Abstract
Nucleophosmin (NPM1) is a nucleocytoplasmic shuttling phosphoprotein, mainly localized at nucleoli, that plays a key role in ribogenesis, centrosome duplication, and response to stress stimuli. Mutations at the C-terminal domain of NPM1 are the most frequent genetic lesion in acute myeloid leukemia and cause the aberrant and stable translocation of the protein in the cytoplasm. The NPM1 C-terminal domain was previously shown to bind nucleic acids. Here we further investigate the DNA binding properties of the NPM1 C-terminal domain both at the protein and nucleic acid levels; we investigate the domain boundaries and identify key residues for high affinity recognition. Furthermore, we demonstrate that the NPM1 C-terminal domain has a preference for G-quadruplex forming DNA regions and induces the formation of G-quadruplex structures in vitro. Finally we show that a specific sequence found at the SOD2 gene promoter, which was previously shown to be a target of NPM1 in vivo, is indeed folded as a G-quadruplex in vitro under physiological conditions. Our data extend considerably present knowledge on the DNA binding properties of NPM1 and suggest a general role in the transcription of genes characterized by the presence of G-quadruplex forming regions at their promoters.
Keywords: Cell Cycle, Nucleolus, Protein DNA-Interaction, Protein Structure, Surface Plasmon Resonance (SPR), Acute Myeloid Leukaemia, B23, G-quadruplex, NPM1
Introduction
Nucleophosmin (also known as NPM1, B23, numatrin, and hereby termed NPM1)3 is an abundant phosphoprotein that was originally identified as a non-ribosomal nucleolar protein playing a key role in ribosome biogenesis: NPM1 is able to bind RNA as well as DNA, and has intrinsic RNase activity that preferentially cleaves pre-rRNA (1–4). NPM1 is also involved in the regulation of the important tumor suppressors p53 (5) and p14arf (6, 7) and therefore mutations at the NPM1 locus or aberrant localization of NPM1 may interfere with p53 or p14arf transcriptional programs and activities. Furthermore, NPM1 plays more than one role outside the nucleolus, including the control of centrosome duplication (8). This function is confirmed by analysis of NPM1 knock-out mice showing unrestricted centrosome duplication, genomic instability, and mid-gestation embryonic lethality (9). The multiple cellular functions of NPM1 inside and outside the nucleolus are due to its chaperone activity and ability to shuttle between the nucleus and cytoplasm (10, 11). These properties are in turn dictated by its modular organization. In fact, several distinct, although partially overlapping, functional domains and signatures have been identified in NPM1 (12, 13).
NPM1 was first identified in 2005 as the most frequently mutated gene in acute myeloid leukemia, its mutations being observed in about 30% of cases (14) that display distinctive molecular and clinical features (13, 15, 16). Mutations are all localized at the C-terminal end of the protein (exon 12) and invariably result in the aberrant cytoplasmic localization of NPM1 (17). More than 40 alterations have been detected and were found to be mutually exclusive with major chromosomal abnormalities (13, 14). NPM1 mutations, which are typically heterozygous, have similar consequences on the mutated protein. The reading frame is altered, due to duplication of short base sequences leading to a protein longer by 4 residues and with a sequence different from the wild type in the last 7 residues. As a result, the novel C-terminal sequence in the mutated proteins lacks the wild-type nucleolar localization signal and acquires a newly formed, Crm1-dependent, nuclear export signal (LXXXVXXVXL) (17, 18). Furthermore, the protein C-terminal domain appears greatly destabilized and partially or totally unfolded, due to alteration of the protein hydrophobic core (19–21). Both of these consequences at the protein level concur to determine the aberrant and stable cytoplasmic localization of the protein in the cytosol (17, 18).
The NPM1 C-terminal domain is responsible for the nucleic acid binding activity of the protein (Fig. 1A). NPM1 was shown to bind both DNA and RNA oligos with a preference for single-stranded structures over double-stranded DNA B structures, in a sequence-unrelated manner (3, 22, 23). Therefore, a role for NPM1 as a single-stranded binding protein was proposed (22), a property that may be linked to the export of ribosome subunits from the nucleus (17). Recently, however, the protein was also identified as a cofactor in the transcriptional activation of the mitochondrial superoxide dismutase 2 (SOD2) gene; in particular it was shown that full-length NPM1 binds a G-rich region at the SOD2 promoter (24). Because leukemic mutant NPM1 is stably localized in the cytoplasm, we decided to further investigate the DNA binding properties of the protein, assuming that some important function related to nucleic acid binding in the nucleus might be impaired in these mutants.
FIGURE 1.
Protein constructs and DNA oligos used in this study. A, sequence of the C-terminal domain of NPM1. The region differentiating NPM1-C70 from NPM1-C53 (whose three-dimensional structure is shown in the inset) is underlined. Mutated lysines are shown in bold. The PSIPRED prediction is also shown (C = coil, H = α-helix, E = β-strand). B, oligos used in this study are shown both in sequence and in putative structure.
In this work we use different DNA sequences, protein constructs, and site-directed mutants to investigate the NPM1 C-terminal domain boundaries involved in DNA binding, assess the role of individual residues in binding, and analyze the effect of DNA and stabilizing salts on NPM1 folding. Importantly, we show that NPM1, whereas being able to recognize with low affinity any DNA oligo tested, binds with high affinity and is able to induce the folding of G-rich sequences that form three-dimensional structures known as G-quadruplexes. These include the above mentioned G-rich sequence at the SOD2 gene promoter, which we demonstrate here is indeed folded as a G-quadruplex under physiological conditions. Taken together these data extend our knowledge on the DNA binding properties of NPM1 and suggest that future efforts should be paid to identify G-quadruplex forming DNA regions that are recognized by NPM1 in vivo.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
NPM1-C53 was expressed and purified as previously reported (20).
