Abstract
Natural tissues are built of metabolites, soluble proteins and solid extracellular matrix components (largely fibrils) together with cells. These are configured in highly organized hierarchies of structure across length scales from nanometre to millimetre, with alignments that are dominated by anisotropies in their fibrillar matrix. If we are to successfully engineer tissues, these hierarchies need to be mimicked with an understanding of the interaction between them. In particular, the movement of different elements of the tissue (e.g. molecules, cells and bulk fluids) is controlled by matrix structures at distinct scales. We present three novel systems to introduce alignment of collagen fibrils, cells and growth factor gradients within a three-dimensional collagen scaffold using fluid flow, embossing and layering of construct. Importantly, these can be seen as different parts of the same hierarchy of three-dimensional structure, as they are all formed into dense collagen gels. Fluid flow aligns collagen fibrils at the nanoscale, embossed topographical features provide alignment cues at the microscale and introducing layered configuration to three-dimensional collagen scaffolds provides microscale- and mesoscale-aligned pathways for protein factor delivery as well as barriers to confine protein diffusion to specific spatial directions. These seemingly separate methods can be employed to increase complexity of simple extracellular matrix scaffolds, providing insight into new approaches to directly fabricate complex physical and chemical cues at different hierarchical scales, similar to those in natural tissues.
Keywords: collagen fibril, nanoscale, microscale, mesoscale, tissue alignment, plastic compression
1. Introduction
There is an increasingly successful sector of tissue engineering (TE) focused on ‘bottom-up’ fabrication of tissues by direct assembly using basic component parts. Here, ‘direct’ TE is where known three-dimensional natural structures and cells are put together as part of a controlled man-made process (Jawad & Brown submitted). In contrast, indirect TE involves construction of a (commonly synthetic polymer) temporary template containing suitable cells. In this case, it is the cells that fabricate the natural tissue structures, ideally in the image of the synthetic template which eventually is lost. The first is a classical, direct fabrication, comparable to the production of mobile phones, while the second relies on the biosynthetic and assembly apparatus of the selected cells and is more comparable with agricultural processing.
There are many potential advantages of direct TE but it inevitably requires a much greater understanding of the three-dimensional spatial organization (or at least the functionally minimal structures) of the natural materials we hope to fabricate. Examples include production of silk-like materials, fibrin networks or native collagen fibrillar materials (Vollrath & Knight 2001; East et al. 2009; Zhang et al. 2009; King et al. 2010).
Importantly, our understanding of such biostructural questions characteristically progresses incrementally and in strict compartments, defined in the main by technical size scales, nano-, (meso-), micro-, milli- (gross). This is largely due to how we focus our scientific and technical efforts. However, it is becoming clear that these artificial divisions of what are, in fact, fully interconnecting rubrics can restrict how we understand the operation of biological hierarchies. In other words, spatial organization should be viewed as intimately linked at all levels. But when we engineer these structures to produce ‘tissues’, their functions must shift depending on the dimensions (size-scale) of the tissue component we wish to control.
This concept of the role of length scale in controlling structure is key to the tissue–engineering interface and is best illustrated by its effects on the movement of tissue elements. For example, (i) molecular mobility (rate and vector) in tissues is controlled by packing, density orientation of collagen nanofibrils of the extracellular matrix, at the nanoscale (perhaps 1–500 nm tracks, depending on molecular radius), (ii) movement of cells is directed by three-dimensional structural microfeatures of approximately 100 nm–50 µm (with outliers), and (iii) movement of bulk fluids (blood, urine, air) by gross tube structures of approximately 50 µm–50 mm.
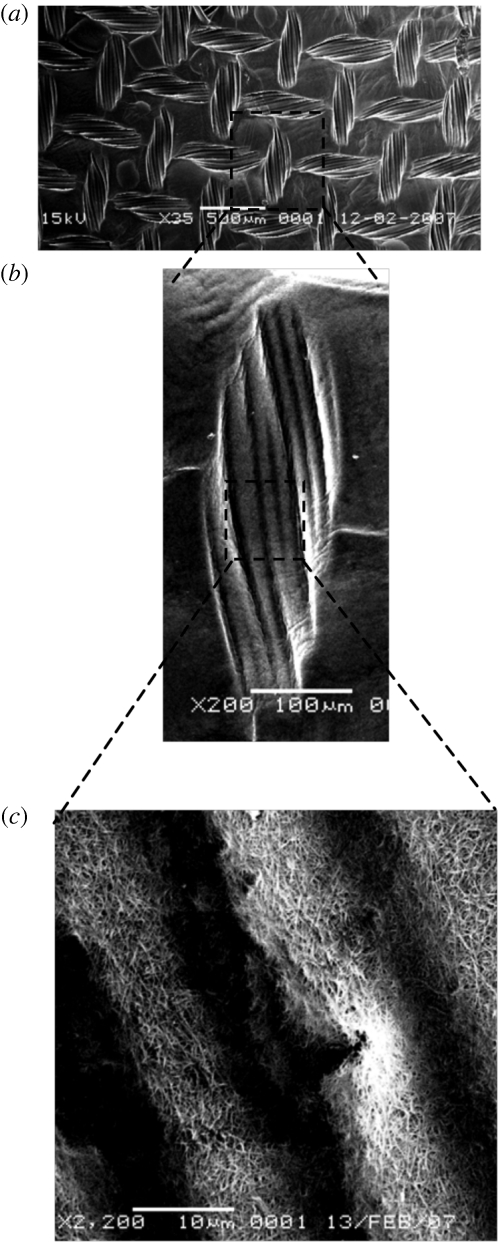
The fundamental nature of this hierarchical interdependency is illustrated in figure 1 by three views of the same nanofibrillar collagen material, but at three widely different magnifications in the scanning electron microscope (SEM). This shows topography embossed into a compressed collagen gel surface by a standard nylon mesh. The first low-power image (figure 1a) shows parallel alignment over the 0.5–1 mm length scale, with the implication that over several centimetres (gross scale) we would perceive this to be an orthogonal alignment (i.e. comprising right angles). Indeed, at the human scale, this is what we see as a net. At a scale down (figure 1b), 1–100 µm, an attached cell might perceive the pattern as a series of strictly parallel fibres of a few micrometres in diameter, and nothing else for many times its length in either direction. In other words, to a cluster of cells, this becomes a highly aligned, uniaxial topography.
