Abstract
We demonstrate a methodology that utilizes the specificity of enzyme-substrate biomolecular interactions to trigger miniaturized tools under biocompatible conditions. Miniaturized grippers were constructed using multilayer hinges that employed intrinsic strain energy and biopolymer triggers, as well as ferromagnetic elements. This composition obviated the need for external energy sources, and allowed for remote manipulation of the tools. Selective enzymatic degradation of biopolymer hinge components triggered closing of the grippers; subsequent reopening was achieved with an orthogonal enzyme. We highlight the utility of these enzymatically triggered tools by demonstrating the biopsy of liver tissue from a model organ system and gripping and releasing an alginate bead. This strategy suggests an approach for the development of smart materials and devices that autonomously reconfigure in response to extremely specific biological environments.
An important area of engineering is the development of functional materials and devices that have the ability to actuate under specific environmental cues. 1–3 Chemical reactions can be used to facilitate changes in material properties and consequently generate mechanical force, thereby triggering actuation. Such chemo-mechanical actuation4–9 can enable an autonomous response from these structures without the need for any wires or tethers. Chemically triggered actuation does not require any external signaling or batteries, potentially lowering fabrication costs and enabling actuation in hard to reach places such as confined organs and vasculature. In addition, rapid diffusion of chemicals in aqueous solutions allows a stimulus to reach distant areas and trigger actuation of multiple tetherless devices simultaneously.10 While chemo-mechanical actuation is widely observed in nature, it has yet to be fully realized in human engineering which predominantly relies on mechanical actuation via pneumatic, hydraulic or electrical signals.11, 12
One considerable challenge is the development of chemo-mechanical structures that actuate only in response to specific cues under biocompatible conditions. Such a capability would offer the possibility for the creation of smart materials and devices that autonomously reconfigure when exposed to diseased locations within the body. This functionality would enable wide-ranging applications such as excision of tissue, stenting, clamping and removal of blockages.3
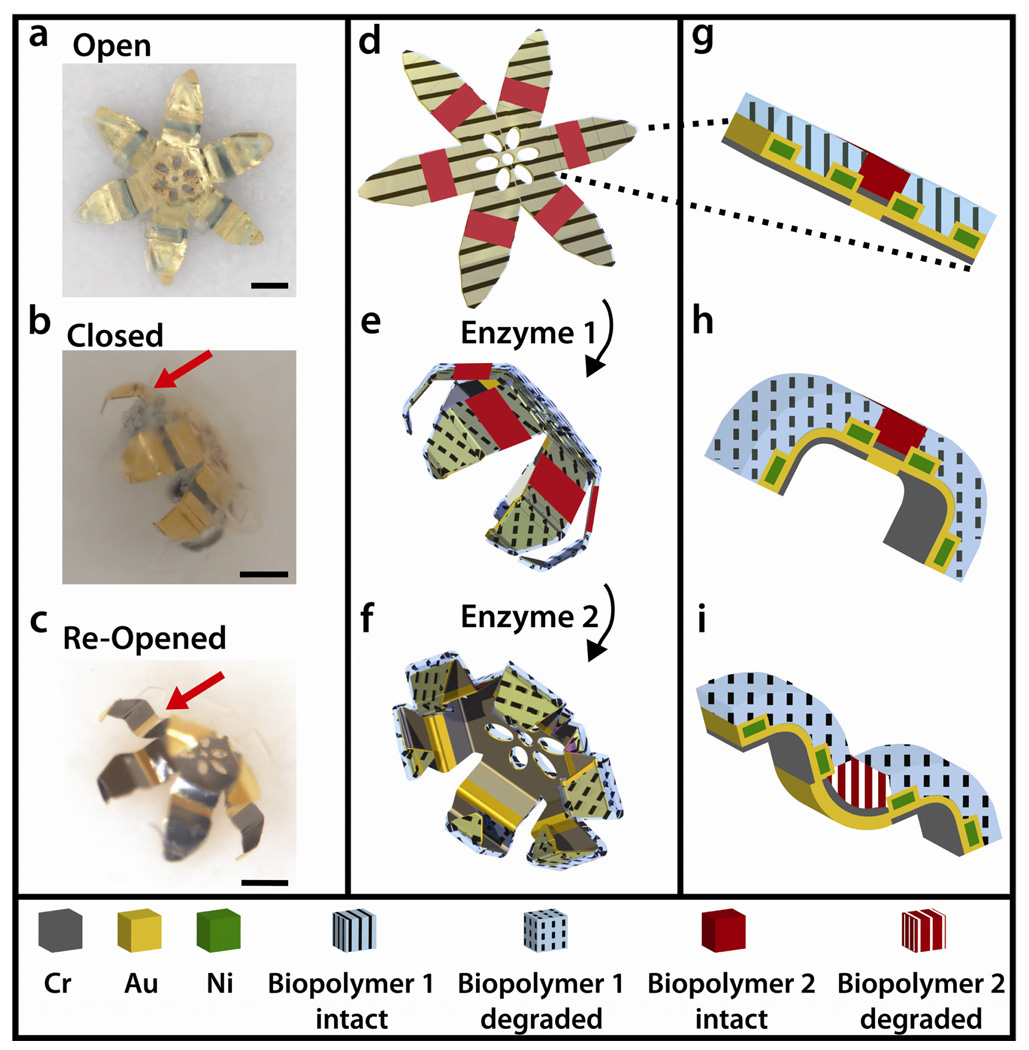
A well-known class of biochemical reactions which has naturally evolved with great selectivity is that of enzymes with their substrates. Biopolymers composed of polypeptides, polysaccharides and polynucleotides are selectively degraded by specific enzymes such as proteases, glycoside hydrolases, and nucleases, respectively. Here, we created multilayer grippers with hinges composed of either gelatin, a polypeptide, or carboxymethylcellulose (CMC), a polysaccharide. These hinges, also containing pre-stressed and structural metal films, were patterned using photolithography and combined with rigid segments to create a gripper. These tools closed and re-opened when exposed to proteases and glucosidases respectively. They were approximately 1.1 mm in diameter when open and approximately 600 µm in diameter when closed (Figure 1). The fabrication process was highly parallel and approximately 1600 grippers could be fabricated simultaneously on a three inch diameter wafer.
