Abstract
Genome-wide RNA interference (RNAi) screens have identified near-complete sets of genes involved in cellular processes. However, this methodology has not yet been used to study complex developmental processes in a tissue-specific manner. Here we report the use of a library of Drosophila strains expressing inducible hairpin RNAi constructs to study the Notch signalling pathway during external sensory organ development. We assigned putative loss-of-function phenotypes to 21.2% of the protein-coding Drosophila genes. Using secondary assays, we identified 6 new genes involved in asymmetric cell division and 23 novel genes regulating the Notch signalling pathway. By integrating our phenotypic results with protein interaction data, we constructed a genome-wide, functionally validated interaction network governing Notch signalling and asymmetric cell division. We used clustering algorithms to identify nuclear import pathways and the COP9 signallosome as Notch regulators. Our results show that complex developmental processes can be analysed on a genome-wide level and provide a unique resource for functional annotation of the Drosophila genome.
Genome-wide RNAi screens have been performed in cultured cells1-4 or by ubiquitous gene silencing in worms5,6 or planarians7. To study complicated developmental processes, however, genes need to be inactivated in a tissue-specific manner in intact animals. This has become possible through the creation of a transgenic RNAi library targeting 88% of the Drosophila8 protein-coding genes. To test the feasibility of this new approach, we focused on the Notch pathway, one of the most important regulators of development9,10. Notch is activated by binding to its ligands Delta or Serrate. After ligand binding, Notch is cleaved by Presenilin and the intracellular domain acts in the nucleus as a transcriptional co-activator. Several important Notch regulators were identified due to their characteristic phenotypes in Drosophila external sensory organs11,12. Adult external sensory organs are formed within the notum (dorsal thorax) during larval and pupal stages of Drosophila development (Fig. 1a). Within a group of epithelial cells called the proneural cluster, a single sensory organ precursor (SOP) is selected in a Notch-dependent process termed lateral inhibition. The SOP cell divides into pIIa and pIIb (Fig. 1a). While pIIa forms the two outer cells (the hair and socket), pIIb gives rise to an apoptotic glial cell and then to the two inner cells (the neuron and sheath) of the organ. During each division, the protein determinants Numb and Neuralized segregate into one of the two daughter cells where Numb inhibits the Notch receptor while Neuralized activates the Notch ligand Delta10,13. The resulting differences in Notch signalling establish unequal fates in the two daughter cells. Thus, Notch acts both in lateral inhibition and in asymmetric cell division (ACD) during external sensory organ development. Although the core components of the Notch pathway act in both processes, other components act specifically during lateral inhibition (E(spl)14) or ACD (Numb15, Sanpodo16,17).
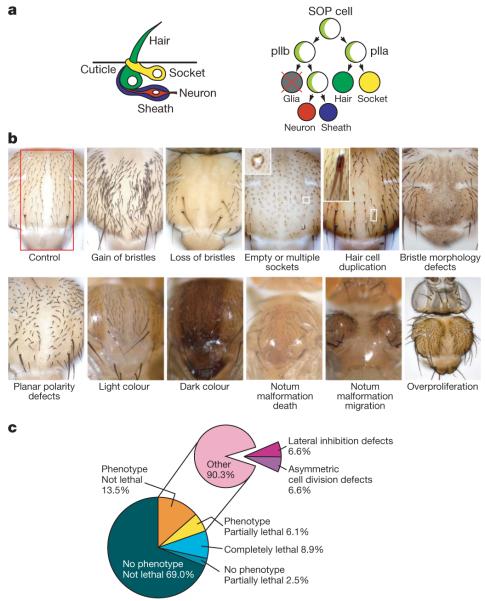
Figure 1. Phenotypic categories and screen results.
a, Schematic view of external sensory organ morphology and lineage (Numb and Neuralized, light green). b, Typical examples of phenotypic categories. The pannier-GAL4 expression area is indicated (red) in the control panel. Examples are Delta (gain of bristles), O-fut1 (loss of bristles), numb (empty or multiple sockets), bazooka (hair cell duplication), CG31961 (bristle morphology defects), starry night (planar polarity defects), CG18095 (light colour), CG34331 (dark colour), CG5462 (notum malformation death), CG7904 (notum malformation migration) and mob as tumor suppressor (overproliferation). Original magnification, ×10 (except overproliferation, ×5). c, Relative frequency of phenotypes in the entire primary genome-wide screen (left) and among visible phenotypes (right).
Genome-wide RNAi screen
Hairpin constructs in the RNAi library are expressed under UAS/GAL4 control18. We tested scabrous-GAL418, pannier (pnr)-GAL419 and fzIII-GAL4 (also known as MS248-GAL4 or P{GawB}MS248)20 using a set of 40 RNAi lines targeting 21 genes involved in external sensory organ development (Supplementary Table 1). Consistently, phenotypes were stronger and lethality lower with pnr-GAL4, and this line was selected for large-scale analysis.