Figure 1.
Scales of hierarchy. Three magnifications of the same image show how systems using topographic cues at the (a) millimetre, (b) micrometre, (c) nanometre scales will receive quite distinct information (orthogonal, parallel, random). This is typical of biological hierarchies in fibrous collagen materials.
However, the hierarchy paradox becomes clearer with the next hierarchy step as we look at the mesoscale (figure 1c: 100 nm–10 µm). At this level, it becomes evident that what appeared to be parallel multi-microfibres are in fact composed of randomly aligned collagen nanofibrils (mean diameter approximately 30 nm; Cheema et al. 2007). Indeed, we can extend this example below the magnification of the SEM as it is known from the classical collagen structural studies that at the level of a few nanometres, single collagen molecules (1.5 nm diameter helix) are packed into each of these 30 nm fibrils. So once again, at this nanometre scale, the collagen molecules are parallel in alignment to the fibril axis, and systems operating at this level may be guided.
To give a more geometric example; to a cluster of fibroblasts, a cube structure of 100 µm side represents 12 separate alignment edges in three planes. However, to a diffusing oxygen molecule, the shape of the cube is completely irrelevant, but the pore size/orientation of the cube material can be critical. While inside a cardiac vessel, a single 100 µm cube is near irrelevant. However, 1000 cubes aggregated together will equate to a blockage or, in a line, will split the blood flow.
In figure 1, we have a continuum of three-dimensional structures, all in the same material, where ‘alignment’ is scale-sensitive. In other words, the material does not have one alignment. Rather, it has a number of patterns of alignment, depending on the length scale at which it is perceived or used. In our collagen gel example, the human eye sensing system (gross) perceives an orthogonal pattern; cells and cell clusters see uniaxial, parallel alignment (multi-micrometres); integrin and cytoskeletal assembly systems see random fibre patterns (sub-micrometres); while protein or glucose mass transports are influenced by uniaxial, parallel nanometre-scale alignment. The system is essentially simple (a single collagen gel) but it is made complex by overlying a simple embossing process onto collagen fibril self-aggregation.
The presence of hierarchies in tissue structures provides us, as tissue engineers, with invaluable information and clues for direct fabrication of tissues (Lakes 1993). The potential benefit here is that understanding how to fabricate three-dimensional structural alignment across two or more size hierarchies may allow us to generate relatively complex biological signalling structures, using simple systems.
In this study, we have tested three systems for generating alignment and direction at different size-scales (note: this is not about cell alignment, though cell alignment can be one aspect). However, this represents a difficult scientific paper to prepare, as it would conventionally appear as three separate reports, immediately destroying the central point of the importance of scale-hierarchy of that alignment. The present format, though unorthodox, allows us to discuss and illustrate the hierarchy itself, as well as its isolated components.
In the first system, we test the ability of directional fluid shear and tension to align nanoscale collagen fibrils over the multi-micrometre length scale. Mechanical alignment of collagen fibrils has been described in ultra-soft low-density collagen gels (Kostyuk & Brown 2004), but is difficult to generate once the collagen-fibril mesh is dense and entangled (D. Karamichos, A. Kureshi & R. A. Brown 2008, unpublished data). With the second system, we demonstrate a technique to generate molecular-scale alignment (protein diffusion gradients) using the three-dimensional spatial organization of the nanofibril collagen gel (random nanofibril alignment to restrict–permit the planes of protein movement). Controlled local cell hypoxia was used here to generate production of angiogenic signalling proteins (including VEGF) in a dense, hypoxic cell depot (Cheema et al. 2008; Hadjipanayi et al. 2010). Movement of growth factor proteins was aligned, forming gradients by constraining layers of dense collagen in an attempt to mimic natural gradients of angiogenic factors, directing vascular cells during vessel development (Fraisl et al. 2009). In the final system, we demonstrate the ability to emboss high-resolution micrometre scale channel patterns into the same random nanofibril collagen material such that resident cells orient to the channel-scale, rather than the fibril-scale (random) pattern.
