Abstract
Many ion channels are modulated by phosphatidylinositol 4,5-bisphosphate (PIP2), but studies examining the PIP2 dependence of channel activity have been limited to cell expression systems, which present difficulties for controlling membrane composition. We have characterized the PIP2 dependence of purified human Kir2.1 and Kir2.2 activity using 86Rb+ flux and patch clamp assays in liposomes of defined composition. We definitively show that these channels are directly activated by PIP2 and that PIP2 is absolutely required in the membrane for channel activity. The results provide the first quantitative description of the dependence of eukaryotic Kir channel function on PIP2 levels in the membrane; Kir2.1 shows measureable activity in as little as 0.01% PIP2, and open probability increases to ∼0.4 at 1% PIP2. Activation of Kir2.1 by phosphatidylinositol phosphates is also highly selective for PIP2; PI, PI(4)P, and PI(5)P do not activate channels, and PI(3,4,5)P3 causes minimal activity. The PIP2 dependence of eukaryotic Kir activity is almost exactly opposite that of KirBac1.1, which shows marked inhibition by PIP2. This raises the interesting hypothesis that PIP2 activation of eukaryotic channels reflects an evolutionary adaptation of the channel to the appearance of PIP2 in the eukaryotic cell membrane.
Keywords: Membrane Lipids, Membrane Reconstitution, Phosphatidylinositol Signaling, Phospholipid Vesicle, Potassium Channels
Introduction
Ion channels are modulated by their lipid environment (1–6), and perhaps the best characterized lipid modulator of ion channel activity is phosphatidylinositol 4,5-bisphosphate (PIP2)3 (7). Inward rectifying potassium channels (Kir) are just one of a host of ion channels that are regulated by PIP2 (7), and a number of genetic diseases, including Andersen-Tawil syndrome (8, 9), Bartter syndrome (9), and neonatal diabetes (10), result from Kir channel mutations that alter PIP2 sensitivity. Structure-function studies and molecular simulations suggest a PIP2 binding site at the top of the cytoplasmic domain, just below the surface of the inner membrane leaflet (9, 11–15). These studies, however, have provided limited and largely qualitative information on how such lipids influence Kir channel function because they all utilize cell-based systems, in which the membrane composition is unknown and cannot be precisely controlled. Alternative approaches using recombinant systems in which the components can be fully controlled are needed to expand our current knowledge (7).
Until recently, high yield recombinant expression and purification of functional ion channels have been technically prohibitive and restricted to prokaryotic channels (3, 16). We have recently demonstrated the feasibility of purifying functional eukaryotic Kir channel after recombinant expression in Saccharomyces cerevisiae (17). To quantitatively examine the PIP2 dependence of eukaryotic Kir channel activity, we have purified human Kir2.1 (KCNJ2) and Kir2.2 (KCNJ12) and performed functional studies using both liposomal 86Rb+ flux assays and patch clamp assays of giant liposomes.
EXPERIMENTAL PROCEDURES
Human Kir2.1 (KCNJ2) and Kir2.2 (KCNJ12) were subcloned into the pND-CTFH vector and expressed in the FGY217 strain of S. cerevisiae as described previously (17). Starter cultures were grown in SC − Ura medium + 2% glucose and then scaled up overnight at 250 rpm and 30 °C in Ultrayield shaker flasks (A600 ∼1.5). SC − Ura + 2% galactose/flask were then inoculated with these cultures for induction (A600 ∼0.2) and grown for ∼48 or 72 h respectively. Cells were harvested by centrifugation at 4000 × g, washed in TBS, resuspended in lysis buffer (50 mm Tris, pH 7.5, 10 mm EDTA, 0.6 m sorbitol, Complete EDTA-free tablets, protease inhibitor set IV (Calbiochem)) and ruptured by 10 passes at high pressure (25,000–35,000 p.s.i) using a Microfluidics cell disrupter. KCl was added to cell lysate 500 mm final, and cell debris was removed by 5 min of centrifugation at 500 × g followed by 25 min at 4000 × g. Crude membranes were collected by centrifugation for 1 h at 120,000 × g and homogenized in resuspension buffer (50 mm Tris 7.5, 10 mm EDTA, 500 mm KCl, 2 mm DTT, 10% glycerol, Complete EDTA-free protease inhibitor) to ∼3.5 mg/ml total protein and then solubilized by the addition of 1% fos-choline 14 (Anatrace) for 3 h at 4 °C. Solubilized membranes were clarified by centrifugation at 56,500 × g. M2 anti-FLAG resin (Sigma-Aldrich) was added to the supernatant and spun slowly on a blood wheel for 3 h at 4 °C. The resin was batch-washed in 0.5% fos-choline 14, packed into a drip column, washed with 25 column volumes of wash buffer with progressively reduced detergent concentrations to 0.03% fos-choline 14, and eluted with 0.2 mg/ml 3×FLAG peptide (Sigma-Aldrich). Elution fractions were combined, concentrated using a 100,000 molecular weight cut-off concentrator (Vivaspin), and separated by gel filtration on a Superdex 200 10/300 GL column (GE Healthcare). Peak fractions were combined, concentrated to ∼3–6 mg/ml, and used for functional studies.