A gene construct for the expression of NPM1-C70 was obtained through gene synthesis (GeneArt, Regensburg, Germany) and cloned into expression vector pET28a(+) (Novagen, San Diego, CA) between restriction sites NdeI and BamHI. Escherichia coli cells, strain BL21(DE3), transformed with the expression vector, were grown to A600 0.5 in LB medium supplemented with kanamycin at 37 °C. At this point 1 mm isopropyl 1-thio-β-d-galactopyranoside was added and cells were further grown at 20 °C for 16 h. Cells were harvested, resuspended in lysis buffer (20 mm Hepes, pH 7.0, 150 mm NaCl, 5 mm β-mercaptoethanol, and 20 mm imidazole) and sonicated. After centrifugation, the supernatant was loaded on a nickel-nitrilotriacetic acid column, pre-equilibrated with lysis buffer. Protein was eluted using a linear gradient of lysis buffer plus imidazole (20 mm to 1.0 m). Fractions containing the protein, as judged from SDS-PAGE, were collected, diluted 10-fold with lysis buffer, and incubated with thrombin (0.5 units/mg of protein) for 1.5 h at 4 °C. After thrombin cleavage the reaction mixture was supplemented with protease inhibitors (Roche Applied Science) and loaded on a nickel-nitrilotriacetic acid column, pre-equilibrated with lysis buffer, to remove the thrombin cleaved N-terminal His tag and uncleaved protein. Protein was recovered from the flow-through, diluted 10-fold with 20 mm Hepes, pH 7.0, 5 mm β-mercaptoethanol, and loaded on a SP-Sepharose column pre-equilibrated with the same buffer. Protein was eluted using a NaCl linear gradient, buffer was exchanged to remove the salt, concentrated up to 40 mg/ml in 20 mm Hepes buffer, pH 7.0, 5 mm β-mercaptoethanol, and stored at −80 °C.
Mutagenesis
NPM1-C70 protein variants were obtained through site-directed mutagenesis and using the QuikChange II kit (Stratagene, La Jolla, CA), following the manufacturer's instructions. Forward oligos used for PCR are the following: K229A, 5′-CATATGCAGGAAAGCTTCgcgAAACAGGAAAAAACGCCGAAAACC-3′; K230A, 5′-CATATGCAGGAAAGCTTCAAAgcgCAGGAAAAAACGCCGAAAACC-3′; K229A/K230A, 5′-CATATGCAGGAAAGCTTCgcggcgCAGGAAAAAACGCCGAAAACCCCGAAAGGCCCG-3′; K233A, 5′-CAGGAAAAAACGCCGgcgACCCCGAAAGGCCCGAGC-3′; K236A, 5′-CCGAAAACCCCGgcgGGCCCGAGCAGCGTG-3′; and K239A, 5′-GAAAGCTTCAAAAAACAGGAAgcgACGCCGAAAACCCCGAAAGGC-3′. NPM1-C70 protein variants were expressed and purified as the wild-type construct.
Oligonucleotides
Oligonucleotides used in this study (Fig. 1B) were purchased from PRIMM s.r.l (Milan, Italy) and purified by HPLC. Oligonucleotides used for SPR analysis were biotinylated at the 5′-end. Prior to use, lyophilized oligos were resuspended in the appropriate buffer (20 mm Hepes, pH 7.0, with or without 100 mm NaCl or KCl according to the different experiments), quantified spectrophotometrically and annealed. For annealing, oligos were heated to 95 °C for 15 min and slowly cooled down at RT, overnight.
Surface Plasmon Resonance
The interactions between biotinylated DNA constructs (ligands) with purified proteins NPM1-C70 or NPM1-C53 (analytes) as well as with the porphyrin TmPyP4 (analyte), were all measured using the SPR technique and a Biacore X100 instrument (Biacore, Uppsala, Sweden). Each biotinylated DNA construct was immobilized on a Sensor Chip SA, pre-coated with streptavidin from Biacore AB. The capturing procedure on the biosensor surface was performed according to the manufacturer's instructions and setting the aim for ligand immobilization to 1000 response units. Running buffer was Hepes-buffered saline-EP, which contains 10 mm Hepes, pH 7.4, 0.15 m NaCl, 3 mm EDTA, 0.005% (v/v) Surfactant P20 (Biacore AB). Analytes were dissolved in running buffer, and binding experiments were performed at 25 °C with a flow rate of 30 μl/min. The association phase (kon) was followed for 180 s, whereas the dissociation phase (koff) was followed for 300 s. The complete dissociation of active complex formed was achieved by addition of 10 mm Hepes, 2 m NaCl, 3 mm EDTA, 0.005% (v/v) P20, pH 7.4, for 60 s before each new cycle start. Analytes were tested in a wide range of concentrations to reach at least a 2-fold increase from lower concentration tested. When experimental data met quality criteria, kinetic parameters were estimated according to a 1:1 binding model using Biacore X100 Evaluation Software. Conversely, an affinity steady state model was applied to fit the data. In this latter case two possible situations were exploited: (a) single binding site, using the equation y = Rmax × [analyte]/([analyte] + KD); and (b) double independent binding site, using the equation y = Rmax × [analyte]/([analyte] + KD) + Rmax1 × [analyte]/([analyte] + KD1).