2. Material and methods
2.1. Long-range, cell-free alignment of collagen fibrils
2.1.1. Preparation of collagen gels
Acellular collagen gels were prepared from 8 ml of native acid-soluble type I rat tail collagen (2.06 mg ml−1; first link, West Midlands, UK) with 2 ml of 10× concentration of minimum essential medium (10× MEM) (Gibco, Invitrogen Corporation) and neutralized with 1 M NaOH. Ten millilitres of the collagen solution was poured into a rectangular-shaped mould (75 × 25 × 15 mm). These were placed into an incubator at 37°C and 5 per cent CO2 for 30 min allowing for the phase transition of the collagen solution into a collagen gel.
2.1.2. Vertical shear flow set-up
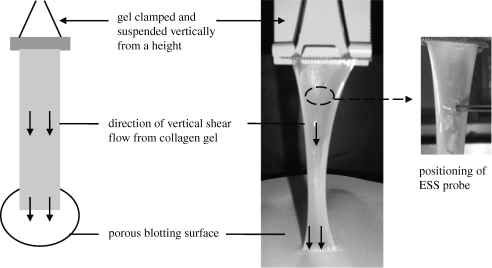
Once set, the gel was released from the mould and clamped with metal mesh at one of its short ends for secure anchorage during suspension. The clamped end was hung up to suspend the gel vertically above three sheets of Whatman no. 1 filter paper (figure 2). The lower edge of the gel was positioned so as to just touch the paper, allowing blotting of fluid from the gel to the paper, generating an increasing vertical stress (gravitational force) over time. Six gels were suspended vertically for 75 min.
Figure 2.
‘Vertical shear flow’ model to produce anisotropic collagen gels. The figure depicts vertical suspension of gel, basal attachment to blotting paper and positioning of ESS probe to monitor alignment of fibres. At the start of the experiment, gel is fully hydrated, but by 30 min there is significant gel compaction and dehydration indicated by thinning and ‘wasting’ of gel.
2.1.3. Preparation of gels without directional shear flow
Six acellular collagen gels were prepared and allowed to set in an identical fashion to test gels. Since these are normally isotropic and were not subjected to a directional shear flow, these were suitable as a control. Removal of the gel from its mould caused some fluid to exude. Therefore, it was found that placing the gel horizontally on a non-absorbent, plastic mat avoided directional fluid loss and slowed the rate of gel compaction and dehydration.
2.1.4. Elastic scatter spectroscopy
Collagen fibril alignment in gels was measured over time using elastic scatter spectroscopy (ESS). This enabled dual readings of backscatter intensity, which were parallel and perpendicular to the vertical strained axis. The optical probe collected data at a fixed point, approximately 2 cm from the top of the gel over an experimental period of 75 min. A dark background behind the collagen gel was essential to limit scatter of any light lost, which may be detected by the optical probe.
The optical probe (Knight Optical Technologies, Leatherhead, UK) is attached to a xenon light source and spectrometer (Ocean Optics, Dunedin, FL). For monitoring the backscattered light, it is placed perpendicular to the surface of the collagen gel. The probe holds two optic fibres encased in a metal sheath—the illumination fibre (diameter 400 µm) delivers short pulses of white light (320–860 nm), while the detection fibre (diameter 200 µm) collects any backscattered light, which is subsequently analysed by the spectrometer. A computer is attached to this system which records and displays the data. At the start of each measurement session, a spectrum of the diffuse reflectance standard (Ocean Optics, Dunedin, FL; a white, diffusing, reflectance material made of polytetrafluoroethene that is almost uniformly reflective (>95%) across a wavelength range of 250–2000 nm) was measured and recorded as a reference to take into account the spectral characteristics and overall intensity of the light source (Kostyuk & Brown 2004). The spectra of test gels were collected at regular intervals during the 75 min experimental period. Measurements were taken at the start of the experiment (0 min) and then every 10 min until 60 min and then every 5 min until 75 min and the end of the experiment. To monitor the alignment of fibres, two readings were taken from the optical probe—parallel (0°) and perpendicular (90°) to the vertical-strained axis on the same site of the collagen gel. A spectrum of backscattered light in the range of 320–860 nm was recorded.
Anisotropic factor (AF) was calculated to indicate levels of anisotropy in the tissue. AF was calculated by the formula: AF = intensity 90°/intensity 0° at a particular wavelength. The AF values were calculated at a wavelength of 600 nm as this wavelength displayed the largest disparity between backscatter intensities recorded at each angular position of the probe.
2.1.5. Scanning electron microscopy analysis
Following 75 min of ‘vertical shear flow’, the collagen gels were fixed in glutaraldehyde solution for SEM processing and analysis. The gel was cut so that only the region where the ESS probe was placed and alignment measurements taken was isolated for SEM analysis. After 60 min of ‘vertical shear flow’ of collagen gels, backscatter intensity was recorded. Following this, two drops of fixative solution, comprised of 0.1 M sodium cacodylate of pH 7.4 and 2.5 per cent glutaraldehyde, was applied to the surface of the gel for a further 15 min. Recordings of backscatter intensity were made at 65, 70 and 75 min, following which the gel was released (untethered). The upper clamp edge of the gel was cut and separated from the rest of the gel, and the 2 cm region where the ESS probe recorded backscatter intensity (i.e. upper quarter of the gel) was divided from the remainder of the gel. A diagonal cut was made in the upper left corner of each specimen to establish the absolute orientation of each sample, before placing into cold fixative. This was stored at 4°C overnight prior to beginning SEM processing of samples, which involved a series of alcohol dehydrations and coating with gold–palladium using a ‘sputter coater’. Dry, sputter-coated specimens were viewed on a JEOL 5500LV SEM.