86Rb+ flux assay was performed as reported previously (18) using liposomes made of 3:1 1-palmitoyl-2-oleoyl-3-phosphatidylethanolamine (POPE) and 1-palmitoyl-2-oleoyl-3-phosphatidylglycerol (POPG), respectively (mass percentage), and the desired phosphatidylinositol phosphates (PIPs) purchased from Avanti. Lipids were solubilized in buffer A (450 mm KCl, 10 mm HEPES, 4 mm N-methyl-d-glucamine, pH 7) with 35 mm CHAPS at 10 mg/ml and mixed with 5 μg of Kir2.1 or Kir2.2 protein/1 mg of lipid. Liposomes were formed by spinning through partially dehydrated Sephadex G-50 columns, first in buffer A and then in buffer B (450 mm sorbitol, 10 mm HEPES, 4 mm N-methyl-d-glucamine, pH 7). Uptake was initiated by adding 400 μl of buffer B with 1–5 μl of 1 μCi of 86Rb+ (PerkinElmer Life Sciences). At various time points, aliquots were flowed through 0.5-ml Dowex cation exchange columns, mixed with scintillation fluid, and counted on a liquid scintillation counter. Each sample was normalized to maximum uptake in valinomycin.
For patch clamping of giant liposomes, 30 μg of protein was solubilized with 2 mg of lipids, and liposomes were formed as described for flux assay. Giant liposomes were formed by a dehydration-rehydration cycle procedure (19). All recordings were done in symmetrical conditions of K-MOPS buffer. Membrane patches were voltage-clamped using a CV-4 headstage, an Axopatch 1-D amplifier, and a Digidata 1322A digitizer board (Axon Instruments). Patch pipettes were pulled from soda lime glass microhematocrit tubes (Kimble) to a resistance of ∼2–3 megaohms. Single channel data were digitized at a sampling rate of 10 kHz, and a low pass analog filter was set to 1 kHz. Analysis of single channel data was performed on the pClamp 9.2 software suite (Axon Instruments).
Tests for statistical significance were performed using an unpaired t test or one-way analysis of variance as appropriate, and statistical significance, reported by an asterisk in Fig. 2, indicates p < 0.05. Structural alignment of Kir2.2 and KirBac1.1 was performed in PyMOL.
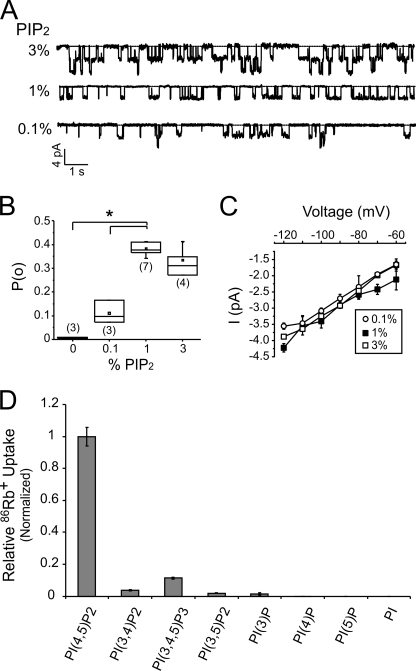
FIGURE 2.