Circular Dichroism
All circular dichroism experiments were performed using a Jasco J710 instrument (Jasco Inc., Easton, MD) equipped with a Peltier apparatus for temperature control. Static spectra of G10-loop and c-MYC oligos were collected at 25 °C, using oligos annealed in the appropriate buffer and concentrated to 20 μm. Spectra were collected using a quartz cell with 1-mm optical path length (Hellma, Plainview, NY) and a scanning speed of 100 nm/min. The reported spectra are the average of five scans. To measure NPM1-C70 induced G-quadruplex formation, oligos dissolved in buffer (without additional salts) but not annealed, and concentrated to 20 μm, were incubated for 1 h with appropriate amounts of NPM1-C70 and spectra collected as above. To monitor NPM-C70 induced unfolding, oligos were annealed in Hepes buffer supplemented with 100 mm NaCl and incubated with NPM1-C70 increasing amounts. The spectral contribution of buffers and proteins was subtracted as appropriate. To measure the effect of the c-MYC and G10-loop oligos on NPM1-C70 structure, CD spectra of the protein (20 μm) in 20 mm Hepes, pH 7.0, alone or incubated with pre-annealed oligos (20 μm) were recorded. The spectral contribution of the oligo alone was then recorded and subtracted as appropriate. Thermal denaturation experiments were performed using a quartz cell with 1-mm optical path length and monitoring the variation of CD signal at 260 nm. Temperature was progressively increased, in 1 °C/min steps, from 25 to 105 °C. The Kaleidagraph software was used for CD spectra analysis and representation.
Urea-induced Protein Denaturation
NPM1-C70 and NPM1-C53 urea-induced denaturations were followed monitoring the change in CD signal at 222 nm, in the presence of increasing amounts of urea. The same experiments were performed in the absence or presence of NaCl 0.25, 0.5, and 1.0 m, respectively. Data were plotted and analyzed using Kaleidagraph software. A simple two-state model was used in all cases to fit the data according to Equation 1,
where ΔGd is the free energy of folding at a concentration D of denaturant, mD-N is the slope of the transition (proportional to the increase in the surface-accessible area on going from the native to the denatured state), and D½ is the midpoint of the denaturation transition. An equation that takes into account the pre- and post-transition baselines was used (25).
RESULTS
Surface Plasmon Resonance Analysis of NPM1-DNA Interaction
Full-length NPM1 is able to interact with a G-rich sequence found at the SOD2 promoter and to function as a cofactor in transcriptional activation (24). This region is predicted to form a hairpin with a five-paired base stem and a stretch of 10 consecutive guanines in the loop (hereby named G10-loop; Fig. 1B). Our first aim was to determine whether this DNA binding activity could be mapped to the C-terminal domain of the protein. To this end, a biotinylated version of the G10-loop was immobilized on a streptavidin chip and used as bait in SPR analysis. Two different constructs of the NPM1 C-terminal domain were used as the analytes: NPM1-C70 comprising the last 70 C-terminal residues, and NPM1-C53 comprising the last 53 residues (Fig. 1A). The first construct was chosen because previous binding data on truncated proteins mapped the NPM1 nucleic acid binding activity to this segment (3), whereas the NPM1-C53 construct was prepared because it is the minimal folding unit of the C-terminal domain (20) that, as established by NMR data, is made of three helices (Fig. 1A) (19).
When testing NPM1-C70 as the analyte we obtained a KD = 7.2 μm (Fig. 2A, Table 1), whereas with NPM1-C53 we obtained a KD = 169 μm (Fig. 2, B and C, Table 1). Interestingly, both the association and dissociation rate constants could be determined when NPM1-C70 was the analyte (Fig. 2A), whereas they were too rapid to be determined with NPM1-C53 (Fig. 2B). This result suggests that the nucleic acid binding site is altered in the shorter protein construct and that, in this case, binding is mainly dictated by electrostatic interactions of the positively charged protein domain (Fig. 1A) with the negatively charged G10-loop.
FIGURE 2.
SPR sensorgrams of the interaction between G10-loop and NPM1 constructs. The oligo, biotinylated at its 5′ end, was immobilized on a SA (streptavidin) sensor chip. A, experimental curves (dashed lines with circles) represent different concentrations of NPM1-C70 used as the analyte and were fitted according to a single exponential binding model with 1:1 stoichiometry (continuous lines). B, experimental curves represent different concentrations of NPM1-C53. Both on and off rates were too rapid to be fitted. C, Scatchard plot used to determine the dissociation constant with NPM1-C53.
TABLE 1.
Dissociation constants and kinetic parameters as determined by SPR analysis
| Ligand | Analyte | KD | kon | koff |
|---|---|---|---|---|
| μm | m−1s−1 | s−1 | ||
| G10-loop | NPM1-C70 | 7.2 ± 0.2 | 2.8 ± 0.1 × 102 | 1.98 ± 0.03 × 10−3 |
| NPM1-C53 | 169 ± 5 | |||
| T10-loop | NPM1-C70 | 307 ± 7 | ||
| NPM1-C53 | 1170 ± 40 | |||
| G5-loop | NPM1-C70 | 40 ± 3 | ||
| NPM1-C53 | 224 ± 6 | |||
| Poly(T) | NPM1-C70 | 120 ± 26 | ||
| NPM1-C53 | 520 ± 30 | |||
| Poly(G) | NPM1-C70 | 5.8 ± 0.8 | ||
| NPM1-C53 | 31 ± 4 | |||
| c-Myc | NPM1-C70 | 1.9 ± 0.1 | 7.4 ± 0.3 × 103 | 1.37 ± 0.03 × 10−2 |
| NPM1-C53 | 82 ± 15 |
Next, we wanted to clarify the structural properties of the DNA hairpin necessary for binding NPM1. To this end we first tested a so-called T10-loop, where the 10 guanines at the loop of the G10-hairpin are replaced by 10 thymines, whereas all the other bases are conserved (Fig. 1B). With NPM1-C70 we obtained a KD = 307 μm that increased to KD = 1.17 mm with NPM1-C53 (Table 1). This experiment indicates that a sequence that forms a hairpin structure resembling that of the native G10-loop but with thymines instead of guanines at the loop is poorly recognized, with a 42-fold lower affinity. NPM1-C53 is poorly competent for binding.