2.2. Mesoscale guidance of protein factors and cell migration
2.2.1. Cell culture
Primary human dermal fibroblasts (HDFs) were maintained in high glucose (4000 mg l−1) Dulbecco's modified Eagle's medium (DMEM, Gibco, Paisley, UK), supplemented with 10 per cent foetal calf serum (First Link, West Midlands, UK), 2 mM glutamine and penicillin/streptomycin (1000 U ml−1; 100 µg ml−1; Gibco Chemicals). The endothelial cells (ECs) used in this study were primary human umbilical vein endothelial cells (HUVECs), which were cultured in complete EC growth medium (Promo Cell, Germany). For removal of cells from the monolayer culture (all <70% confluence), flasks were washed with 0.1 M phosphate-buffered saline, and incubated with trypsin (0.5%) for 5 min at 37°C.
2.2.2. Segmented gel casting
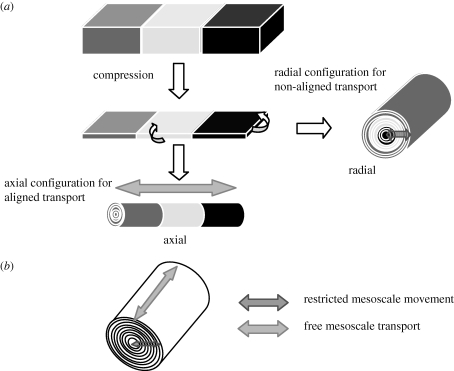
Type I collagen gels containing a specific cell type were cast at the correct densities: 2 × 106 HDFs in a 1.5 ml gel, and 1 × 105 HUVECs (ECs) in a 1.5 ml gel (0.7 × 22 × 10 mm). Following 30 min of setting at 37°C, these were placed in a fresh mould at either end (10 × 22 × 33 mm), and an acellular gel (5 ml solution) was poured into the mould to set around the cellular ends. Following a further 30 min of setting time, the entire construct was compressed for 5 min, and resulted in a 100 µm sheet, which could be spiralled in either a radial or axial manner depending upon the experiment (figure 3). The constructs were laid on flat Petri dishes, and wound using the blunt end of a scalpel blade either along the short (radial model) or long (axial model) axis.
Figure 3.
(a) Scheme showing segmental gel-setting, compression, and axial and radial configuration. Dark grey, EC; light grey, acellular gap; black, HDF. (b) Scheme illustrating the direction of transport of cells and cell-generated macromolecules in the axial (low resistance to movement) and radial (restricted transport) directions.
2.2.3. Immunohistochemical staining
In vitro-cultured spiral (axial and radial) constructs were unrolled, washed in 5 ml phosphate-buffered saline and fixed in 100 per cent ice-cold methanol for 1 h. An incision was made on the unrolled corner, which represented the core HDF region to allow for spatial identification of core/surface or HDF/EC regions. Routine immunostaining was then used to stain the entire construct. The primary antibody used was mouse anti-human CD31 (Dako, USA) and the secondary antibody was anti-mouse IgG-FITC (R&D, UK) followed by DAPI (Sigma, UK) nuclear staining.
2.2.4. Image analysis
Micrographs of immunofluorescent-stained gels were captured with a fluorescent microscope (Olympus BX61) using a 10× objective. As the gels were three-dimensional, this microscope allowed focusing of constructs through the 100 µm depth. A minimum of 10 random fields were photographed per sample region. The distance travelled was determined by assigning each cluster group an average distance travelled. The total numbers of clusters were counted per compartment and each of these was assigned an average distance travelled (axial model average distance: EC compartment = 0, ACELL compartment = 11 mm, HDF compartment = 22 mm; radial model average distance: EC compartment = 0; ACELL compartment = 480 µm; HDF compartment = 720 µm). The average distance was taken to be the mid-way distance of the specific region. Imaging software (Image J, USA) was used to determine the area of EC clusters (aggregates greater than 2 ECs). Total number of EC clusters was counted manually.
2.3. Embossing micrometre-scale channel patterns
2.3.1. Collagen gels with embossed features
Collagen gels were prepared as described previously; 5 ml of neutralized solution were allowed to set in a stainless steel circular mould (diameter 22 mm) for 30 min at 37°C. A custom-made stainless steel embossing mask (thickness 50 µm, each bar approximately 70 µm wide, 420 µm spacing) was placed onto the blotting paper (Whatman N1, UK), set gel was transferred on top of the mask and load applied for 5 min (figure 4). For the purpose of investigating cell behaviour on the embossed grooves, cell suspension (lung fibroblast-like cell line, W1 38, HPA Culture Collections, UK, courtesy of R. Potter) was pipetted onto the embossed surface of the constructs (100 cells per construct) and cultured for 72 h. Cells were visualized using live cell marker (calcein). Embossed collagen constructs with or without cells on the surface were fixed and processed for SEM analysis (as described in §2.1) and images were taken to determine surface morphology and cell behaviour.
Figure 4.
Scheme of embossing topographical features onto the fluid-leaving surface of collagen gel using directional fluid flow.