Figure 1.
Schematic of multilayered thin film gripper with integrated biopolymer layers that can close and re-open on exposure to enzymes. (a–c) Optical images of the grippers in the flat, closed, and re-opened states respectively. The red arrow indicates the second set of hinges. Scale bars represent 200 µm (a, b, c). (d–f) Schematic representations of the grippers in the three corresponding states above. (d) The gripper is held flat by the thick, crosslinked biopolymer (hatching). When this biopolymer is selectively degraded by enzyme 1 the modulus decreases and the gripper closes. (f) The second trigger, a rigid biopolymer insensitive to enzyme 1, is preventing a second set of hinges from actuating, keeping the gripper closed. (f) Subsequently this trigger can be actuated by enzyme 2 to re-open the gripper. (g–i) Cross section magnified view of a single hinge illustrating behavior of biopolymer trigger. (g) Both biopolymer layers are stiff, preventing all bending. (h) On degrading the biopolymer with enzyme 1, the second hinge remains flat. (i) The second polymer is degraded by enzyme 2 and this hinge bends in the opposite direction.
Grippers were designed with alternating rigid segments and flexible hinges. Rigid segments remained flat during the entire cycle of closing and re-opening, providing the mechanical strength required for secure gripping. Flexible hinges were initially flat, and curved only on exposure to the appropriate enzyme. Closing and re-opening was achieved using a gripper design with two kinds of hinges which bent with either concave or convex curvatures.13 The concept behind this two-stage actuation is as follows: On the appropriate trigger, all hinges covered with one biopolymer actuate, while the entire second set of hinges remain flat, causing the gripper to close. As the modulus of the second biopolymer is reduced, all the second hinges also bend, but in the opposite direction, thus re-opening the gripper (Figure 1b).
The grippers were microfabricated in two dimensions (2D) on a silicon wafer using conventional photolithography techniques and were subsequently released from the substrate. The first step involved depositing a sacrificial copper (Cu) layer14 on the wafer by thermal evaporation. Next, flexible and rigid components consisting of chromium (Cr), nickel (Ni) and gold (Au) were deposited and patterned by lift-off metallization and electrodeposition. Ni, which is ferromagnetic, was incorporated to allow for remote magnetic manipulation. The gripper was designed such that only Cr and Au surfaces were exposed to render it bioinert.15 Aqueous biopolymer solutions were dispensed onto the features and patterned by exposure to ultraviolet (UV) light through a quartz photomask. The uncrosslinked biopolymer was washed away, and the sacrificial layer was dissolved to release untethered grippers.