A total of 20,262 transgenic RNAi lines were screened; these are predicted to target 11,619 of the 14,139 protein-coding genes (82.2%) in release 5.7 of the Drosophila genome21. Ten flies each were analysed and phenotypic abnormalities were recorded in a database (http://bristlescreen.imba.oeaw.ac.at). Because pnr-GAL4 is only expressed in a central region of the notum (Fig. 1b), lateral areas were unaffected and served as internal controls. Phenotypes were described using controlled vocabulary (Fig. 1b, Supplementary Table 2). Phenotypic strength (Px) was expressed on a scale of 0 (not affected) to 10 (completely affected) as the fraction of the pnr-GAL4 expression area (or the fraction of external sensory organs present in that area) that displayed the respective phenotype. External sensory organ phenotypes were classified into ‘gain’ (PGain), ‘loss’ (PLoss), ‘empty or multiple sockets’ (PSockets), ‘hair cell duplication’ (PDuplication) and ‘bristle morphology defects’ (PBMorph). In addition, orientation defects were recorded as ‘planar polarity defects’ (PPolarity). Morphological notum defects were classified into ‘notum malformation death’ (PMDeath; loss of tissue), ‘notum malformation migration’ (PMMigration; thorax closure defects), ‘overproliferation’ (POverprolif; notum enlargement) and ‘colour’ (PColour). GO term analysis showed that phenotype groups are significantly enriched for genes known to act in appropriate processes (Supplementary Table 3). The group of 211 genes for which POverprolif is >0 contains tumour suppressors such as Pten and 5 of the 7 Hippo pathway members represented in the library. The group of 71 genes with PPolarity of ≥4 contains four of the six core planar cell polarity (PCP) components, and four other PCP effectors. Visible phenotypes were scored for 19.6% of all RNAi lines (3,973 lines, 3,004 genes). A total of 1,803 lines (8.9% of all lines, 1,339 genes) were lethal whereas 496 lines (2.5% of all lines, 475 genes) were semi-lethal with no associated morphological defects. Sixty-nine per cent of lines (13,990 lines, 8,681 genes) gave no phenotype (Fig. 1c). Thus, our screen has assigned potential loss-of-function phenotypes to 21.2% (3,004 of 14,139) of the protein-coding genes in the Drosophila genome, more than half of which (1,597 genes, 2,067 lines) had not been characterized before.
Quality control
To determine false-negative rates, we assembled a list of genes required for Notch signalling and/or ACD based on Flybase and more recent published information. Forty-two of the 50 selected genes are represented in the RNAi library (Supplementary Table 4) and 24 gave the expected phenotype. Eight genes could not be analysed owing to early lethality (six genes) or strong notum malformations (cdc2 and bre1). The resulting false-negative rate of 29.4% (10 of 34 genes) is in good agreement with previously reported data for transgenic RNAi8.
Long double-stranded RNA hairpins are known to cause off-target effects22. Nonspecific phenotypes can also be generated by overexpression of genes near the transgene insertion site. We therefore generated a second hairpin construct or obtained a second RNAi line from the NIG-FLY stock centre for 73 genes (136 lines) that had resulted in visible phenotypes (Supplementary Table 5). Similar or identical phenotypes were observed for 63.0% of the genes (46 genes, 82 lines). Interestingly, however, we could confirm 88% (22 of 25 genes) of those phenotypes that were supported by more than one line in the library, although most of these lines are independent insertions of the same transgene. We also selected 43 non-essential genes (72 lines) with known adult phenotypes (or the absence thereof; Supplementary Table 6). For 93% of these (40 genes, 67 lines), our screen identified the correct phenotype. In three cases, the observed phenotypes were different from what has been described, but could be explained by the described cellular function (endocytosis for orange, Ras activity for Neurofibromin 1, and cAMP signalling for dunce) if RNAi causes phenotypes stronger than the described alleles. In any case, these data indicate that the false-positive rate of our screen is less than 7%.
Phenotypic categories
Notch acts twice during external sensory organ development23. During lateral inhibition, loss and gain of Notch signalling increases and decreases the number of external sensory organs, respectively. During ACD, increased Notch signalling causes hair duplications or multiple sockets, whereas decreased Notch signalling causes all daughter cells to assume an internal fate and results in the apparent absence of external sensory organs. Thus, Notch regulators can be in the loss, gain, socket and duplication categories. We therefore used the phenotypes of genes known to be involved in Notch signalling and ACD (data not shown) to create compound categories that include all or most of these positive-control genes. The asymmetric cell division (ACD) category (PLoss ≥7 or PDuplication >0 or PSockets >0) is made up of 226 genes (262 lines) and includes bora, bazooka, Delta, musashi, Notch, numb, Presenilin, sanpodo, tramtrack and twins. The lateral inhibition category includes all genes for which PLoss is ≥7. In addition, we re-screened all images of genes where PGain is >0 and retained only those where the extra external sensory organs are grouped together, indicating the formation of multiple SOP cells from one proneural cluster. The resulting lateral inhibition category contains 233 genes (264 lines). Analysis of all genes in these categories revealed (Supplementary Table 3) that the GO terms ‘asymmetric cell division’ and ‘nervous system development’ are significantly enriched in the ACD category. The lateral inhibition category over-represents the GO terms ‘neurogenesis’, ‘Notch signalling pathway’ and ‘cell fate commitment’. We therefore used these categories in various secondary assays to identify genes acting globally in the Notch pathway or more specifically during ACD.
Identification of Notch regulators
During wing development, Notch loss of function leads to wing notching or broader wing veins24. To identify general Notch regulators, we expressed RNAi hairpins in the wing imaginal disc for 201 genes (226 lines) in the ‘lateral inhibition’ category. We analysed ten flies each for wing vein and wing margin phenotypes (Fig. 2, Supplementary Fig. 1 and Supplementary Table 7). All other wing abnormalities were also recorded and separated into phenotypic classes (Supplementary Fig. 2). In addition to seven known Notch pathway members (Notch, mind bomb 1, Delta, groucho, extra macrochaetae, Presenilin and O-fucosyltransferase 1 (O-fut1)), this secondary analysis identified 23 genes not previously implicated in Notch signalling.