Relationship between PIP2 and Kir2.1 open probability. A, representative continuous recordings of Kir2.1 currents at −100 mV from giant liposome inside-out excised patches composed of the indicated percentages of PIP2 and 25% POPG on a POPE background. B, box-and-whisker plot of measured Kir2.1 open probabilities in liposomes with the same compositions as in A. The mean open probabilities, represented by the square box, are 0.11 ± 0.03 for 0.1% PIP2, 0.38 ± 0.01 for 1% PIP2, and 0.34 ± 0.03 for 3% PIP2 (± S.E.). The number of recordings for each condition is indicated in brackets, the whiskers indicate data range, the box shows the upper and lower quartile values and median, and the asterisk indicates statistical significance (p < 0.05). C, current-voltage plot of Kir2.1 analyzed from the similar recordings as in A. The derived slope conductances are 35.0 ± 0.8 pS for 0.1% PIP2, 34.5 ± 1.5 pS for 1% PIP2, and 36.8 ± 1.5 pS for 3% PIP2 (± S.E.). D, 86Rb+ uptake of Kir2.1 in liposomes composed of 25% POPG, 74% POPE, and 1% of the indicated PI species. All counts were taken at 30 min in the time course and normalized to PIP2 (n = 3, ± S.E.).
RESULTS AND DISCUSSION
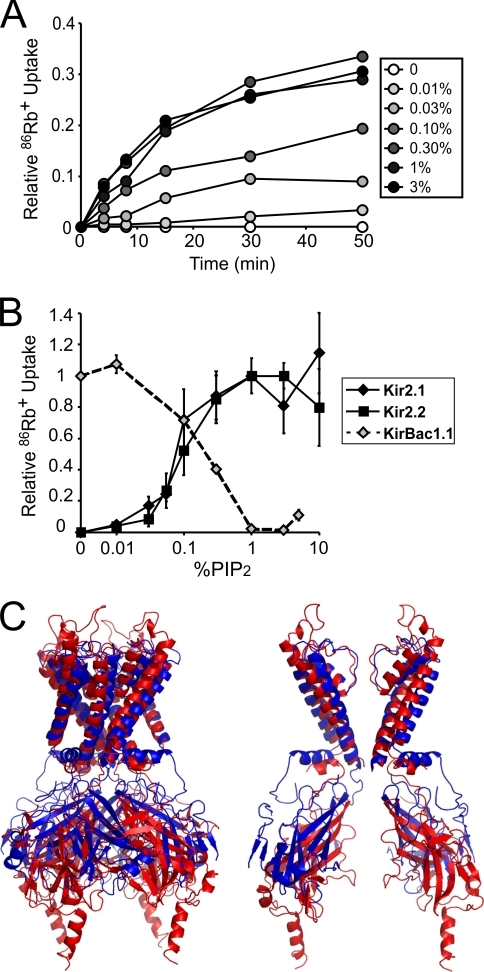
Human Kir2.1 and Kir2.2 proteins were expressed in S. cerevisiae, purified in milligram quantities as mono-dispersed tetrameric complexes, reconstituted in liposomes, and assayed for activity by 86Rb+ flux assay (17). All experiments were performed on liposomes composed of 3:1 POPE:POPG with replacement of POPE by varying amounts of PIPs, reported as a mass:mass ratio. Without PIP2 incorporated in the membrane, we detected no activity from either Kir2.1 or Kir2.2 channels, but there was a robust dose-dependent increase in channel-dependent 86Rb+ flux with incorporation of PIP2 (Fig. 1, A and B). Kir2.1 and Kir2.2 activity is detectable in as little as 0.01% PIP2 and appears to plateau at ∼1–3% (Fig. 1B). These data demonstrate that low levels of PIP2 are sufficient and necessary for channel activity on the POPE:POPG background. We have shown previously that the prokaryotic Kir channel, KirBac1.1, is also directly modulated by PIP2 in reconstituted liposomes, except that PIP2 has the opposite effect and inhibits the channel (Fig. 1B) (16, 19). Interestingly, the dynamic ranges of PIP2 modulation of Kir2.1, Kir2.2, and KirBac1.1 activity are almost identical (Fig. 1B), although Kir2.1 and Kir2.2 appear to be more sensitive. Indeed, Kir2 channels may be more sensitive to PIP2 than other Kir channels such as Kir3.1 (2).