We chose commercially processed derivatives of natural biopolymers to allow for aqueous handling: gelatin, derived from collagen, and CMC, from cellulose. The two biopolymers are targeted by different families of enzymes - without overlapping activity - so that each set can be actuated selectively. Additionally, gelatin is degraded by enzymes which occur in disease states, such as proteases in cancer.16, 17 This offers the possibility for autonomous actuation in response to a disease marker. The other biopolymer, CMC, degrades on exposure to non-mammalian enzymes that do not interact with animal tissue.
Both raw biopolymers were synthetically modified by chemical grafting of methacrylate groups to the polymer backbone18–21 (Supporting Figure S1). This modification enabled crosslinking22 under UV light in the presence of molecular crosslinkers and free-radical photoinitiators.23 The highlight of this process is that the crosslinked material retained the necessary accessible monomer groups to allow for enzymatic recognition and cleavage.
The biopolymers were patterned sequentially atop the metal layers, while underlying multilayer hinges were constructed by layering thin films with specific levels of tension. The bending angles of the hinges were designed using a multilayer mechanics model (See Supporting Information). The magnitude of the strain differential across the thickness of this multilayer stack leads to a specific bending angle at equilibrium.24–27 Controlled bending of the multilayer hinge was achieved by altering the mechanical properties of the biopolymer. A relatively stiff crosslinked biopolymer on top of a pre-stressed bilayer stack arrested its bending, causing it to remain flat. Removal or softening of this biopolymer allowed the hinge to bend (Figure 2).
Figure 2.
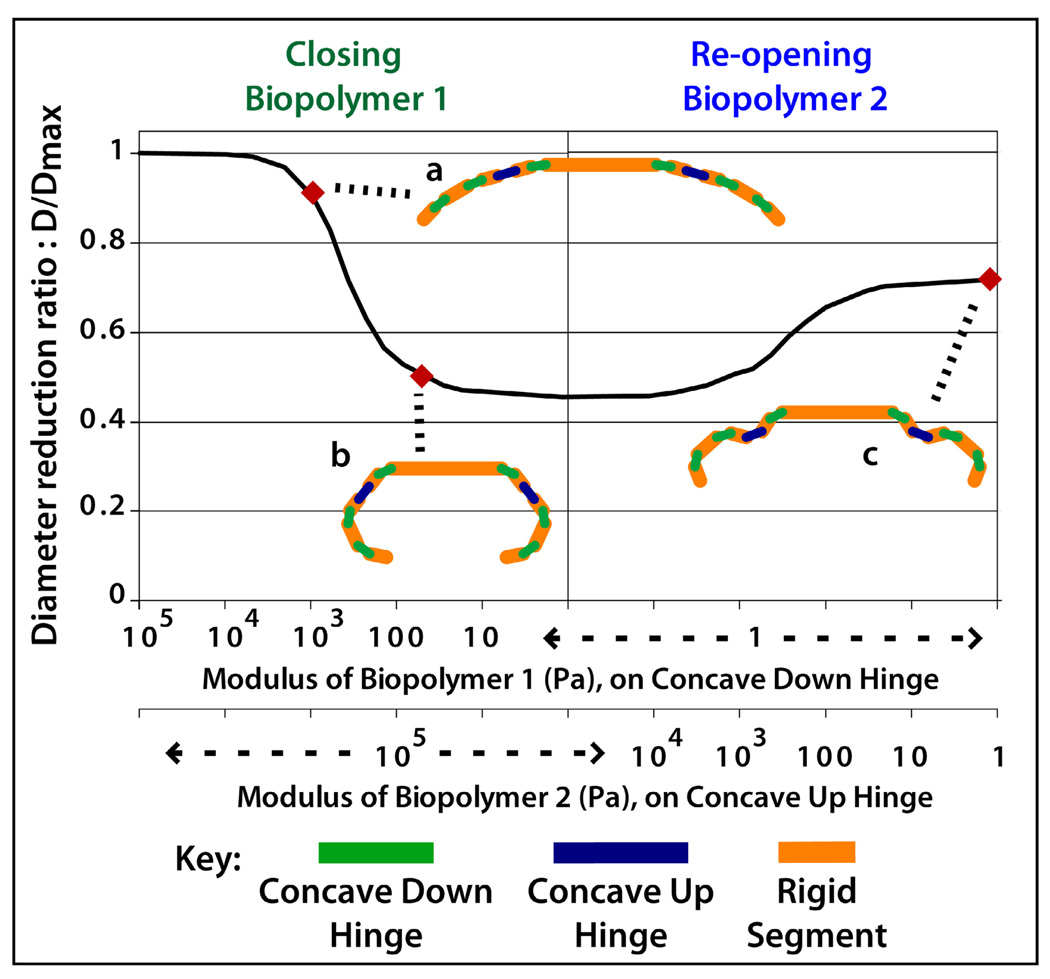
Mathematical modeling of the actuation of grippers as a function of moduli of biopolymers. (a) In the initial state both polymers are stiff (moduli higher than 104 Pa) and the entire gripper is flat. (b) When the first biopolymer is degraded and its modulus decreases to approximately 100 Pa, the first set of six hinges bends causing the gripper to close. (c) The remaining 2 hinges bend when the modulus of the second biopolymer trigger is decreased.