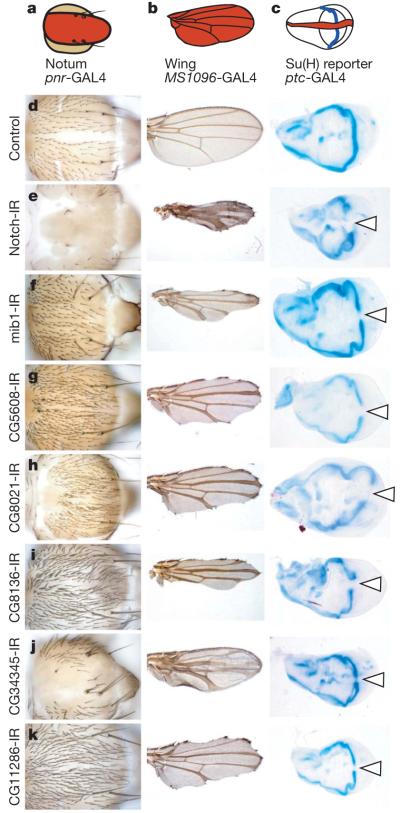
Figure 2. Secondary assays for general components of Notch signalling.
Genes with a Notch-like phenotype on the notum were re-screened for wing morphology or expression of a transcriptional Notch reporter. a–c, Schematics with GAL4 expression domains shown in red. d–k, Appropriate controls and new genes scoring positive in all assays are shown. Transformant IDs (TIDs) for each UAS-RNAi line are as follows: 1112 (Notch), 27526 (mib1), 45569 (CG5608), 23675 (CG8021), 23748 (CG8136), 20105 (CG34345) and 17541 (CG11286). Arrowheads mark loss of β-galactosidase staining. IR, inverted repeat. Original magnification, ×10 (nota, left column), ×2.5 (wings, middle column) and ×5 (wing discs, right column).
Notch signalling activates the transcription factor Suppressor of Hairless (Su(H)). The genes identified could participate in this signal transduction cascade, act in parallel to Su(H) or act as Su(H) targets. To distinguish these, we used a lacZ reporter for Su(H) activity25 (Supplementary Table 7). In the developing wing disc, this reporter is expressed along the dorso–ventral compartment boundary (Fig. 2c). When we inactivated the potential Notch regulators along the anterior–posterior compartment boundary using patched (ptc)-GAL4, 8 genes (11 lines) caused a loss of β-galactosidase staining in the area where the boundaries overlap (Fig. 2d–k). Besides Notch itself and the known Notch regulators mind bomb 1 and O-fut1, this identified five genes that function upstream of Su(H)-mediated transcriptional activation and have not previously been implicated in Notch signalling.
Notch signalling is tightly connected to endocytic trafficking13. Interestingly, one of the newly identified Notch regulators (CG5608) is homologous to VAC14, a yeast gene that controls trafficking to multivesicular bodies26. Vac14 is an upstream activator of the PtdIns(3)P 5-kinase Fab1 (also known as PIKfyve)26. Although the function of Drosophila CG5608 has not been analysed, Notch accumulates with Delta in intracellular vesicles in fab1 mutant Drosophila cells27. Further analysis of the CG5608 RNAi phenotype might help to understand the connection between phosphoinositide synthesis and Notch signalling. This is particularly interesting because Vac14 loss results in neurodegeneration in mice28, and multiple lines of evidence have connected the Notch pathway to neurodegeneration29 before.
Genes regulating asymmetric cell division
Loss of external sensory organs can be caused either by increased Notch signalling during lateral inhibition or by reduced Notch signalling during ACD23 (Fig. 3a). To resolve this complication, we assayed the presence of SOP cells by expressing green fluorescent protein (GFP) from the phyllopod (phyl) promoter (Fig. 3b). phyl is expressed in the proneural cluster and then restricted to the SOP30. GFP was fused to the asymmetric localization domain of Partner of numb (Pon) so that the construct can also be used to analyse asymmetric protein segregation. We imaged at least 3 pupae each for 80 genes (91 lines) in the ACD category (Fig. 3b, Supplementary Fig. 3 and Supplementary Table 8). Thirty-three genes (33 lines) caused a complete loss of SOPs within the pnr expression domain whereas 6 genes (8 lines) caused a partial SOP loss. Eight genes (13 lines) increased SOP number, indicating that they are required for lateral inhibition. Importantly, 7 of these genes were tested with MS1096-GAL4 (also known as P{GawB}BxMS1096), and 5 caused Notch loss-of-function wing phenotypes (Supplementary Table 7), demonstrating that they are general regulators of Notch signalling. Finally, we identified 28 genes (31 lines) that may have a role in ACD because SOP cells were normal but no differentiated external sensory organs developed.
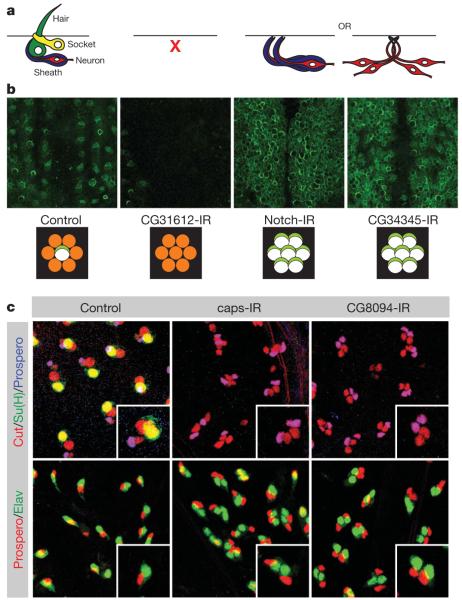
Figure 3. Secondary assays for asymmetric cell division genes.
a, Loss of bristle phenotypes can be caused by defects in SOP specification or survival, or by cell fate transformations. b, Decreases (CG31612) and increases (Notch, CG34345) in SOP cell number were observed. Schematics below each image show interpretations for the number of SOP cells (white with light green crescent) formed from proneural clusters (orange). c, Antibody lineage staining identified cell fate transformation phenotypes. All SOP daughter cells are marked by Cut, socket cells by Su(H), sheath cells by Prospero and neurons by Elav. TIDs are 27228 (Notch), 20105 (CG34345), 30545 (CG31612), 3046 (caps) and 35338 (CG8094). Original magnification, ×40.