3. Results
3.1. Long-range, cell-free alignment of collagen fibrils
3.1.1. Measurement of fibril alignment using elastic scatter spectroscopy
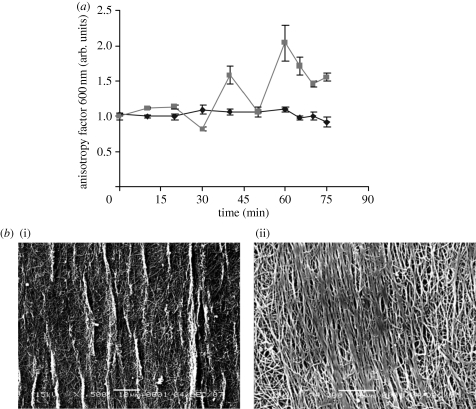
A total of 12 collagen lattices were monitored for fibril alignment using ESS; six were subjected to the vertical shear flow technique and six which were kept horizontal to avoid directional fluid loss remained as control gels. Control gels maintained an AF of approximately 1 (i.e. random) throughout the experimental period. Figure 5a shows the mean AF values plotted against time for test (n = 6) and control (n = 6) gels. Gels subjected to vertical shear flow demonstrated a mean AF of 2 ± 0.2 at 60 min. This was the time point AF values were maximal, indicating that maximum fibril alignment was achieved. Control gels at this time point produced an AF of 1.1 ± 0.03, which is effectively isotropic (zero alignment AF = 1). The difference at 60 min was statistically significant (p < 0.0001, unpaired t-test). This was not a linear increase in AF but decreases in mean AF at 30 and 50 min occurred in all replicates (figure 5a). A modest fall in AF, to 1.55 ± 0.05 by 75 min, is thought to be the result of local relaxation of fibrils as the total volume of aligned gel increased.
Figure 5.
(a) Graph demonstrating AF changes over time for control (filled diamonds) and vertical shear flow (filled squares) collagen gels over a 75 min experimental period. Each data point is a mean AF value (±s.d.) calculated from six individual AF values, which are each calculated as a ratio of backscatter intensities obtained with the probe at two orthogonal positions: parallel (0°) and perpendicular (90°) to the long axis of the tissue. AF = intensity 90°/intensity 0° (three backscatter intensities were recorded for each angular position at every time point for each gel; n = 6 control gels, n = 6 vertical shear flow gels). (b) SEM images of a gel that has been subjected to vertical shear flow for 75 min: (i) low (1500×) magnification and (ii) high (4300×) magnification. Regions of alignment of collagen fibrils are parallel to the direction of shear flow.
3.1.2. Scanning electron microscopy imaging of collagen fibrils
SEM analysis showed that control gels maintained a random (isotropic) organization of fibres until the end of the experiment (data not shown). Gels that were subjected to the vertical shear flow technique developed a high degree of collagen fibril alignment in the direction of shear flow (figure 5b). However, this was seen to occur in patches across the gel rather than uniformly, throughout the material.
3.2. Mesoscale guidance of protein factors and cell migration
Triple segment gels were formed where two cell populations (HDFs and ECs) were seeded into the two of the three gels, separated by a third, acellular, compartment, in two spatial arrangements—radial and axial. The basic collagen structure is the same in both models and at all size scale levels; only the relative position of the cell types (the ‘compartments’) differs. In the axial model, the spiral, multi-layer compartments lie along a single axis, parallel to the construct itself. The spiral layers are approximately 50 µm thick, with each layer being made of coaxial collagen lamellae of 1–5 µm. In the axial model, all these levels of collagen layering are coaxial (parallel) and interconnected. In effect, in the second model, the same three compartments are radial, lying perpendicular to the axis. In this biaxially asymmetric structure, movement between compartments, of macromolecules and cells with effective radii in the mesoscale range, will be restricted much more in one model than the other. The density of the collagen nanofibre mesh of each spiral layer will restrict movement radially but coaxial movement between these lamellae and layers will be unrestricted at this scale.1
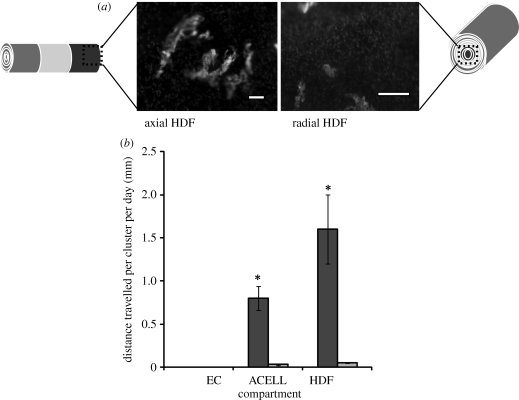
EC cluster formation was measured in the different compartments of each model following two weeks of culture. The total distance travelled, i.e. multiplying the number of clusters by average distance travelled, is shown in figure 6. Distance travelled was measured in terms of EC clusters identified in the HDF compartment and in this case migration of EC clusters in the axial model travelled an average of 1.6 mm d−1, as opposed to a significant, 28-fold lower rate of 0.057 mm d−1 in the radial model (p < 0.001; figure 6). There was little evidence of HDF migration into the EC compartments in either model, as very few CD31(−)ve cells were seen in the EC compartment. This indicates that in this model, the ECs were migratory and the HDFs were not, consistent with the idea that the ECs were migrating in response to an angiogenic factor gradient produced by hypoxic HDFs, as demonstrated previously. This is important to the model as it suggests that both growth factor diffusion and EC movement between the compartments are involved, though in this case the proportional contribution of each to overall measured cell movement cannot be determined.