Using the model, we determined that for a typical gel modulus of 104 Pa, a 150 µm thick patterned gel would be sufficient to ensure a flat state28, which was verified experimentally. In order to create a miniaturized integrated tool we used a computer simulation to model serial linkages of rigid segments and various hinge types as a 2D cross section, so that the folding state of any tool could be visualized in silico before experiments. We first simulated grippers with a single set of hinges that simply closed on actuation.
Using this model, we observed that a reduction in modulus to 100 Pa would cause the gripper to close, and observed a similar actuation profile in experiments (Figure 3). We then modeled grippers with two sets of hinges that closed and re-opened. Closing occurred when the modulus of the first biopolymer (CMC) was reduced to 100 Pa while the second biopolymer (gelatin) remained stiff (104 Pa) (Figure 2a–e). Re-opening occurred when the modulus of gelatin was reduced in turn. During the actuation of the gelatin-triggered hinge, the modulus of CMC remained low and was not affected.
Figure 3.
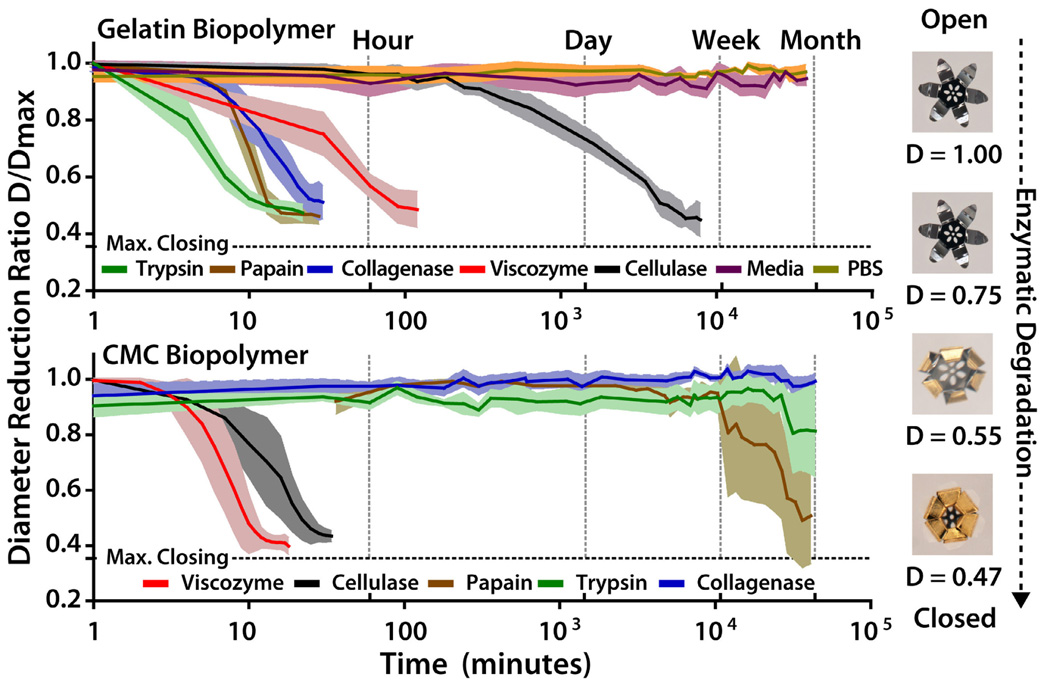
Kinetics of enzymatic triggering. Plots of the experimentally measured diameter reduction ratio (D/Dmax) versus time on exposure to a variety of enzymes. The line denotes the average value measured over 5 trials and the shaded region denotes the standard deviation. The curves demonstrate sensitivity to enzyme class. The grippers that use a gelatin based trigger actuated in less than 30 minutes in trypsin, papain and collagenase, actuated in less than an hour in viscozyme, actuated in less than a week by cellulase, and did not actuate in PBS or cell culture media with serum after several months. The grippers with a cellulase biopolymer layer are actuated in less than ten minutes in viscozyme and 1 hour in cellulase. There is some actuation within a month in papain and trypsin; in over a month there is no significant actuation in collagenase, PBS, or media. (PBS and media results are included in the Supporting Figure S5)
Selectivity of actuation was studied using grippers with either a gelatin or a CMC trigger. Several enzymes were screened (See Supporting Information): proteases from animal pancreatic origin (trypsin), plant origin (papain), and bacterial origin (collagenase), which are specific to the polypeptide (gelatin) grippers, and carbohydrate degrading enzymes from fungal origin (cellulase) that are specific to the polysaccharide (CMC) trigger. We also tested a commercial mixture of many enzymes used for plant cell wall lysis (Viscozyme), as well as phosphate buffered saline (PBS) and cell culture media with serum. For these experiments, special care was taken to ensure that all grippers were fabricated simultaneously on a single wafer and differed only in the application of gelatin or CMC hinge triggers as a final step.