To explore the dimensional requirements of the hairpin for high affinity recognition, we next immobilized a G5-loop, which maintains the same hairpin arrangement of the G10-loop but with only five guanines in the loop (Fig. 1B). With this oligo, we obtained a KD = 40 μm with NPM1-C70 and 224 μm with NPM1-C53. These experiments indicate that both the presence of guanines at the hairpin loop and their number contribute to the global affinity, suggesting that the three-dimensional structure of the G10-loop plays an important role.
To further assess the DNA binding properties of NPM1, we next tested an oligo made only of T-bases (38-mer), hereby named poly(T) (Fig. 1B), and obtained a KD = 120 μm with NPM1-C70 and a KD = 520 μm with NPM1-C53 (Table 1). This suggests that the increased flexibility of the poly(T) linear oligo with respect to the T10-loop can partly compensate for the absence of guanines.
A further experiment was designed to establish the role played by guanines in a DNA sequence that does not natively form hairpin structures. We thought that if the protein preferentially recognizes with high affinity a hairpin loop made of guanines, it might also be able to induce such loop formation in a poly(G) oligo and recognize it with good affinity (Fig. 1B). Thus with poly(G) we expected to find a KD for NPM1-C70 higher than that found with the G10-loop but lower than that found with the poly(T) oligo.
To our surprise, with NPM1-C70 we obtained a KD = 5.8 μm, comparable with that of the physiological substrate G10-loop; with NPM1-C53 we obtained instead a KD = 31 μm, the lowest measured so far with this protein construct (Table 1). These results suggest that the recognition of a poly(G) oligo is far more specific than we might have anticipated. This may be rationalized by hypothesizing that this oligo is not linear but has the potential to form a structure that resembles that of the G10-loop. Indeed, by inspecting the literature, we realized that a poly(G) oligo, if long enough, such as our 38-mer (Fig. 1B), has the potential to form three-dimensional structures known as G-quadruplexes (26, 27).
NPM1 C-terminal Domain Binds a Sequence Known to Form a G-quadruplex
G-quadruplexes are formed by sequences displaying at least four stretches of at least three guanines, with no sequence requirements for the intervening loops that are usually one to seven nucleotides long (G3N1–7G3N1–7G3N1–7G3) (26, 28). The guanines interact with each other in an arrangement different from the classical B DNA pairing, forming planar tetrads stabilized by the so-called Hoogsteen type H-bonds (27, 28). These structures are greatly stabilized by Na+ or K+ ions that intercalate in the rings formed by the four guanines in the tetrad. Repetitive G-rich sequence stretches are highly represented in the human genome and are clustered at gene promoters, suggesting their functional importance (27, 28).
We decided to investigate whether one well characterized G-quadruplex might interact with our NPM1 constructs. We focused our attention on a sequence contained in the NHEIII (nuclease hypersensitive element III) region of the c-Myc promoter that is known to regulate up to 90% of total c-MYC expression. This is a well characterized example of a parallel G-quadruplex forming region, both in vitro and in vivo (29). An oligo representing the G-quadruplex forming region of this promoter (hereby c-MYC oligo) was therefore immobilized on a chip for SPR analysis (Fig. 1B).
Interestingly, we found that NPM1-C70 binds the c-MYC quadruplex with a KD = 1.9 μm (Fig. 3A and Table 1), confirming that NPM1-C70 has high affinity for a sequence that adopts a G-quadruplex structure. Similarly to the G10-loop, and contrary to the other oligos, both the association and dissociation rates could be determined, suggesting that c-MYC G-quadruplex recognition is specific. Moreover, the experiment performed using NPM1-C53 as the analyte led to a KD = 82 μm, confirming reduced affinity with this shorter domain (Fig. 3, B and C, and Table 1).
FIGURE 3.
SPR sensorgrams of the interaction between c-MYC and NPM1 constructs. A, experimental curves (dashed lines with circles) represent different concentrations of NPM1-C70 used as the analyte and were fitted according to a single exponential binding model with 1:1 stoichiometry (continuous lines). B, experimental curves represent different concentrations of NPM1-C53. C, Scatchard plot referring to curves in panel B for dissociation constant determination.