FIGURE 1.
Reconstituted human Kir2.1 and Kir2.2 require PIP2 for activity. A, representative 86Rb+ uptake time courses of Kir2.1 in liposomes composed of 25% POPG and increasing percentages of PIP2 on a POPE background. B, activity/percentage PIP2 relationship for Kir2.1 (black diamonds, n = 4, ± S.E.), Kir2.2 (black squares, n = 3, ± S.E.), and KirBac1.1 (gray diamonds, n = 2, ± S.E.) in liposomes composed of 25% POPG and increasing percentages of PIP2 on a POPE background. 86Rb+ uptake counts were taken at 15 min in the time course for Kir2.1, 20 min for Kir2.2, and 4 min for KirBac1.1. Counts were normalized to the percentage of PIP2 of highest uptake for Kir2.1 and Kir2.2 and to uptake in no PIP2 for KirBac1.1. C, structural alignment of Kir2.2 (3JYC, red) and KirBac1.1 (1P7B, blue) based on the backbone atoms of the transmembrane domains. Left, complete tetramer. Right, for clarity of the pore, chain A and C transmembrane domains and chain B and D cytoplasmic domains.
To permit detailed voltage clamp analysis of the PIP2 dependence of channel activity, Kir2.1 was reconstituted in giant liposomes (19). Measured single channel currents have a unitary conductance (∼35 pS slope conductance) similar to Kir2.1 currents measured from heterologous or native expression systems (Fig. 2A) (20, 21). Increasing PIP2 over the range of concentrations expected in cell membranes increases open probability (Fig. 2B), with no effect on unitary conductance (Fig. 2C), consistent with previous reports of the effects of exogenous PIP2 incorporation into cellular membranes (21, 22). The PIP2 activity relationship plateaus at ∼1–3% PIP2 in both patch clamp and 86Rb+ flux assays (Figs. 1C and 2B). Interestingly, the open probability reaches a maximum of ∼0.4 at 1% PIP2 (Fig. 2B) over the range of −80 to −120 mV (data not shown), much lower than the high open probabilities (>0.9) previously reported in heterologous expression systems (21). This discrepancy might suggest that other lipid/protein modulators may be involved in activating Kir2.1 in cellular membranes.
We tested the effect of all PIP variants on Kir2.1 activity; Kir2.1 activation is highly selective for PIP2 (Fig. 2D). At 1%, PI(3,4,5)P3 activated Kir2.1 to ∼10% of activity induced by PIP2. PI(3,5)P2, PI(3,4)P2, and PI(3)P had very weak activating effects, whereas PI, PI(4)P, and PI(5)P did not activate the channel above background (Fig. 2D). These results are consistent with previous studies that suggested some of these PIP variants can activate Kir2.1 channels only at high concentrations, whereas others were unable to activate Kir2.1 channels (23).
Prior studies of PIP2 modulation of eukaryotic Kir channels have relied on cell-based systems, which pose a number of limitations. First, changing the lipid composition typically requires diffusing micelles onto a membrane patch, a poorly controlled method that provides no quantification of lipid incorporation into the membrane (24). Even perfusing water-soluble dioctanoyl PIP derivatives onto membranes permits only relative quantification of the effects of different lipids (25) because partitioning is not easily quantified. Second, because the exact composition is unknown, the influence of other protein or lipid modulators cannot be excluded (26). We have overcome these limitations by using reconstituted liposomes of defined lipid composition, permitting us to describe, for the first time, the dependence of a eukaryotic channel activity on membrane PIP2 level. The minimum range in which reconstituted Kir2.1 and Kir2.2 are modulated by PIP2 (Fig. 1B) lies well within physiological levels of PIP2 in eukaryotic plasma membranes (∼0.25–1%) (27, 28). Furthermore, this result definitively shows that Kir2.1 and Kir2.2 activity (and by extension probably activity of all eukaryotic Kir channels) can be directly stimulated by PIP2 in the membrane and that activation occurs without intermediary proteins. Kir channel activity may also be modulated by other lipids and cholesterol (29), and this can now be quantitatively addressed using the approach described.