In Figure 3, we show the response of the tools to different hydrolytic enzymes. Using optical microscopy, the diameter (D) of grippers exposed to different enzymes was recorded over time. We utilized a parameter, D/Dmax, which allowed us to quantify closing and re-opening in various enzymes, and plot average data for 5 grippers. Open grippers at maximum spread have a defined edge-to-edge ratio D/Dmax of 1, and in this specific design, completely closed grippers have a theoretical minimum ratio of 0.355. As an example, we experimentally observed that polypeptide grippers in trypsin fold to 90% of maximum (D/Dmax = 0.42) in approximately 10 minutes.
We observed that none of the grippers closed in the presence of PBS or mammalian cell media over 60 days. We observed a selectivity of 1:189 or greater for all individual enzymes screened (trypsin, cellulase, collagenase, papain). Enzyme-gripper selectivity was gauged by comparing time to achieve a diameter reduction ratio of 0.6. For example, cellulase closed CMC microgrippers in 18 minutes, and gelatin microgrippers in 3400 minutes (two days), a ratio of 1:189. This ratio of over two orders of magnitude demonstrates high enzyme-substrate specificity in microgripper actuation. Both CMC and gelatin grippers closed in the presence of Viscozyme, which contains a mixture of enzymes, with CMC grippers closing more quickly due to higher activity of carbohydredases in Viscozyme.
Sensitivity of the grippers to decreasing concentrations of enzyme activity was also examined. We took serial dilutions of collagenase covering three orders of magnitude (5400 Units/mL to 21 Units/mL) and incubated UV sterilized gelatin grippers in the solutions. While the grippers in the highest concentrations closed in minutes, they took several weeks to close in the lowest enzyme concentrations. Similarly, CMC grippers exposed to a 2250 Unit/mL cellulase solution closed in under 5 minutes as compared to approximately 18 hours in a 27 Unit/mL dilution. Decreased enzyme activity is expected to increase closing time as both bond cleavage and diffusion will be reduced. However, the ability to actuate in varying enzyme concentrations suggests possible activity in vivo.
We note that the gripper actuation was sensitive to the appropriate enzyme class for rapid actuation. This allowed a time window for differential actuation of orthogonal enzymes. We demonstrated actuation of hinges in series by placing the grippers in solutions of papain and then cellulase, or vice versa. We observed orthogonal actuation of specific grippers in only the corresponding enzymes (Supporting Figure S3).