The G10-loop Forms a G-quadruplex Structure in Vitro
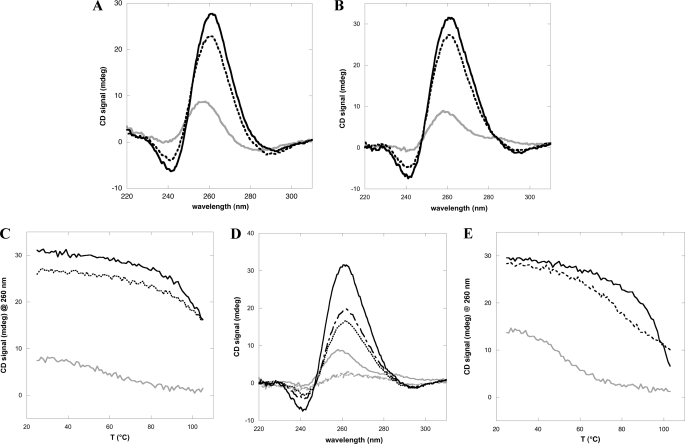
The results reported so far suggest that the NPM1 C-terminal domain recognizes with particularly high affinity sequences known to form G-quadruplex three-dimensional arrangements. Under this light, it is interesting to note that the G10-loop sequence, which is predicted to form a hairpin according to conventional Watson-Crick pairing (Fig. 1B), also matches the above mentioned folding rule for G-quadruplexes and, accordingly, is predicted to form a G-quadruplex by the Quadparser algorithm (26). Therefore, when annealing, this oligo has the potential to form at least two alternative structures. To infer which of these two structures is the most likely to be populated, we first collected the CD spectra of G10-loop and c-MYC, for comparison. It is well known that circular dichroism is diagnostic of G-quadruplex formation (28). In particular, by comparing the spectra of the c-MYC oligo annealed in the absence or presence of 100 mm NaCl or 100 mm KCl, respectively, we observe a red-shift and increase in intensity of the peak at around 260 nm and the formation of a through at 240 nm (Fig. 4A). Importantly the same features are observed in the case of the G10-loop spectra (Fig. 4B). These variations are both considered hallmarks of parallel G-quadruplex formation (28, 30). A second indication of a G-quadruplex structure for the G10-loop is derived from denaturation experiments. In Fig. 4C the thermal melting profiles of G10-loop in the 25–105 °C interval are shown, whereas their corresponding static spectra are reported in Fig. 4D. These data indicate that the melting transition is still not complete at 105 °C for the G10-loop in the presence of 100 mm KCl or NaCl, whereas a poorly cooperative transition with a Tm centered at around 65–70 °C is observed when the same experiment is performed in the absence of monovalent cations. The predicted melting temperature for a G10-loop adopting hairpin structure is Tm = 69.1 °C in 100 mm monovalent cations, according to the mfold server. Conversely the Quadpredict algorithm predicts higher Tm values, i.e. 94.9 and 77.1 °C, in the presence of 100 mm KCl or NaCl, respectively. These higher values are in better agreement with our experimental data, suggesting a G-quadruplex structure in these conditions. Thermal melting profiles were also collected for the c-MYC oligo (Fig. 4E), as a control, and found to be similar to those obtained with the G10-loop oligo (Fig. 4C).
FIGURE 4.
G10-loop adopts a G-quadruplex fold in vitro. A, CD spectra of the c-MYC oligo annealed in the absence (gray continuous line) or presence of 100 mm NaCl (dashed line) or 100 mm KCl (black continuous line), respectively. A red shift of the peak at 260 nm and the formation of a through at 240 nm is observed. B, CD spectra of the G10-loop oligo. The same features observed with c-MYC oligo are observed. The line format is the same as in A. C, thermal denaturation profiles of G10-loop in the 25–105 °C interval and in the absence (gray continuous line) or presence of 100 mm NaCl (dashed line) or 100 mm KCl (black continuous line). D, CD spectra of the G10-loop in the absence (gray continuous, gray dashed dotted, and gray dotted at 25, 95, and 105 °C, respectively) or presence of 100 mm KCl (black continuous, black dashed dotted, and black dotted at 25, 95, and 105 °C, respectively). Spectra at 95 and 105 °C in the absence of salts are the same, whereas in the presence of 100 mm KCl, they are different, indicating that the melting transition is not yet complete in the latter case. E, thermal denaturation profiles of c-MYC oligo in the 25–105 °C interval, collected in the absence (gray continuous line) or presence of 100 mm NaCl (dashed line) or 100 mm KCl (black continuous line).
Finally we analyzed the binding of the porphyrin TmPyP4 to our oligos, by means of SPR. This molecule is known to bind G-quadruplex structures with high affinity and a complex stoichiometry involving at least two binding sites with different affinities (31–33). The interaction of TmPyP4 with poly(T) oligo (Fig. 5A) determines a series of sensorgrams that, in the concentration range explored, always reach equilibrium before the end of the contact time between analyte and ligand, allowing both kinetic analysis, with a simple 1:1 model interaction (Fig. 5A), and the construction of a Scatchard plot (Fig. 5B). The average value for this double determination is KD = 345 ± 15 nm and is compatible with the presence of a single binding site (Table 2). When analyzing the T10-loop data, again we observed a single binding site with a lower dissociation constant (KD = 35 ± 15 nm) (Fig. 5, C and D, and Table 2). Conversely, when analyzing TmPyP4 binding to the c-MYC oligo (Fig. 5, E and F) a more complex behavior was observed, depending on the TmPyP4 concentration. At low concentrations (bottom curves of Fig. 5E) the dissociation phase is characterized by a single process corresponding to the off-rate constant of a high affinity binding site. By increasing the TmPyP4 concentration, the off-rate becomes clearly biphasic (upper curves of Fig. 5E) reflecting the titration of a second binding site with lower affinity. Moreover, not all the traces reach the equilibrium, so that the corresponding Scatchard plot is determined for a subpopulation of the porphyrin concentrations studied. As a result, the equilibrium analysis performed via the Scatchard plot is not compatible with a single binding site (see the dashed line in Fig. 5F), whereas an excellent agreement is obtained assuming two independent binding sites (solid line in Fig. 5F and see Table 2 for the corresponding KD values). Importantly, the experiments performed with the G10-loop indicated that TmPyP4 binding to this oligo follows the same behavior as observed with c-MYC, with the same stoichiometry and similar dissociation constants for the high and low affinity sites, respectively (Fig. 5, G and H, and Table 2). In conclusion, TmPyP4 binding data obtained with the G10-loop are in agreement with those obtained with c-MYC and with previous work on G-quadruplex forming oligos (31–33), whereas data obtained with the hairpin T10-loop are not. Taken together, our results suggest that, in the presence of physiological amounts of monovalent cations, the G10-loop folds as a G-quadruplex in vitro and might also have this structure when recognized in vivo by NPM1 (24).
FIGURE 5.
