020592t. [DOI] [PubMed] [Google Scholar]
- 21.(a) Bollinger JM, Jr, Krebs C. Curr Opin Chem Biol. 2007;11:1151–158. doi: 10.1016/j.cbpa.2007.02.037. [DOI] [PubMed] [Google Scholar]; (b) Maiti D, Lee DH, Gaoutchenova K, Wurtele C, Holthausen MC, Sarjeant AAN, Sundermeyer J, Schindler S, Karlin KD. Angew Chem Int Ed Engl. 2008;47:82–85. doi: 10.1002/anie.200704389. [DOI] [PubMed] [Google Scholar]
- 22.(a) Chen P, Solomon EI. J Am Chem Soc. 2004;126:4991–5000. doi: 10.1021/ja031564g. [DOI] [PubMed] [Google Scholar]; (b) Crespo A, Marti MA, Roitberg AE, Amzel LM, Estrin DA. J Am Chem Soc. 2006;128:12817–12828. doi: 10.1021/ja062876x. [DOI] [PubMed] [Google Scholar]; (c) Maiti D, Sarjeant AAN, Karlin KD. Inorg Chem. 2008;47:8736–8747. doi: 10.1021/ic800617m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francisco WA, Knapp MJ, Blackburn NJ, Klinman JP. J Am Chem Soc. 2002;124:8194–8195. doi: 10.1021/ja025758s. [DOI] [PubMed] [Google Scholar]
- 24.(a) Klinman JP. Philos Trans R Soc Lond B Biol Sci. 2006;361:1323–1331. doi: 10.1098/rstb.2006.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nagel ZD, Klinman JP. Nat Chem Biol. 2009;5:543–550. doi: 10.1038/nchembio.204. [DOI] [PubMed] [Google Scholar]; (c) Klinman JP. Chem Phys Lett. 2009;471:179–193. doi: 10.1016/j.cplett.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knapp MJ, Rickert K, Klinman JP. J Am Chem Soc. 2002;124:3865–3874. doi: 10.1021/ja012205t. [DOI] [PubMed] [Google Scholar]
- 26.(a) Sutcliffe MJ, Scrutton NS. Phys Chem Chem Phys. 2006;8:4510–4516. doi: 10.1039/b609622k. [DOI] [PubMed] [Google Scholar]; (b) Hay S, Pudney C, Hothi P, Johannissen LO, Masgrau L, Pang J, Leys D, Sutcliffe MJ, Scrutton NS. Biochem Soc Trans. 2008;36:16–21. doi: 10.1042/BST0360016. [DOI] [PubMed] [Google Scholar]
- 27.Hay S, Scrutton NS. Biochemistry. 2008;47:9880–9887. doi: 10.1021/bi8005972. [DOI] [PubMed] [Google Scholar]
- 28.Bauman AT, Jaron S, Yul ET, Burchfiel JR, Blackburn NJ. Biochemistry. 2006;45:11140–11150. doi: 10.1021/bi060905a. [DOI] [PubMed] [Google Scholar]
- 29.Wilcox BJ, et al. Biochemistry. 1999;38:3235–3245. doi: 10.1021/bi982255j. [DOI] [PubMed] [Google Scholar]
- 30.Merkler DJ, et al. Bioorg Med Chem. 2008;16:10061–10074. doi: 10.1016/j.bmc.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller DA, Sayad KU, Kulathila R, Beaudry GA, Merkler DJ, Bertelsen AH. Arch Biochem Biophys. 1992;298:380–388. doi: 10.1016/0003-9861(92)90425-v. [DOI] [PubMed] [Google Scholar]
- 32.(a) Mesaik MA, Rahat S, Khan KM, Ullah Z, Choudhary MI, Murad S, Ismail Z, Rahman A, Ahmad A. Bioorg Med Chem. 2004;12:2049–2057. doi: 10.1016/j.bmc.2004.02.034. [DOI] [PubMed] [Google Scholar]; (b) Katz J, Lieberman I, Barker HA. J Biol Chem. 1953;200:431–441. [PubMed] [Google Scholar]; (c) Jones WD. J Chem Soc Perkin I. 1981:344–348. [Google Scholar]
- 33.Morrison TJ, Billet F. J Chem Soc. 1952. pp. 3819–3822. [Google Scholar]
- 34.Davison AJ, Kettle AJ, Fatur DJ. J Biol Chem. 1986;261:1193–1200. [PubMed] [Google Scholar]
- 35.The observed kinetic isotope effects reported here encompass both the primary and α-secondary deuterium effect. Because the α-secondary deuterium kinetic isotope effect is relatively small (~1.2) in relationship to the intrinsic primary effect (10–11), the contribution of the α-secondary effect to our measurements are within the error (≤12%).