We used the enzymatically triggered grippers to demonstrate medically relevant tasks. It is difficult to reach closed lumina in the body, such as the biliary tree, with tethered tools. One alternative is to manipulate untethered devices using magnetic forces, also permitting visualization via magnetic resonance imaging.29 CMC-gelatin grippers that could be closed and reopened were used to securely grip a 700 µm alginate bead (Figure 4a–b). Closing was actuated using a cellulase trigger. We were then able to move the gripper with the bead securely in its grasp using a magnetic stylus and subsequently release the bead using a collagenase trigger. This demonstration highlights possible applicability in pick-and-place operations and on-demand drug delivery.7
Figure 4.
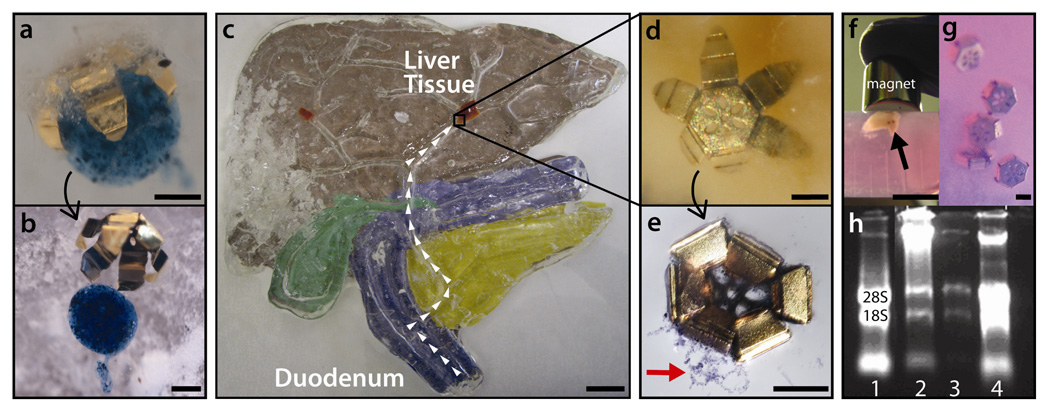
Pick-and-place, external biopsy in a model of the human biliary system, and RNA retrieval from cells after capture. (a–e) Procedures were conducted in an acrylic fluidic channel model representation of the human biliary tree. The common bile duct of the model was 5 mm, which approximates adult human size. (a–b) We used a gripper that closed in response to cellulase to retrieve a 700 µm alginate bead outside the model. After placing the gripper in the duodenum we guided it deep into the liver with a magnetic stylus. (b) We then added collagenase and agitated to release the bead. (d–e) In a separate experiment, a gripper with a CMC biopolymer trigger was placed in the duodenum and magnetically guided (arrows) through the bile ducts to a piece of liver tissue. (d) After insertion of cellulase, the gripper actuated in minutes and clamped on tissue. Magnetic manipulation both generated sufficient force to biopsy tissue and subsequently guide the loaded gripper out for extraction. (e) After staining with trypan blue, retrieved tissue was visible (arrow). (f–h) In experiments performed in round bottomed tubes, HuCCT1 cholangiocarcinoma cells, (f), and H69 bile duct cells, (g), were cultured and formed into pellets by centrifugation. After grippers were deposited and guided to the H69 cell mass, cellulase was added and the grippers burrowed into the tissue under magnetic force. (f) Less than 10 grippers could lift a mass of cells and guide it out of the vial (arrow). (H) RNA was extracted from both samples and is shown in a gel: Lane 1 is a control from H69 cells, Lanes 2 and 3 are two samples from H69 cell retrievals and Lane 4 is from a HuCCT1 cell retrieval. Scale bars represent 200 µm (a,b,d,e,g), 2 cm (c) and 1 cm (f). Larger views of (d,f,g) are shown in Figure S7 in the supporting information.
Avian liver tissue was biopsied from a model organ (cast from acrylic resin) with size scales approximately that of an adult human (Figure 4c). For this experiment we utilized CMC grippers that closed in response to cellulase. After placing the grippers in the duodenum, we remotely piloted them through the ampulla of Vater, through the common bile duct (5mm diameter lumen30), and into the liver. Cellulase solution was then added via syringe and the gripper closed around the tissue. Magnetic manipulation was used to extract the gripper and excised tissue, which was then stained (Figure 4c–e).