Figure 6.
(a) CD31(+)ve-stained EC clusters found in the human dermal fibroblast (HDF) compartment of the axial- and radial-configured models. DAPI-stained nucleus of CD31(−)ve cells are HDFs. Scale bars = 100 µm. (b) Mean distance travelled per EC cluster per day within the axial and radial model. Cultures were incubated for two weeks. Black bars, axial; grey bars, radial. *p < 0.001.
3.3. Embossing micrometre-scale channel patterns for alignment of cells
3.3.1. Plastic compressed (PC) constructs with embossed features
Forty micrometre-wide grooves were embossed into the surface of compressed collagen gels (figure 4). The standardized embossing template was placed at the lower fluid-leaving surface of the collagen gel at the time of casting, such that all of the fluid was forced to pass around the bars of the embossing template. This has been shown to be necessary for stable deep-pattern embossing, into the stiff collagen layer at the fluid-leaving surface (T. Alekseeva, E. Hadjipanayi & R. A. Brown 2010, unpublished data).
SEM images (figure 7) show that this technique produced a well-defined regular pattern of parallel 40 µm wide grooves, embossed onto the fluid-leaving surface of the collagen gel. Dimensions of the grooves reflected those of the template. Fibroblasts seeded onto surfaces embossed at this scale showed good alignment, parallel with that of the grooves, particularly the groove edges. This cell alignment was identified using calcein vital staining (figure 8) to confirm that aligned cells were viable, consistent with the idea that alignment was cell-attachment and/or migration-dependent.
Figure 7.
Scanning electron micrographs demonstrating the surface of PC collagen construct with regular, parallel embossed grooves 40 µm wide and 30 µm deep, set within a collagen material made up of nanoscale (30–50 nm diameter) fibrils with random x–y orientation in this image.
Figure 8.
Effect of the topographical features on the fluid-leaving surface of PC collagen gel on cell behaviour. Cells seeded onto the surface of collagen constructs with parallel 40 µm grooves were visualized using live-cell dye, calcein (grey). Cells became aligned to the groove features after 3 days in culture. For comparison, note the lack of alignment to the grooves on the collagen surface between the grooves. (a) General view; (b,c) higher magnifications. Arrows indicate grooves, ovals outline aligned cells in the groove. Scale bars, (a) 200 µm and (b,c) 100 µm.
4. Discussion
The emergence of new methods to generate controlled shape, topology and three-dimensional architecture in native protein materials (Elsdale & Bard 1972; Lanfer et al. 2008) is rapidly opening up new possibilities for biomimetic/biorelevant engineering of tissues. This draws on a rich literature on the effects of structural alignment in synthetic materials on cell-molecular guidance (Wilkinson 2004; Shah et al. 2005). However, such studies have so far tended to concentrate on the alignment and guidance properties of systems operating within relatively narrow size ranges. The use of native biological materials has begun to highlight the complex manner in which alignment in natural systems operates differently in different ranges of their structural hierarchy. In effect, this is a feature of the inherently anisotropic nature of self-aggregating protein nanofibres in many natural materials and the new approaches to their fabrication under physiological conditions, retaining the natural hierarchies. This study of the formation and properties of alignment hierarchies across three length scales uses the example of anisotropic native collagen nanofibre matrix constructs, fabricated as defined three-dimensional structures by the plastic compression process of cell-seeded collagen gels.
At the (tens) nanometre level, collagen fibril alignment has been reported by allowing fibroblast-populated collagen gels to contract under uniaxial restraint (Eastwood et al. 1996; Kostyuk & Brown 2004). This is an indirect, slow and poorly controlled technique but alternatives using high-intensity magnetic fields (Tranquillo et al. 1996) or flow (Lanfer et al. 2008) are equally difficult to apply. Furthermore, alignment under tension was even more difficult to achieve at high collagen fibril densities (O. Kostyuk & R. A. Brown 2004, unpublished data). In this study, we describe a model system for generating nanofibrillar alignment by application of flow and uniaxial tension during the gel compaction process, where fluid is expelled from the gel as it hangs vertically from a frame. Development of alignment at this scale (average fibril diameter 30 nm; Cheema et al. 2007) was followed by its changing effect on the scattering of white light (ESS, AF), as demonstrated previously in cell-driven alignment (Kostyuk & Brown 2004).
Collagen gels compressed under vertical shear flow gradually developed a modest fibril alignment over 60 min with the measured AF rising from 1 (non-aligned) to a maximum AF of 2, parallel with loading. This alignment of AF is comparable with that generated by fibroblast-populated gels and externally tensioned (20% strain) acellular collagen gels (Kostyuk & Brown 2004), which developed maximum AF values close to 3. Not only is this a simple and rapid technique of producing limited fibrillar alignment, but it is also independent of cell action, hence suitable for a direct fabrication process that does not damage resident cells.