To distinguish cell fate transformations from differentiation defects, we analysed 23 of these genes (26 lines) using cell-type-specific markers (Fig. 3c and Supplementary Table 9). Clear transformations of outer into inner cells were observed for six genes (six lines) whereas the others showed either normal lineages or abnormalities that can not be explained by cell fate transformations. Taken together with the 128 additional genes (146 lines) for which loss of function caused PSockets of >0 or PDuplication of >0, we have identified 134 regulators of ACD.
One of these is capricious (caps), a transmembrane protein implicated in cell–cell interactions during compartment boundary formation31,32 and neuronal targeting33. On caps RNAi, SOP cells generate two pIIb cells, a phenotype typically observed for genes involved in Notch activation (Fig. 3c). Indeed, capsc18fs mutant clones show phenotypes indicating a loss of Notch signalling (Supplementary Fig. 4), suggesting that Caps might be involved in generating cell adhesion between signal-sending and -receiving cells during Notch/Delta signalling.
A genome-wide network for Notch signalling
Genes or proteins in a signalling pathway often interact genetically or physically. Several attempts have been made to map such interactions on a genome-wide level34-36, but these lack functional validation. To create a functionally validated map for Notch signalling, we first combined all genes in the ‘lateral inhibition’ or ‘asymmetric cell division’ categories and added genes previously implicated in Notch signalling that had not scored as positive in our screen. For each gene we searched for yeast two-hybrid, biochemical, genetic and other interaction data from various databases (see Methods). We only maintained interactions with partners also contained in our candidate set and removed all genes without interaction partners. The resulting gene network (http://bristlescreen.imba.oeaw.ac.at) includes 780 interactions between 177 genes, only 42 of which were previously known to act in Notch signalling during external sensory organ development (Fig. 4 and Supplementary Fig. 5).
Figure 4. An interaction network for Notch signalling.
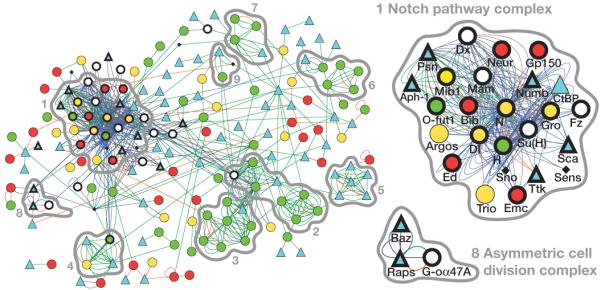
‘Lateral inhibition’ and ‘asymmetric cell division’ category genes with interactions are displayed as a network map. Genes are shown as nodes (node shape/colour denotes phenotype: gain, red circle; loss, green circle; both gain and loss, yellow circle; socket or hair duplication, blue triangle; completely lethal, black diamond; control genes with no phenotype in gain, loss, socket or duplication categories, white circle; positive control genes, bold outline), and relationships between nodes as edges (edge colour denotes interaction type: genetic, blue; interolog, green; yeast two-hybrid, orange; other, grey). MCODE-identified complexes are outlined in grey and rank is indicated. Enlargements of positive control complexes for the Notch pathway and asymmetric cell division are shown.
To identify putative complexes, we used the Molecular Complex Detection (MCODE) and found nine clusters of highly interconnected nodes (Fig. 4). In protein–protein interaction networks, such clusters represent protein complexes and/or genetic pathways. Complex one is centred on Notch and Delta. It includes Notch-modifying enzymes, such as O-fucosyltransferase, and proteins in the Notch signalling pathway, such as the Presenilin complex and the nuclear protein Hairless. It also includes the guanine nucleotide exchange factor protein Trio, which has been shown to interact with Notch37 but had not been functionally implicated in Notch signalling before. A second complex includes the known ACD regulators G-oα47A, Rapsynoid (also known as Pins) and Bazooka. Thus, network analysis of genetic data can successfully predict pathways that mediate even complex multicellular processes.
To validate the predictive capacity of our network, we tested complexes in which most members were not known to regulate Notch signalling before. Complex nine contains the importin α protein Karyopherin α3 (Kap-α3) as well as the importin α export receptor Cas (Fig. 5a). Cas mutants were previously shown to cause ACD defects38.In Kap-α3 mutant clones (Fig. 5b, c), we observed strongly penetrant cell fate transformations similar to Cas. Neighbouring genes to complex nine include the nuclear pore components Nup358 and Nup50 (CG2158) (Fig. 5a). In all cases, RNAi causes cell fate transformations characteristic of increased Notch activity suggesting that nuclear import of an inhibitory component is rate limiting for the Notch pathway. Such a component has been postulated before38 and the identification of the critical importin-α might facilitate its biochemical identification. Because the Kap-α3–Cas complex also contains nuclear Lamin (Fig. 5a), our data might provide explanations for certain aspects of laminopathies. Interestingly, deregulated Notch signalling has previously been suggested to be crucially involved in certain laminopathies based on their stem cell phenotype39.