Additionally, the ability of these tools to retrieve cells for further diagnostic analysis was tested. We cultured a line of normal, SV-40 transformed, bile duct cells (H69) and a line of cholangiocarcinoma cells (HuCCT1). After collecting the cells in a pellet we introduced CMC triggered grippers and closed them with cellulase (Figure 4f–g). The force exerted by an external magnet used to move the grippers was sufficient to hold up a large (9 mm) cell clump with eight grippers. RNA was extracted from the cells retrieved with the grippers. The quality of the RNA, and by extension, the integrity of the cells that were retrieved, was verified via running extracts on a denaturing agarose gel (Figure 4h). We also included an RNA positive control (lane 1) obtained from H69 cells that were not retrieved with the grippers. The similarity of the 28S and 18S bands in the control and gripper retrievals demonstrates that this technique enables the collection of high quality RNA and clinically relevant data.
In conclusion, we have demonstrated grippers triggered by specific enzyme substrate interactions. These hybrid metal/polymer tools are a step toward the creation of miniaturized devices and materials that respond autonomously to specific biochemicals and disease markers. For example, by matching the biopolymer to proteolytic enzymes that are naturally secreted from cancer cells it should be possible to facilitate a tool that responds only to cancerous environments.31 It should also be possible to delay actuation by crosslinking protease inhibitors32 into the biopolymer or accelerate it using protease zymogens such as trypsinogen.33 Our methodology may be extended to other enzyme-biopolymer pairs such as the degradation of DNA based biopolymers34–36 using nucleases. In principle, the process is also compatible with nanoscale patterning techniques such as electron beam or direct write techniques37, 38 which suggests the possibility of further miniaturization.
Supplementary Material
Acknowledgements
The authors thank George M. Stern and Nana Atuobi for valuable discussions. This work was supported by the NIH Director's New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD004346-01 and DP2-OD004346-01S1, and the Medical Scientist Training Program. Information about the NIH Roadmap can be found at http://nihroadmap.nih.gov. We also acknowledge support from the Camille-Dreyfus and Beckman Foundations.
Footnotes
Supporting Information Available: Information about methods, degradation & kinetics experiments, details of the in vitro simulated liver biopsy, cancer cell biopsy & RNA extraction, imaging, and thin film modeling. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pfeifer R, Lungarella M, Iida F. Science. 2007;318:1088–1093. doi: 10.1126/science.1145803. [DOI] [PubMed] [Google Scholar]
- 2.Madden JD. Science. 2007;318:1094–1097. doi: 10.1126/science.1146351. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes R, Gracias DH. Materials Today. 2009;12:14–20. [Google Scholar]
- 4.Jager EWH, Smela E, Inganäs O. Science. 2000;290:1540–1545. doi: 10.1126/science.290.5496.1540. [DOI] [PubMed] [Google Scholar]
- 5.Aliev AE, Oh J, Kozlov ME, Kuznetsov AA, Fang S, Fonseca AF, Ovalle R, Lima MD, Haque MH, Gartstein YN, Zhang M, Zakhidov AA, Baughman RH. Science. 2009;323:1575–1578. doi: 10.1126/science.1168312. [DOI] [PubMed] [Google Scholar]
- 6.Leong TG, Randall CL, Benson BR, Bassik N, Stern GM, Gracias DH. Proc. Natl. Acad. Sci. U. S. A. 2009;106:703–708. doi: 10.1073/pnas.0807698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phua K, Leong KW. Nanomedicine (Lond) 2010;5:161–163. doi: 10.2217/nnm.09.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi J, Schmidt JJ, Montemagno CD. Nat. Mater. 2005;4:180–184. doi: 10.1038/nmat1308. [DOI] [PubMed] [Google Scholar]