Importantly, SEM analysis of gels subjected to this shear flow alignment confirmed the ESS analysis of collagen fibril alignment, but with a key caveat. The fibril alignment which was approximately parallel with the load-flow axis was heterogeneous. The structure produced comprised strips or bundles of well-aligned collagen fibrils parallel with the loading set between areas of poorly or non-aligned fibres. This is consistent with the modest AF values suggesting some orientation, as the ESS technique gives an average response over some hundreds of micrometre surface (Kostyuk & Brown 2004), comprising both highly and poorly aligned areas.
The localized development of fibril alignment is also consistent with changes in AF with time (figure 5a). In fact, it is likely that the dynamic process of fibril alignment and bundling under load may occur as a series of mini-cycles of stress shielding, fibril failure and remodelling, generating more and more aligned bundles. As the first few collagen fibrils are loaded and align with the load, they will effectively stress-shield those surrounding areas of fibrils that are not parallel. As loading and gel dehydration continues, other fibril bundles will contribute to carrying load and at times the load will shift as some of the aligned bundles fail and transfer load to non-aligned areas. This model for generation of non-homogeneous fibril alignment, then, is important as a practical technique for directly generating alignment in collagen constructs. However, it also demonstrates how the natural fibril hierarchy structure can propagate alignment from the nanometre (approx. 30 nm diameter fibril) scale up to the multi-micrometre (fibril bundle) scale, under tension, to produce tissue-like heterogeneous structure.
Importantly, the ESS measurement would average this scatter by fibrils ± alignment right through the collagen layer (not just at the surface which is seen in the SEM). Consequently, this alternate parallel bundling and poor alignment mix of structures (in the x–y plane) is likely to be present at all levels (across the z-plane).
In the mesoscale hierarchy, based on cell-macromolecular angiogenic factor guidance, it is clear that the three-dimensional structure of the collagen spiral produces a biaxial alignment of cell movement. However, it is also clear that (based on the lack of fibroblast mobility to match that of ECs) the hypoxia-generated angiogenic factors have a role. Unstimulated HDFs have been previously shown to migrate very little in this density of collagen (Hadjipanayi et al. 2009), whereas ECs do migrate strongly under angiogenic gradients in this system (Hadjipanayi et al. 2010). In other words, protein growth factor gradients have developed in the axial and radial models, between the hypoxic HDF compartment and the EC compartment. As discussed, it seems likely that some degree of asymmetric restriction of macromolecule diffusion is present as proteins should pass more slowly through than between/along the dense collagen layers. However, in this study, this remains undemonstrated as only cell movement was directly shown to be affected by the model type. Direct measurement of protein diffusion gradients was outside the scope of this work.
As discussed previously, this model is a particularly important example of an alignment hierarchy not only as it operates so strongly at the cell (mesoscale) level. It is also formed so simply from the three-dimensional assembly of naturally size-selective, anisotropic collagen meshworks. Indeed, this form of guidance is likely to be extremely common in natural as well as engineered tissues. However, it also demonstrates very well the selective effect of the scale-hierarchy on the response it produces. Particles that are significantly above or below the size exclusion of the collagen network will not be guided, as the difference between the radial and axial would disappear, either to uniformly high permeability, or uniformly zero.
The final level and example of induced alignment in the collagen materials used the plastic nature of the collagen compression process to generate surface-embossed patterns, i.e. extended parallel surface voids to an approximate depth of 30 µm. Grooves ran for the full (millimetre) length of the constructs and as such mirror surface topographies, which are commonly used in spatial biology and TE for cell guidance. Alignment of fibroblasts in this case was most marked at the edges of each groove. Indeed, it is likely that the declivity of the groove edge is a major source of cell cues, as for this groove dimension it is unlikely that any single cell could traverse, and so use, the full shape including both edges. Equally, it appears unlikely that cells were able to align to collagen fibril alignment cues, at the scale hierarchy level below this (approx. 30 nm fibril diameter) as there was no known fibril alignment at that scale. This is despite evidence that topographic cues in the low nanometre scale, acting at the integrin receptor level, are well documented. It is possible that increased surface stiffness along the groove edge contributes to this cell response as deflection of the fluid outflow during compression owing to the presence of the embossing template could produce such local effects.
Interestingly, in the context of alignment at different hierarchies, these embossed ridges (and their edges) are a special case of three-dimensional structure, being at the surface of the construct. If it were possible by this technique to produce an embossed ‘groove’ running through the body of the collagen gel (i.e. a channel), this structure would by definition have no edges. This would test the possibility that cells are taking primary cues from, and at the scale of, the groove edges. If this were the case, cell alignment seen here for edge-rich grooves would not be expected within the same diameter of channel.
To conclude, the core message here is that there is a pressing need to distinguish the scale at which any given material can provide functional three-dimensional guidance cues in an engineered tissue. In crude terms, functions are commonly distinct at the molecular, cellular and bulk/gross levels, and architectural features can be built into materials at each of these levels. Yet, the nature of biological materials means that it is often possible to achieve two or more of these scales in a single material (in this case dense collagen gels). Furthermore, these are relatively simple, scale-sensitive processing technologies. Through this understanding it now seems increasingly feasible to fabricate quite complex, multi-hierarchy, biomimetic ‘tissues’, directly, using natural matrix materials.
Acknowledgements
U.C. is a BBSRC David Phillips Fellow and is funded through this route. Johnson & Johnson Medical GmbH and the BBSRC additionally funded this work. We would like to thank Hector Hadjipanayi for help with images.