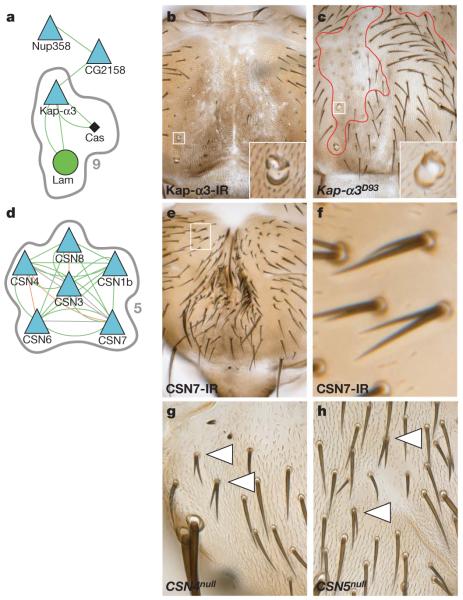
Figure 5. Nuclear import and the COP9 signallosome regulate Notch signalling and asymmetric cell division.
a, MCODE complex nine contains Cas, Kap-α3 and Lam (grey outline). Selected neighbouring nodes from the main interaction network have been included. b, c, Both Kap-α3 RNAi and Kap-α3 mutant clones show multiple socket phenotypes. d, MCODE complex five shows nodes for the COP9 signallosome. e–h, Knockdown of CSN7 (e, close up in f) and both CSN4 (g) and CSN5 (h) mutant clones cause hair cell duplication phenotypes. Original magnification, ×10 (b, c, e) and ×20 (g, h).
The predicted complex five (Fig. 5d) contains subunits of the COP9 signallosome (CSN). RNAi of six of the seven CSN subunits screened resulted in occasional generation of two pIIa cells (1 ≤ PDuplication ≤ 3), consistent with an inhibitory function in the Notch pathway (Fig. 5e, f and Supplementary Fig. 6). The phenotypes are also observed in mitotic clones mutant for CSN4 or CSN5 (Fig. 5g, h). CSN is an isopeptidase that removes Nedd8 conjugates from Cullin-RING class E3 ubiquitin ligases and prevents their degradation40. Indeed, defects in lateral inhibition are also observed in Cullin-3 (gft) mutants41 suggesting that Cullin-3 might be the relevant CSN target during Notch signalling.
Conclusions
Our data show that genome-wide loss-of-function analysis by RNAi can be used to study complex developmental processes. From a comprehensive phenotypic database we extracted genes involved in Notch signalling by secondary screening or integration with other types of genome-wide analysis. Vac14 may regulate PtdIns(3,5)P2 synthesis and trafficking of Notch or its ligand Delta into multivesicular bodies. Kap-α3 is a cargo for Cas and might control the nuclear import of a rate limiting inhibitory signalling component. Finally, the COP9 signallosome regulates Cullin-1 (lin19) and Cullin-3 (ref. 42), which in turn could regulate ubiquitylation of Notch or essential Notch pathway members. We describe an additional 23 uncharacterized genes and 5 putative protein complexes for which the mechanistic connection to Notch signalling is less clear. Most of those act downstream or in parallel to Su(H). So far, only a few functionally important nuclear targets of Notch have been identified. Our analysis could considerably expand the list of nuclear Notch targets and provide insight into how Notch regulates the various cell fate and morphology decisions in which it has been implicated.
In addition, we report a wide range of morphological phenotypes that can be used to derive groups of genes potentially involved in planar polarity, proliferation control, cell migration and apoptosis. The possibility to perform secondary screens on those groups illustrates the enormous power of transgenic RNAi now available to study development on a genome-wide level.
METHODS SUMMARY
For genome-wide and second RNAi line screening, crosses between UAS-hairpin RNAi males and pnr-GAL4 females were raised at 22 °C and shifted to 29 °C during larval stages. Ten flies were examined visually and all morphological abnormalities were recorded. Phenotypic classes are defined in Methods.
For secondary assays of wing morphology or Notch reporter activity, MS1096-GAL4 or ptc-GAL4;Gbe+Su(H)-lacZ were used, respectively (both at 25 °C). Lineage analysis was essentially as described15 using pnr-GAL4 at 29 °C. For live imaging43, at least three UAS-RNAi;pnr-GAL4,phyllopod>>EGFP::Pon.LD pupae were imaged at 14 h APF (aged at 29 °C) on a Zeiss LSM510 confocal microscope. phyllopod>>EGFP::Pon.LD was generated by fusing enhanced GFP (EGFP) to the amino terminus of the Pon localization domain and placing it downstream of a 3.4 kb phyllopod promoter fragment30. Mutant clones were generated using the Ubx-FLP/FRT system with (Kap-α3D93, CSN4null and CSN5null clones) or without (capsc18fs clones) a cell lethal on the non-mutant chromosome arm17.
The Notch interaction network was generated by combining interaction data from the DroID, STRING44 and BioGRID45 databases. Interactions were only maintained if both interactors were contained in the ‘lateral inhibition’ or ‘asymmetric cell division’ categories or a list of previously characterized Notch regulators. The resulting network was drawn using Cytoscape46. The MCODE47 algorithm was used to identify sub-network complexes.
A list of fly stocks and antibodies used as well as further methodological details are in the Supplementary Information.
Supplementary Material
Acknowledgements
Antibodies, plasmids and fly stocks were provided by the Vienna Drosophila RNAi Center, the Bloomington Drosophila Stock Center, the National Institute of Genetics Fly Stock Center (NIG-FLY), the Developmental Studies Hybridoma Bank, C.-T. Chien, R. J. Fleming, T. Klein, R. Lehmann, A. Nose, G. S. Suh and F. Wirtz-Peitz. We thank C. Cowan for comments on the manuscript, V. Rolland for help with secondary analysis, E. Kleiner, G. Haas, S. Reiter, T. Pritz, Z. Topalovic, S. Farina Lopez, S. Wculek and Ö. Copur for technical assistance in constructing the custom second RNAi line collection, C. Gallagher for generating the Su(H) antibody, P. Pasierbek for bio-optics support, F. Stocker for graphics assistance and P. Serrano Drozdowskyj for creating the online database. M.Y. was supported by a European Union Marie Curie Mobility Fellowship. Work in J.A.K.’s laboratory is supported by the Austrian Academy of Sciences, the Wiener Wissenschafts-, Forschungs- und Technologiefonds (WWTF), the Austrian Science Fund (FWF) and the EU network ONCASYM.