SPR sensorgrams of the interaction between the porphyrin TmPyP4 and different oligos (panels A, C, E, and G for poly(T), T10-loop, c-MYC, and G10-loop, respectively) and their corresponding Scatchard plots (panels B, D, F, and H). A, poly(T), from bottom to top of the panel [TmPyP4] = 1600, 800, 400, 200, 100, 50, and 25 nm. Experimental data are reported as dashed lines with circles (where the concentration was measured twice, also as dashed line with squares); best fit assuming a 1:1 interaction is reported as a solid line. B, poly(T), Scatchard plot, open circles are experimental data points and the solid line is the fit assuming a single binding site. C, T10-loop, from bottom to top of the panel [TmPyP4] = 1600, 800, 400, 200, 100, 50, and 25 nm. Symbols used are the same as in panel A. D, T10-loop, Scatchard plot, the continuous line represents the best fit according to 1:1 interaction model. E, c-MYC, from bottom to top of the panel [TmPyP4] = 1600, 800, 400, 200, 100, 50, 25, 12.5, 6.25, and 3.125 nm. Experimental data are reported as solid lines. F, c-MYC, concentrations used in the Scatchard plot are [TmPyP4] = 1600–25 nm. Open circles are experimental data points and the dashed line represents the best fit assuming a single binding site, whereas the solid line is the best fit assuming two independent binding sites. G, G10-loop, from bottom to top of the panel [TmPyP4] = 1600, 800, 400, 200, 100, 62.5, 31.25, 25, and 15.625 nm. Experimental data are reported as solid lines. H, G10-loop, the concentrations used in the Scatchard plot are [TmPyP4] = 1600–31.25 nm. Symbols used are the same as in panel F.
TABLE 2.
SPR analysis of TmPyP4 binding to selected DNA oligos
| Ligand | KD | kon | koff |
|---|---|---|---|
| m | m−1s−1 | s−1 | |
| Poly T | 330.9 ± 0.4 × 10−9 (Kinetics) | 1.013 ± 0.001 × 108 | 33.52 ± 0.02 |
| 360 ± 30 × 10−9 (Scatchard) | |||
| 345 ± 15 × 10−9 (average value) | |||
| T10-loop | 20 ± 10 × 10−9 (Kinetics) | 2.9 ± 0.8 × 1010 | 6 ± 2 × 102 |
| 50 ± 8 × 10−9 (Scatchard) | |||
| 35 ± 15 × 10−9 (average value) | |||
| c-Myc | Site 1: 8 ± 14 × 10−9 Site 2: 1.3 ± 0.2 × 10−6 | ||
| G10-loop | Site 1: 26 ± 3 × 10−9 Site 2: 1.9 ± 0.9 × 10−6 |
NPM1-C70 Stimulates the Formation of G-quadruplex Structures
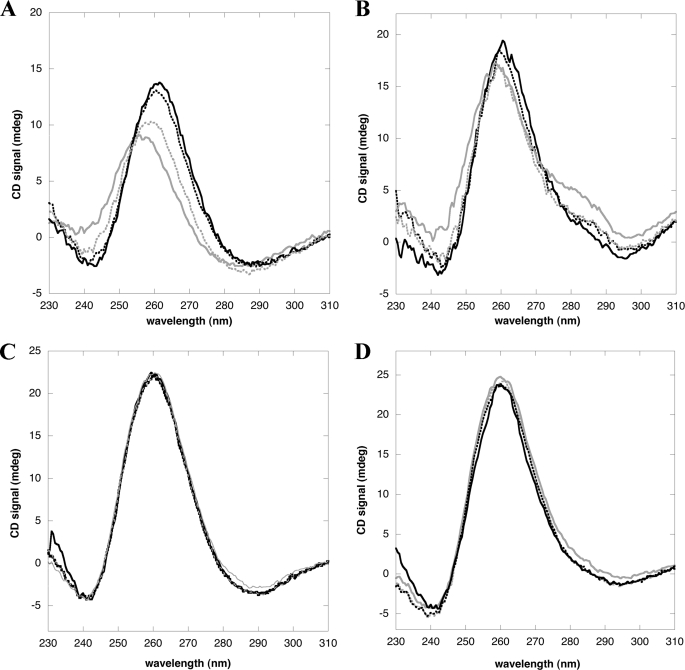
Having established that NPM1-C70 binds with high affinity preformed G-quadruplex structures, we next investigated whether it is able to induce G-quadruplex formation in vitro. In Fig. 6A we report CD spectra of the not annealed c-MYC oligo titrated with NPM1-C70, in the absence of monovalent ions. By progressively increasing the amount of NPM1-C70 we observe, once again, a red-shift and increase in signal of the 260 nm peak and the progressive formation of a through at 240 nm, which indicates that a G-quadruplex is formed upon protein binding. The same effect, albeit less evident, is obtained when using the G10-loop oligo (Fig. 6B). In a mirror experiment, using pre-annealed c-MYC or G10-loop oligos (Fig. 6, C and D, respectively) we checked whether NPM1-C70 might have the property to destabilize pre-formed G-quadruplex structures. In both cases increasing amounts of protein had no effect on the CD signal of oligos pre-annealed in the presence of monovalent cations. Thus NPM1-C70 is able to induce G-quadruplex formation in unstructured oligos, whereas it does not unwind pre-structured oligos.
FIGURE 6.
NPM1-C70 is able to promote the formation of G-quadruplex arrangement in unstructured oligos, whereas it does not unfold G-quadruplexes. A, not annealed c-MYC oligo (20 μm) in the absence (gray solid line) and presence of increasing amounts of NPM1-C70 (gray dots, black dots, and black solid line for 10, 20, and 30 μm, respectively); the red shift and increase in amplitude at 260 nm and the formation of a through at 240 are indicative of G-quadruplex formation. B, not annealed G10-loop oligo (20 μm) in the absence (gray solid line) or presence of increasing amounts of NPM1-C70 (gray dots, black dots, and black solid line for 70, 100, and 150 μm, respectively). C, spectra of c-MYC oligo (20 μm), pre-annealed in the presence of 100 mm NaCl, and incubated with increasing amounts of NPM1-C70 (same amounts as in panel A). D, same experiment as in panel C performed using the G10-loop (20 μm). Amounts of NPM1-C70 are the same as in panel B.