It is striking that both human Kir channels (Kir2.1 and Kir2.2) and the prokaryotic KirBac1.1 are directly modulated by PIP2 when reconstituted in liposomes yet exhibit completely opposite effects; Kir2.1 and Kir2.2 are activated by PIP2 (Figs. 1, A and B, and 2), whereas KirBac1.1 is inhibited (Fig. 1B) (16, 19). We have suggested that such a marked difference in PIP2 action may be attributed to the absence of three highly conserved residues in the transmembrane-cytoplasmic linkers of prokaryotic KirBacs as compared with eukaryotic Kir channels (16, 19). This structural difference may result in intracellular displacement of the cytoplasmic domain of Kir2.2 as compared with KirBac1.1, as apparent in crystal structure alignments (Fig. 1C) (30, 31), and may affect interactions between the slide helix and cytoplasmic domain and perhaps therefore gating in these channels (32). The marked difference in PIP2 modulation between eukaryotic and prokaryotic Kir channels raises the interesting hypothesis that membrane lipids can drive the evolution of protein structure and function (32). PIP2 and other phosphoinositide species are typically absent in prokaryotes (33–35), an environment in which KirBac1.1 is active (18). The sensitivity of KirBac1.1 to inhibition by PIP2 may have been a byproduct of the evolution of regulatory domains for soluble ligands such as ATP. In eukaryotic membranes, where PIP2 is one of the most abundant PIPs and is primarily localized at the plasma membrane (27, 36), Kir channels could not retain the same dependence on PIP2 because KirBac1.1 would be almost completely inhibited at cellular PIP2 levels (16). Rather, because Kir channel activity either needed to be reduced, silenced during trafficking in intracellular organelles, or activated by localized changes in plasma membrane PIP2 (28, 37), eukaryotic Kir channels evolved to be highly sensitive to PIP2 for activation by relatively minor changes in channel sequence and structure. As multiple ion channels and transporters are regulated by PIP2 (7), it is possible that this same pattern and potential evolutionary mechanism is present between other proteins and their prokaryotic counterparts.
This work was supported, in whole or in part, by National Institutes of Health Grant HL54171 (to C. G. N.). This work was also supported by American Heart Association Fellowship 0810196Z (to W. W. L. C.).
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- POPE
- 1-palmitoyl-2-oleoyl-3-phosphatidylethanolamine
- POPG
- 1-palmitoyl-2-oleoyl-3-phosphatidylglycerol
- pS
- picosiemens.
REFERENCES
- 1.Fan Z., Makielski J. C. (1997) J. Biol. Chem. 272, 5388–5395 [DOI] [PubMed] [Google Scholar]
- 2.Huang C. L., Feng S., Hilgemann D. W. (1998) Nature 391, 803–806 [DOI] [PubMed] [Google Scholar]
- 3.Heginbotham L., Kolmakova-Partensky L., Miller C. (1998) J. Gen. Physiol. 111, 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma H. P., Saxena S., Warnock D. G. (2002) J. Biol. Chem. 277, 7641–7644 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt D., Jiang Q. X., MacKinnon R. (2006) Nature 444, 775–779 [DOI] [PubMed] [Google Scholar]
- 6.Suh B. C., Inoue T., Meyer T., Hille B. (2006) Science 314, 1454–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suh B. C., Hille B. (2008) Annu. Rev. Biophys. 37, 175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donaldson M. R., Jensen J. L., Tristani-Firouzi M., Tawil R., Bendahhou S., Suarez W. A., Cobo A. M., Poza J. J., Behr E., Wagstaff J., Szepetowski P., Pereira S., Mozaffar T., Escolar D. M., Fu Y. H., Ptácek L. J. (2003) Neurology 60, 1811–1816 [DOI] [PubMed] [Google Scholar]