- 9.Seo W, Phillips ST. J. Am. Chem. Soc. 2010;132:9234–9235. doi: 10.1021/ja104420k. [DOI] [PubMed] [Google Scholar]
- 10.Paxton WF, Sundararajan S, Mallouk TE, Sen A. Angew. Chem. Int. Ed Engl. 2006;45:5420–5429. doi: 10.1002/anie.200600060. [DOI] [PubMed] [Google Scholar]
- 11.De Volder M, Reynaerts D. J. Micromech. Microeng. 2010;20:43001. [Google Scholar]
- 12.Shahinpoor M, Kim KJ. Smart Mater. Struct. 2005;14:197–214. [Google Scholar]
- 13.Bassik N, Stern GM, Gracias DH. Appl. Phys. Lett. 2009;95:91901. doi: 10.1063/1.3212896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamal M, Bassik N, Cho J, Randall CL, Gracias DH. Biomaterials. 2010;31:1683–1690. doi: 10.1016/j.biomaterials.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merchant B. Biologicals. 1998;26:49–59. doi: 10.1006/biol.1997.0123. [DOI] [PubMed] [Google Scholar]
- 16.Koblinski JE, Ahram M, Sloane BF. Clin. Chim. Acta. 2000;291:113–135. doi: 10.1016/s0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 17.Zucker S, Wieman JM, Lysik RM, Wilkie D, Ramamurthy NS, Golub LM, Lane B. Cancer Res. 1987;47:1608–1614. [PubMed] [Google Scholar]
- 18.Peng HT, Martineau L, Shek PN. J. Mater. Sci. Mater. Med. 2008;19:997–1007. doi: 10.1007/s10856-007-0167-5. [DOI] [PubMed] [Google Scholar]
- 19.Reis AV, Fajardo AR, Schuquel IT, Guilherme MR, Vidotti GJ, Rubira AF, Muniz EC. J. Org. Chem. 2009;74:3750–3757. doi: 10.1021/jo900033c. [DOI] [PubMed] [Google Scholar]
- 20.van Dijk-Wolthuis WNE, Franssen O, Talsma H, van Steenbergen MJ, Kettenes-van den Bosch JJ, Hennink WE. Macromolecules. 1995;28:6317–6322. [Google Scholar]
- 21.Baier Leach J, Bivens KA, Patrick CW, Jr, Schmidt CE. Biotechnol. Bioeng. 2003;82:578–589. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 22.Lin OH, Kumar RN, Rozman HD, Noor MAM. Carbohydr. Polym. 2005;59:57–69. [Google Scholar]
- 23.Cha C, Kohman RH, Kong H. Adv. Funct. Mater. 2009;19:3056–3062. [Google Scholar]
- 24.Nikishkov GP. J. Appl. Phys. 2003;94:5333. [Google Scholar]
- 25.Arora WJ, Nichol AJ, Smith HI, Barbastathis G. Appl. Phys. Lett. 2006;88:053108. [Google Scholar]
- 26.Abermann R, Martinz HP. Thin Solid Films. 1984;115:185–194. [Google Scholar]
- 27.Hoffman RW, Daniels RD, Crittenden EC. Proc. Phys. Soc. B. 1954;67:497–500. [Google Scholar]
- 28.Henderson GVS, Jr, Campbell DO, Kuzmicz V, Sperling LH. J. Chem. Educ. 1985;62:269–272. [Google Scholar]
- 29.Gimi B, Artemov D, Leong T, Gracias DH, Gilson W, Stuber M, Bhujwalla ZM. Cell Transplant. 2007;16:403–408. doi: 10.3727/000000007783464803. [DOI] [PubMed] [Google Scholar]
- 30.Avisse C, Flament JB, Delattre JF. Surg. Clin. North Am. 2000;80:201–212. doi: 10.1016/s0039-6109(05)70402-3. [DOI] [PubMed] [Google Scholar]
- 31.Tutton MG, George ML, Eccles SA, Burton S, Swift RI, Abulafi AM. Int. J. Cancer. 2003;107:541–550. doi: 10.1002/ijc.11436. [DOI] [PubMed] [Google Scholar]
- 32.Brew K, Dinakarpandian D, Nagase H. Biochim. Biophys. Acta. 2000;1477:267–283. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- 33.Neurath H, Walsh KA. Proc. Natl. Acad. Sci. U. S. A. 1976;73:3825–3832. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Um SH, Lee JB, Park N, Kwon SY, Umbach CC, Luo D. Nat. Mater. 2006;5:797–801. doi: 10.1038/nmat1741. [DOI] [PubMed] [Google Scholar]
- 35.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 36.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 37.Jhaveri SJ, McMullen JD, Sijbesma R, Tan LS, Zipfel W, Ober CK. Chem. Mater. 2009;21:2003–2006. doi: 10.1021/cm803174e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherzer T, Beckert A, Langguth H, Rummel S, Mehnert R. J. Appl. Polym. Sci. 1997;63:1303–1312. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.