Footnotes
One contribution to a Theme Supplement ‘Translation and commercialization of regenerative medicines’.
Critical to this study, this spiral structure loses its biaxial guidance properties where we consider small nutrient molecules or oxygen (sub-nanoscale) or millimetre-scale organisms (e.g. an arthropod mite). Movement of sub-nanoparticles will be unrestricted in both axes (so both models) and millimetre-scale organisms would be restricted between the layers, making this structure symmetrically permeable or impermeable, at these other two hierarchical levels.
References
- Cheema U., Brown R. A., Alp B., MacRobert A. J. 2008. Spatially defined oxygen gradients and VEGF expression in an engineered 3D cell model. Cell. Mol. Life Sci. 65, 177–186. ( 10.1007/s00018-007-7356-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema U., Chuo C. B., Sarathchandra P., Nazhat S. N., Brown R. A. 2007. Engineering functional collagen scaffolds: cyclical loading increases material strength and fibril aggregation. Adv. Funct. Mater. 17, 2426–2431. ( 10.1002/adfm.200700116) [DOI] [Google Scholar]
- East E., Golding J. P., Phillips J. B. 2009. A versatile 3D culture model facilitates monitoring of astrocytes undergoing reactive gliosis. J. Tissue Eng. Regen. Med. 3, 634–646. ( 10.1002/term.209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood M., Porter R., Khan U., McGrouther G., Brown R. 1996. Quantitative analysis of collagen gel contractile forces generated by dermal fibroblasts and the relationship to cell morphology. J. Cell Physiol. 166, 33–42. () [DOI] [PubMed] [Google Scholar]
- Elsdale T., Bard J. 1972. Collagen substrata for studies of cell behaviour. J. Cell Biol. 54, 626–637. ( 10.1083/jcb.54.3.626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisl P., Mazzone M., Schmidt T., Carmeliet P. 2009. Regulation of angiogenesis by oxygen and metabolism. Dev. Cell 16, 167–179. ( 10.1016/j.devcel.2009.01.003) [DOI] [PubMed] [Google Scholar]
- Hadjipanayi E., Mudera V., Brown R. A. 2009. Guiding cell migration in 3D: a collagen matrix with graded directional stiffness. Cell Motil. Cytoskeleton 66, 121–128. ( 10.1002/cm.20331) [DOI] [PubMed] [Google Scholar]
- Hadjipanayi E., Brown R. A., Mudera V., Deng D., Liu W., Cheema U. 2010. Controlling physiological angiogenesis by hypoxia-induced signalling. J. Control. Rel. 146, 309–317. ( 10.1016/j.jconrel.2010.05.037) [DOI] [PubMed] [Google Scholar]
- Jawad H., Brown R. A. Submitted Meso-scale engineering of collagen as a functional biomaterial. [Google Scholar]
- King V. R., Alovskaya A., Wei D. Y., Brown R. A., Priestley J. V. 2010. The use of injectable forms of fibrin and fibronectin to support axonal ingrowth after spinal cord injury. Biomaterials 31, 4447–4456. ( 10.1016/j.biomaterials.2010.02.018) [DOI] [PubMed] [Google Scholar]
- Kostyuk O., Brown R. A. 2004. Novel spectroscopic technique for in situ monitoring of collagen fibril alignment in collagen gels. Biophys. J. 87, 648–655. ( 10.1529/biophysj.103.038976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakes R. 1993. Materials with structure hierarchy. Nature 361, 511–515. ( 10.1038/361511a0) [DOI] [Google Scholar]
- Lanfer B., Freudenberg U., Zimmermann R., Stamov D., Korber V., Werner C. 2008. Aligned collagen fibrillar matrices obtained by shear flow deposition. Biomaterials 29, 3888–3895. ( 10.1016/j.biomaterials.2008.06.016) [DOI] [PubMed] [Google Scholar]
- Shah R., Sinanan A. C. M., Knowles J. C., Hunt N. P., Lewis M. P. 2005. Craniofacial muscle engineering using a 3-dimensional phosphate glass fibre construct. Biomaterials 26, 1497–1505. ( 10.1016/j.biomaterials.2004.04.049) [DOI] [PubMed] [Google Scholar]
- Tranquillo R. T., Girton T. S., Bromberek B. A., Triebes T. G., Mooradian D. L. 1996. Magnetically oriented tissue-equivalent tubes: application to a circumferentially orientated media-equivalent. Biomaterials 17, 349–357. ( 10.1016/0142-9612(96)85573-6) [DOI] [PubMed] [Google Scholar]
- Vollrath F., Knight D. P. 2001. Liquid crystalline spinning of spider silk. Nature 410, 541–548. ( 10.1038/35069000) [DOI] [PubMed] [Google Scholar]
- Wilkinson C. D. 2004. Making structures for cell engineering. Eur. Cells Mater. 8, 21–26. [DOI] [PubMed] [Google Scholar]
- Zhang X., Reagan M. R., Kaplan D. L. 2009. Electrospun silk biomaterial scaffolds for regenerative medicine. Adv. Drug Deliv. Rev. 61, 988–1006. ( 10.1016/j.addr.2009.07.005) [DOI] [PMC free article] [PubMed] [Google Scholar]