Identification of Key Residues in the NPM1-quadruplex Interaction
Data reported so far indicate that the NPM1 region comprised between the longer NPM1-C70 construct and the shorter NPM1-C53 construct (aa 225–241) is necessary for high affinity binding. This region contains five lysine residues that we hypothesized might play a role in the specific recognition played by NPM1-C70 versus NPM1-C53 (Fig. 1A). To test this hypothesis we mutated each of them into alanine. The K229A/K230A double mutant was also prepared. With these variant proteins we performed a complete set of SPR experiments to measure the binding affinities with the G10-loop and c-MYC oligos. Interestingly, none of the single mutants significantly affected the interaction when the variant proteins were tested against both oligos (Table 3). However, when testing the double mutant K229A/K230A, we observed a marked loss of affinity and obtained a KD = 135 μm for the G10-loop and KD = 78 μm for the c-MYC oligo. These values are very close to those obtained when testing the two oligos with NPM1-C53 (compare Table 3 with Table 1) and suggest that residues Lys229 and Lys230 cooperate to the specific and high affinity recognition played by NPM1-C70 on both oligos. Besides that, they also suggest that the mode of recognition of the c-MYC and G10-loop is likely to be similar, pointing again to a G-quadruplex structure for the G10-loop oligo.
TABLE 3.
Effect of single alanine mutations on NPM1-C70 binding to G10-loop and c-MYC oligos
| G10-loop |
c-MYC |
|||||
|---|---|---|---|---|---|---|
| KD | kon | koff | KD | kon | koff | |
| μm | m−1s−1 | s−1 | μm | m−1s−1 | s−1 | |
| NPM1-C70 | 7.2 ± 0.2 | 2.8 ± 0.1 × 102 | 1.98 ± 0.03 × 10−3 | 1.9 ± 0.1 | 7.4 ± 0.3 × 103 | 1.37 ± 0.03 × 10−2 |
| K229A | 14.7 ± 0.8 | 1.6 ± 0.2 × 102 | 2.30 ± 0.04 × 10−3 | 10.3 ± 0.9 | 7.3 ± 0.4 × 102 | 7.5 ± 0.3 × 10−3 |
| K230A | 10.9 ± 0.3 | 1.50 ± 0.02 × 102 | 1.60 ± 0.02 × 10−3 | 15 ± 1 | 2.8 ± 0.1 × 102 | 4.3 ± 0.1 × 10−3 |
| K233A | 7.5 ± 0.5 | 4.5 ± 0.2 × 102 | 3.4 ± 0.1 × 10−3 | 17.2 ± 0.6 | 3.0 ± 0.1 × 102 | 5.2 ± 0.1 × 10−3 |
| K236A | 8.1 ± 0.2 | 3.3 ± 0.1 × 102 | 2.70 ± 0.02 × 10−3 | 10 ± 1 | 2.8 ± 0.2 × 102 | 2.7 ± 0.2 × 10−2 |
| K239A | 19.8 ± 0.5 | 1.04 ± 0.02 × 102 | 2.05 ± 0.02 × 10−3 | 6.8 ± 0.9 | 8.7 ± 0.7 × 102 | 5.9 ± 0.3 × 10−3 |
| K229A/K230A | 135 ± 8 | 78 ± 7 | ||||
Folding Studies
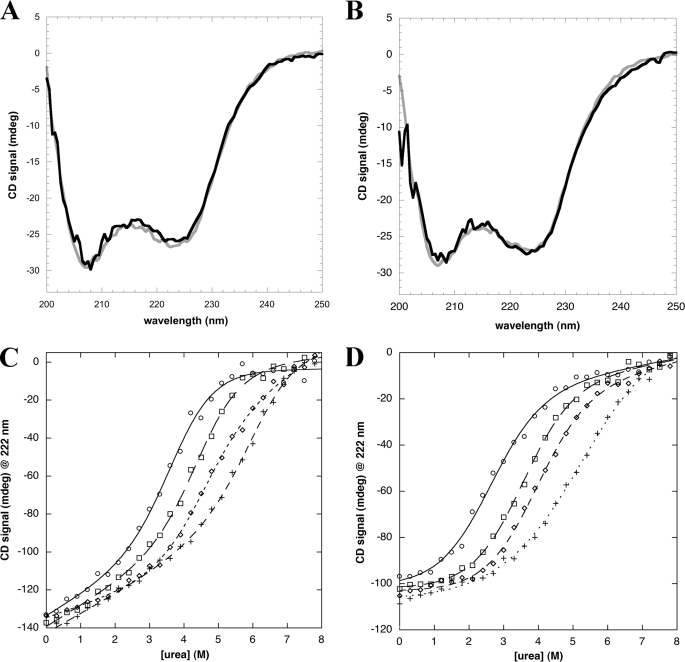
Previous structural studies on the NPM1 C-terminal domain (19) as well as our own folding studies (20, 21) were performed on the NPM1-C53 construct rather than on NPM1-C70 (Fig. 1A). The reason is that the segment comprising the residues that distinguish NPM1-C70 from NPM1-C53 is predicted to belong to the large natively unstructured domain that separates the chaperone N-terminal domain of the protein from the C-terminal domain (12). In Fig. 1A we report the prediction made using the PSIPRED algorithm that shows a high consensus coil prediction for this region (sequence underlined). The same predictions are obtained when using all the algorithms available in the Disprot server (not shown). Because we showed that this region is necessary for high affinity binding, we now wondered if folding predictions were correct and if yes, whether this region folds upon contact with DNA. A first indication comes from CD analysis of the protein in the absence or presence of c-MYC (Fig. 7A) or G10-loop (Fig. 7B) oligos. In both cases, CD spectra are almost completely superimposible, suggesting that the secondary structure content is invariant, with no extra folding induced upon DNA binding.