- 9.Lopes C. M., Zhang H., Rohacs T., Jin T., Yang J., Logothetis D. E. (2002) Neuron 34, 933–944 [DOI] [PubMed] [Google Scholar]
- 10.Ashcroft F. M. (2005) J. Clin. Invest. 115, 2047–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., He C., Yan X., Mirshahi T., Logothetis D. E. (1999) Nat. Cell Biol. 1, 183–188 [DOI] [PubMed] [Google Scholar]
- 12.Shyng S. L., Cukras C. A., Harwood J., Nichols C. G. (2000) J. Gen. Physiol. 116, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soom M., Schönherr R., Kubo Y., Kirsch C., Klinger R., Heinemann S. H. (2001) FEBS Lett. 490, 49–53 [DOI] [PubMed] [Google Scholar]
- 14.Cukras C. A., Jeliazkova I., Nichols C. G. (2002) J. Gen. Physiol. 119, 581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stansfeld P. J., Hopkinson R., Ashcroft F. M., Sansom M. S. (2009) Biochemistry 48, 10926–10933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enkvetchakul D., Jeliazkova I., Nichols C. G. (2005) J. Biol. Chem. 280, 35785–35788 [DOI] [PubMed] [Google Scholar]
- 17.D'Avanzo N., Cheng W. W., Xia X., Dong L., Savitsky P., Nichols C. G., Doyle D. A. (2010) Protein Expr. Purif. 71, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enkvetchakul D., Bhattacharyya J., Jeliazkova I., Groesbeck D. K., Cukras C. A., Nichols C. G. (2004) J. Biol. Chem. 279, 47076–47080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng W. W., Enkvetchakul D., Nichols C. G. (2009) J. Gen Physiol. 133, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu G. X., Derst C., Schlichthörl G., Heinen S., Seebohm G., Brüggemann A., Kummer W., Veh R. W., Daut J., Preisig-Müller R. (2001) J. Physiol. 532, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie L. H., John S. A., Ribalet B., Weiss J. N. (2008) J. Physiol. 586, 1833–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shyng S. L., Nichols C. G. (1998) Science 282, 1138–1141 [DOI] [PubMed] [Google Scholar]
- 23.Rohács T., Lopes C. M., Jin T., Ramdya P. P., Molnár Z., Logothetis D. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilgemann D. W. (2004) Science 304, 223–224 [DOI] [PubMed] [Google Scholar]
- 25.Rohács T., Lopes C., Mirshahi T., Jin T., Zhang H., Logothetis D. E. (2002) Methods Enzymol. 345, 71–92 [DOI] [PubMed] [Google Scholar]
- 26.Suh B. C., Hille B. (2005) Curr. Opin. Neurobiol. 15, 370–378 [DOI] [PubMed] [Google Scholar]
- 27.Nasuhoglu C., Feng S., Mao J., Yamamoto M., Yin H. L., Earnest S., Barylko B., Albanesi J. P., Hilgemann D. W. (2002) Anal. Biochem. 301, 243–254 [DOI] [PubMed] [Google Scholar]
- 28.Hilgemann D. W. (2007) Pflugers Arch. 455, 55–67 [DOI] [PubMed] [Google Scholar]
- 29.Rosenhouse-Dantsker A., Leal-Pinto E., Logothetis D. E., Levitan I. (2010) Channels 4, 63–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo A., Gulbis J. M., Antcliff J. F., Rahman T., Lowe E. D., Zimmer J., Cuthbertson J., Ashcroft F. M., Ezaki T., Doyle D. A. (2003) Science 300, 1922–1926 [DOI] [PubMed] [Google Scholar]
- 31.Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D'Avanzo N., Cheng W. W., Wang S., Enkvetchakul D., Nichols C. G. (2010) Channels 4, 139–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raetz C. R., Dowhan W. (1990) J. Biol. Chem. 265, 1235–1238 [PubMed] [Google Scholar]
- 34.Dowhan W., Bogdanov M., Mileykovskaya E. (2008) Biochemistry of Lipids, Lipoproteins and Membranes, Fifth Ed., Elsevier, Amsterdam [Google Scholar]
- 35.Phung L. V., Tran T. B., Hotta H., Yabuuchi E., Yano I. (1995) Microbiol. Immunol. 39, 105–116 [DOI] [PubMed] [Google Scholar]
- 36.Behnia R., Munro S. (2005) Nature 438, 597–604 [DOI] [PubMed] [Google Scholar]
- 37.Shen W., Tian X., Day M., Ulrich S., Tkatch T., Nathanson N. M., Surmeier D. J. (2007) Nat. Neurosci. 10, 1458–1466 [DOI] [PubMed] [Google Scholar]