METHODS
Fly strains and constructs
The following fly strains were used: pnr-GAL4 (MD237; Bloomington Drosophila Stock Center (BDSC)); sca-GAL4 (G537.4)18; fzIII-GAL4 (MS248)20,48; MS1096-GAL4 (BDSC); ptc-GAL4 (G559.1)18; Gbe+Su(H)-lacZ25 (a gift from T. Klein); FRT82B,Kap-α3D93 (a gift from R. J. Fleming); CSN4null and FRT82B,CSN5null (gifts from G. S. Suh); FRT2A,capsc18fs (a gift from A. Nose); and pnr-GAL4,phyllopod>>EGFP::Pon.LD. All RNAi lines came from the Vienna Drosophila RNAi Center (VDRC) library and their details are published8, except second RNAi lines, which were custom made (see below) or supplied by the National Institute of Genetics Fly Stock Center (NIG-FLY). VDRC UAS-driven inverted repeat (UAS-IR) construct target prediction mapping to release 5.7 of the Drosophila genome (Flybase release FB2008_04) was carried out essentially as described8, except that all genes targeted by ≥80% of 19-base polymers were considered as on-target genes. For this reason, one construct may have more than one target. When no target was identified by automated mapping efforts, constructs were annotated manually and identified as ‘manual mapping’ (Supplementary Table 2 and http://bristlescreen.imba.oeaw.ac.at). Only protein-coding gene targets were considered when calculating phenotypic rates. Previously uncharacterized genes are those identified only with CG numbers in genome release 5.7. Flies were raised on standard media at 25 °C unless otherwise stated. Note that the numbers of lines re-tested in each secondary assay do not always match those identified in the primary screen because certain lines have been replaced after the completion of the RNAi library. To generate phyllopod>> EGFP::Pon.LD flies, using standard cloning techniques, the nuclear GFP and polyA cassette from pStinger-phyl3.4-GFP (a gift from C.-T. Chien30) was removed, an SV40 polyA cassette (PCR amplified from the pUAST vector) was added, and the vector was converted to an attR-site-containing Gateway Destination vector (pStinger-phyl3.4-Gateway) where the first 14 amino acids plus gateway linkers are fused in frame to the coding sequence of interest after an LR clonase (Invitrogen) reaction to generate an expression clone. EGFP::Pon.LD49 was then PCR amplified with primers that add Gateway 5′ and 3′ attB recombination sites and three 3′ in-frame STOP codons from the template pBS-TATA-EGFP::Pon.LD-SV40 (gift from F. Wirtz-Peitz), and inserted into pDONR221 (Invitrogen) using a BP Clonase (Invitrogen) recombination reaction. An LR Clonase (Invitrogen) reaction was used to insert EGFP::Pon.LD from the entry clone into pStinger-phyl3.4-Gateway. The final vector was confirmed by sequencing before injection into w1118 embryos for the production of transgenic flies.
Second RNAi lines
We generated or obtained from the NIG-FLY stock centre second UAS-IR fly lines for 73 genes that showed a phenotype in the primary bristle screen. The second line collection of transgenic fly stocks were independently created and designed in such a way that there was a maximum of 70% overlap between the primary construct in the original library and the new IR sequence. Most IR sequences overlap the original construct by less than 50%. Primer pairs for second custom hairpin constructs were designed as previously published8, with the following additional constraints/exceptions. Two hundred primer pairs were designed for each target using Primer3 (ref. 50). Only those that were predicted to have a single primary target, an s19 score ≥0.9 and a maximum CAN count of five were chosen. Restriction sites of the preferred enzymes EcoRI and XbaI were avoided; in rare cases BglII was used as an alternative. Genomic DNA as template was preferred over cDNA. In case of no suitable primers, PCR conditions submitted to Primer3 were loosened to a product size range of 120 to 600 bp, melting temperature of 62 to 72 °C and higher maximum allowed self complementarity. Detailed primer information is provided in Supplementary Table 10. The UAS-IR constructs were cloned using published methods8 into a custom designed vector, pKC26, and then inserted into a second chromosome site (43.4) using a customized φ31 system51.
RNAi bristle screen
The pre-screen was carried out using 40 UAS-IR lines8 targeting 21 genes known to be involved in asymmetric cell division and the Notch pathway using pnr-GAL4, sca-GAL4 and fzIII-GAL4 at 18, 25 and 29 °C. The GAL4/UAS system is temperature-dependent, and higher temperatures result in stronger expression but also increase the chances of lethality. Phenotypic assessment was basically the same as that in the genome-wide RNAi bristle screen (see below).