- 36.Cho AE, Guallar V, Berne BJ, Friesner R. J Comput Chem. 2005;26:915–931. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.(a) Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]; (b) GLIDE. Schrodinger, LLC; Portland, OR: 2000. [Google Scholar]
- 38.QSITE. Schrodinger, LLC; Portland, OR: 2000. [Google Scholar]
- 39.(a) Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sanbonmatsu KY, Tung CS. J Struct Biol. 2007;157:470–480. doi: 10.1016/j.jsb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) MacKerell AD, et al. J Phys Chem. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Sousa SF, Fernandes PA, Ramos MJ. J Phys Chem A. 2007;111:10439–10452. doi: 10.1021/jp0734474. [DOI] [PubMed] [Google Scholar]
- 41.(a) Lee C, Yang WT, Parr RG. Phys Rev B Condens Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]; (b) Lee C, Parr RG. Phys Rev A. 1990;42:193–200. doi: 10.1103/physreva.42.193. [DOI] [PubMed] [Google Scholar]
- 42.Stevens WJ, Basch H, Krauss M. J Chem Phys. 1984;81:6026–6033. [Google Scholar]
- 43.Rappe AK, Casewit CJ, Colwell KS, Goddard WA, III, Skiff WM. J Am Chem Soc. 1992;114:10024–10035. [Google Scholar]
- 44.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 46.(a) Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577–8593. [Google Scholar]; (b) Cheatham TE, Miller JL, Fox T, Darden TA, Kollman PA. J Am Chem Soc. 1995;117:4193–4194. [Google Scholar]; (c) deSouza ON, Ornstein RL. Biophys J. 1997;72:2395–2397. doi: 10.1016/S0006-3495(97)78884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The VMAX,app at one fixed [O2] and the D(VMAX/KAG) value had to be determined from a replot of VMAX,app vs. [O2] because the rate equation for an equilibrium-ordered kinetic mechanism is unsymmetrical.
- 48.Katopodis AG, May SW. Biochemistry. 1990;29:4541–4548. doi: 10.1021/bi00471a006. [DOI] [PubMed] [Google Scholar]
- 49.Merkler DJ, Kulathila R, Consalvo AP, Young SD, Ash DE. Biochemistry. 1992;31:7282–7288. doi: 10.1021/bi00147a011. [DOI] [PubMed] [Google Scholar]
- 50.Cook PF, Cleland WW. Biochemistry. 1981;20:1790–1796. doi: 10.1021/bi00510a013. [DOI] [PubMed] [Google Scholar]
- 51.Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. Garland Science Publishing; New York, NY: 2007. [Google Scholar]
- 52.Francisco WA, Wille G, Smith AJ, Merkler DJ, Klinman JP. J Am Chem Soc. 2004;126:13168–13169. doi: 10.1021/ja046888z. [DOI] [PubMed] [Google Scholar]
- 53.(a) Cook PF. Isotopes Environ Health Stud. 1998;34:3–17. doi: 10.1080/10256019808036353. [DOI] [PubMed] [Google Scholar]; (b) Evans JP, Blackburn NJ, Klinman JP. Biochemistry. 2006;45:15419–15429. doi: 10.1021/bi061734c. [DOI] [PubMed] [Google Scholar]
- 54.(a) Stone SR, Morrison JF. Biochemistry. 1988;27:5493–5499. doi: 10.1021/bi00415a016. [DOI] [PubMed] [Google Scholar]; (b) Brouwer AC, Kirsch JF. Biochemistry. Vol. 21. 1982. pp. 1302–1307. [DOI] [PubMed] [Google Scholar]
- 55.(a) Kamachi T, Kihara N, Shiota Y, Yoshizawa K. Inorg Chem. 2005;44:4226–4236. doi: 10.1021/ic048477p. [DOI] [PubMed] [Google Scholar]; (b) Yoshizawa K, Kihara N, Kamachi T, Shiota Y. Inorg Chem. 2006;45:3034–3041. doi: 10.1021/ic0521168. [DOI] [PubMed] [Google Scholar]
- 56.Ahn N, Klinman JP. Biochemistry. 1983;22:3096–3106. doi: 10.1021/bi00282a012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.