The genome-wide screen was carried out using a collection of transgenic fly lines carrying UAS-IR constructs8. To drive expression of IRs for each gene on the notum, males from each UAS-IR fly line were crossed with pnr-GAL4 virgin females. Virgin females were obtained in large numbers using a ‘virginizer’ line where the pnr-GAL4 stock also contained an hs-hid transgene (originally from R. Lehmann) on the Y chromosome (w1118/Y, hs-hid;pnr-GAL4/TM3,Sb). Stocks were maintained at 22 °C. Once third instar larvae were observed in the stock bottles, the parents were discarded and the bottles heat-shocked at 37 °C in a water bath for one hour on two consecutive days to kill progeny carrying the hs-hid chromosome (males). A similar strategy was used to obtain appropriate GAL4 virgin females for other assays in this study. Mating to UAS-IR males and egg laying were carried out at 22 °C for two days, then the crosses were shifted to 29 °C to enhance the expression and activity of GAL4. About 14 days after mating, 10 males (or females if not enough males of the correct genotype were available, and in the case of X chromosome UAS-IR insertion lines) were screened for phenotypes on the notum. The number of flies screened for each line was sometimes less than 10 when there was associated lethality. We judged each phenotype under a normal stereo microscope (Stemi 2000, Zeiss) equipped with a digital camera (PowerShot G6, Canon), and if there was any phenotype took several representative pictures for each transgenic line. Each phenotype was scored in a semiquantitative manner based on the affected area within the pnr-GAL4 expression zone, ranging from 0 to 10 (0 means no phenotype; 10 means 90 to 100% of the area was affected by the given phenotype). Areas of the notum where pnr-GAL4 was not expressed served as internal controls for each fly. Phenotypic classes are defined as: ‘gain’ for any number of extra external sensory organs, ‘loss’ for missing external sensory organs, ‘empty or multiple sockets’ for external sensory organs consisting of one or more sockets but no hair, ‘hair cell duplication’ for duplication of hairs, with or without concomitant duplication of the socket, ‘bristle morphology defects’ for other morphological abnormalities of the external sensory-organ hair, ‘planar polarity defects’ for deviations from the posterior orientation of external sensory organs, ‘notum malformation death’ for loss or reduction of the pnr-GAL4-expressing region, ‘notum malformation migration’ for thorax closure defects where the two heminota remain separated, ‘overproliferation’ for enlargement of the pnr-GAL4-expression area and ‘colour’ for any colour difference between the central, pnr-GAL4-expression area and lateral regions. Phenotypes that do not fall into any of these categories were recorded as ‘other’. Adult fly cuticles were prepared using standard methods and mounted in Hoyer’s media. Cuticle preparation images were processed from multiple image stacks using Helicon Focus software (Helicon Soft). All phenotypic information, including stereo microscope and cuticle preparation images, was stored in a custom database. An online database with all results from the primary screen, including images for each phenotype, is available at http://bristlescreen.imba.oeaw.ac.at.
Wing RNAi screen and Su(H) reporter assay
Males for 226 UAS-IR lines with a ‘lateral inhibition’ external sensory organ phenotype were crossed with MS1096-GAL4 virgin females. At least ten wings from ten different adult flies (preferably male, but also female when necessary) were checked under a normal stereo microscope (Stemi 2000, Zeiss). If any phenotype or abnormality was found, the wings were dissected and mounted on a slide, and were observed under a histology microscope to investigate detailed phenotypes. Definitions for phenotypes recorded in the MS1096-GAL4 secondary screen are as follows: notched margin (invagination of the wing margin, excluding invaginations near ectopic vein material), wider vein (wider delta-like-vein phenotypes, considered weak if only wider near the wing margin), bent up (wings bent dorsally), bent up or lethal (wings bent dorsally or lethal (could not distinguish between CyO and non-CyO flies in the same vial)), bent down (wings bent ventrally), wrinkled (general wrinkled structure/appearance), furrow (wrinkled proximal-distal ‘furrows’ but not generally wrinkled), wrinkled margin (wrinkled wing margins, but not generally wrinkled), blistered wings (bubbles between the epithelial layers), necrotic (brown patches of necrotic-looking tissue), tufts of trichomes (multiple wing hairs arranged in clusters), supernumary trichomes (general increase in density of trichomes), ectopic bristles (ectopic structures on the wing blade resembling wing margin bristles), and size (grossly smaller or larger than control wings (subjective, not quantified)). For the lacZ reporter assay, males from UAS-IR lines that showed wing notching or delta-like vein phenotypes in the wing RNAi screen were crossed with ptc-GAL4; Gbe+Su(H)-lacZ/TM6b virgin females. Wing discs from at least five third instar (non balancer) larvae were dissected and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) solution using standard protocols and imaged by light microscopy.
Antibodies
The antibodies used in this study are as follows: mouse anti-cut (2B10, 1:500, DSHB), rat anti-Su(H) (1:100, made after ref. 52), mouse anti-Prospero (MR1A, 1:10, DSHB), rabbit anti-Prospero (1:1,000; ref. 53), mouse anti-Elav (9F8A9, 1:30, DSHB) and rat anti-Elav (7E8A10, 1:100, DSHB).
GO term enrichment
Genes with different single or combinations of phenotypes in the primary screen were grouped together and checked for significant enrichment of associated GO terms. Data were analysed using GOstat (T. Beissbarth, http://gostat.wehi.edu.au/cgi-bin/goStat.pl), searching against the Flybase database of GO and corrected for multiple testing using the False Discovery Rate (Benjamini correct method).
Orthology prediction
All orthology predictions for candidate genes were made using InParanoid (http://inparanoid.sbc.su.se/cgi-bin/index.cgi)54.
Interaction map
Publicly available interaction data were integrated to obtain a unified and more complete Drosophila interactome network with which to work. The data resources used were DroID (www.droidb.org) (version 2 and version 3), STRING44 (version 7.0 and version 7.1) and BioGRID45 (version 2.0.40). The inclusion of redundant information from different data sets was avoided by using the PubMed identifying number, where available, as a unique tag to describe the interaction between two entities. Genetic and interolog links were only imported from one data source per entity pair. STRING text mining data were only included if there was at least one other evidence type to support the suggested interaction. Finally, interactions were included into further consideration only if both interactors belonged to the set of ‘lateral inhibition’ or ‘asymmetric cell fate’ or were contained in a list of previously characterized Notch regulators. The resulting network was drawn using Cytoscape46 (http://www.cytoscape.org). Socket and duplication phenotypes are dominant over loss and gain phenotypes for determining node shape. The MCODE47 algorithm was used to identify subnetwork complexes using default parameters (loops excluded, haircut selected, fluff not selected, K-Core value 2 and maximum depth 100), except for ‘node score cutoff’, which was changed to 0.5. Detailed files for import into Cytoscape are available (http://bristlescreen.imba.oeaw.ac.at).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Kiger AA, et al. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 3.Bettencourt-Dias M, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 4.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 6.Gonczy P, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 7.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 9.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 10.Schweisguth F. Notch signaling activity. Curr. Biol. 2004;14:R129–R138. [PubMed] [Google Scholar]
- 11.Bang AG, Hartenstein V, Posakony JW. Hairless is required for the development of adult sensory organ precursor cells in Drosophila. Development. 1991;111:89–104. doi: 10.1242/dev.111.1.89. [DOI] [PubMed] [Google Scholar]
- 12.Schweisguth F, Posakony JW. Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994;120:1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- 13.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- 14.Nagel AC, Maier D, Preiss A. Su(H)-independent activity of hairless during mechano-sensory organ formation in Drosophila. Mech. Dev. 2000;94:3–12. doi: 10.1016/s0925-4773(00)00319-1. [DOI] [PubMed] [Google Scholar]
- 15.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 16.O’Connor-Giles KM, Skeath JB. Numb inhibits membrane localization of sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell. 2003;5:231–243. doi: 10.1016/s1534-5807(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 17.Hutterer A, Knoblich JA. Numb and α-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 2005;6:836–842. doi: 10.1038/sj.embor.7400500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 19.Calleja M, et al. Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development. 2000;127:3971–3980. doi: 10.1242/dev.127.18.3971. [DOI] [PubMed] [Google Scholar]
- 20.Cavodeassi F, Rodriguez I, Modolell J. Dpp signalling is a key effector of the wing-body wall subdivision of the Drosophila mesothorax. Development. 2002;129:3815–3823. doi: 10.1242/dev.129.16.3815. [DOI] [PubMed] [Google Scholar]
- 21.Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkarni MM, et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nature Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 23.Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev. Biol. 1990;142:13–30. doi: 10.1016/0012-1606(90)90147-b. [DOI] [PubMed] [Google Scholar]
- 24.Blair SS. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- 25.Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- 26.Dove SK, et al. Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 2002;12:885–893. doi: 10.1016/s0960-9822(02)00891-6. [DOI] [PubMed] [Google Scholar]
- 27.Rusten TE, et al. Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol. Biol. Cell. 2006;17:3989–4001. doi: 10.1091/mbc.E06-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl Acad. Sci. USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koo EH, Kopan R. Potential role of presenilin-regulated signaling pathways in sporadic neurodegeneration. Nature Med. 2004;10(Suppl):S26–S33. doi: 10.1038/nm1065. [DOI] [PubMed] [Google Scholar]
- 30.Pi H, Huang SK, Tang CY, Sun YH, Chien CT. phyllopod is a target gene of proneural proteins in Drosophila external sensory organ development. Proc. Natl Acad. Sci. USA. 2004;101:8378–8383. doi: 10.1073/pnas.0306010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milan M, Perez L, Cohen SM. Boundary formation in the Drosophila wing: functional dissection of Capricious and Tartan. Dev. Dyn. 2005;233:804–810. doi: 10.1002/dvdy.20384. [DOI] [PubMed] [Google Scholar]
- 32.Milan M, Weihe U, Perez L, Cohen SM. The LRR proteins capricious and Tartan mediate cell interactions during DV boundary formation in the Drosophila wing. Cell. 2001;106:785–794. doi: 10.1016/s0092-8674(01)00489-5. [DOI] [PubMed] [Google Scholar]
- 33.Shinza-Kameda M, Takasu E, Sakurai K, Hayashi S, Nose A. Regulation of layer-specific targeting by reciprocal expression of a cell adhesion molecule, capricious. Neuron. 2006;49:205–213. doi: 10.1016/j.neuron.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 35.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 36.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 37.Crowner D, Le Gall M, Gates MA, Giniger E. Notch steers Drosophila ISNb motor axons by regulating the Abl signaling pathway. Curr. Biol. 2003;13:967–972. doi: 10.1016/s0960-9822(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 38.Tekotte H, et al. Dcas is required for importin-α3 nuclear export and mechano-sensory organ cell fate specification in Drosophila. Dev. Biol. 2002;244:396–406. doi: 10.1006/dbio.2002.0612. [DOI] [PubMed] [Google Scholar]
- 39.Meshorer E, Gruenbaum Y. Gone with the Wnt/Notch: stem cells in laminopathies, progeria, and aging. J. Cell Biol. 2008;181:9–13. doi: 10.1083/jcb.200802155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mistry H, Wilson BA, Roberts IJ, O’Kane CJ, Skeath JB. Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech. Dev. 2004;121:1495–1507. doi: 10.1016/j.mod.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nature Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- 43.Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nature Cell Biol. 2001;3:50–57. doi: 10.1038/35050558. [DOI] [PubMed] [Google Scholar]
- 44.von Mering C, et al. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev. Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 49.Lu B, Ackerman L, Jan LY, Jan YN. Modes of protein movement that lead to the asymmetric localization of partner of Numb during Drosophila neuroblast division. Mol. Cell. 1999;4:883–891. doi: 10.1016/s1097-2765(00)80218-x. [DOI] [PubMed] [Google Scholar]
- 50.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 51.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gho M, Lecourtois M, Geraud G, Posakony JW, Schweisguth F. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development. 1996;122:1673–1682. doi: 10.1242/dev.122.6.1673. [DOI] [PubMed] [Google Scholar]
- 53.Vaessin H, et al. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 54.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.