FIGURE 7.
Folding analysis of the region differentiating NPM1-C70 from NPM1-C53 (amino acids 225 to 241). A, the CD spectra of NPM1-C70 (20 μm) in the absence (gray line) or presence of c-MYC oligo (20 μm; black line) are almost identical. B, CD spectra of NPM1-C70 (20 μm) in the absence (gray line) or presence of G10-loop oligo (20 μm; black line). C, urea-induced denaturation of NPM1-C70 in the absence (open circles) or presence of 250 mm (open squares), 500 mm (open diamonds), and 1.0 m (crosses) NaCl are reported. Data were fitted according to a two-state model (lines) and folding parameters are reported in Table 4. D, urea-induced denaturation of NPM1-C53 under the same conditions as in panel C. Symbols are also the same.
To determine whether this segment is actually folded or unfolded, we performed comparative urea denaturation experiments on NPM1-C70 and NPM1-C53 in the presence of increasing amounts of NaCl (Fig. 7, C and D, respectively). Two critical values are extrapolated by fitting the denaturation curves according to a simple two-state model, i.e. the midpoint of denaturation Urea1/2 and the mD-N value. The latter is the slope of the transition and gives an estimate of the change in solvent accessible surface when moving from the denatured to the native state (therefore the higher this value the more compact the native state). The product of these two values represents protein stability (ΔG). In Table 4 it is shown that, in the absence of salt, the ΔG value for NPM1-C70 is considerably higher than the ΔG for NPM1-C53, by roughly 1.5 kcal/mol. In fact, whereas the mD-N values are similar, the Urea1/2 value for NPM1-C70 is 3.8 versus 2.6 m for NPM1-C53. If the region differentiating the two constructs were natively unstructured and free to move in the solvent in NPM1-C70, one would expect similar Urea1/2 and ΔG values. On the contrary our data suggest that this segment forms a compact structure with the remaining protein segment and thus contributes to overall stability. To further corroborate this finding we performed the same experiments in the presence of increasing salt amounts (Table 3). It is well known that NaCl is highly effective in stabilizing protein structure and promoting compactness (20, 21). Therefore we reasoned that, if the segment were natively unstructured in the absence of salt, it should progressively acquire compactness when increasing the salt concentration and this should be reflected in increased mD-N values. Our data indicate that this does not occur, with mD-N values remaining roughly the same, within the experimental error, when increasing NaCl concentration up to 1.0 m. Taken together these experiments indicate that, contrary to predictions, NPM1-C70 adopts a compact structure, without an unstructured N-terminal arm, which constitutes the functional DNA binding domain of the protein.
TABLE 4.
Urea-induced denaturation of NPM1-C70 and NPM1-C53 in the presence of increasing salt amounts
Urea1/2 and mD-N were obtained by fitting experimental data. ΔG values are calculated as the product of Urea1/2 and mD-N values.
| mD-N NPM1-C70 | mD-N NPM1-C53 | Urea1/2 NPM1-C70 | Urea1/2 NPM1-C53 | ΔG NPM1-C70 | ΔG NPM1-C53 | |
|---|---|---|---|---|---|---|
| No salt | 0.93 ± 0.16 | 0.787 ± 0.07 | 3.76 ± 0.15 | 2.55 ± 0.07 | 3.49 | 2.02 |
| 0.25 m NaCl | 0.93 ± 0.12 | 0.864 ± 0.08 | 4.51 ± 0.11 | 3.56 ± 0.09 | 4.19 | 3.06 |
| 0.50 m NaCl | 0.82 ± 0.11 | 0.843 ± 0.08 | 4.55 ± 0.14 | 3.91 ± 0.08 | 3.77 | 3.29 |
| 1.00 m NaCl | 0.79 ± 0.11 | 0.747 ± 0.09 | 6.20 ± 0.29 | 5.06 ± 0.17 | 4.9 | 3.79 |
DISCUSSION
In this work, we have investigated the DNA binding properties of NPM1, building on the foundations already established by others (3, 22–24). Our analysis suggests that, besides NPM1-C53 being able to recognize any oligo tested with low affinity, high affinity recognition is achieved only with NPM1-C70. Moreover, rather than being just a single-stranded binding protein as initially suggested (3, 22), the preferential binding of NPM1 is for oligos with a defined three-dimensional structure. This is the case of the G10-loop, a sequence found at the promoter of the SOD2 gene, which binds NPM1 and was predicted to form a hairpin based on conventional Watson-Crick pairing (24). However, a hairpin structure is not sufficient to confer high recognition properties, as shown by the results obtained with the T10-loop. The presence and number (see the G5-loop) of guanines in the loop, and therefore the three-dimensional structure adopted by this particular sequence, is indeed critical (see Table 1).
Further investigations indicate that the expected hairpin might not be the structure adopted by the G10-loop in vitro and possibly also in vivo. First, we showed that NPM1-C70 is able to recognize with high affinity G-quadruplex forming oligos, like the poly(G) oligo and, above all, the G-rich sequence found at the NHEIII1 region of the c-MYC promoter. Then we presented several lines of evidence that the G10-loop, whose sequence obeys the G-quadruplex folding rule (26), is indeed folded as a parallel G-quadruplex, at least in vitro. In fact, we showed by CD spectroscopy that the G10-loop presents all the hallmarks of parallel G-quadruplex formation in the presence of physiological concentrations of K+ and Na+ ions. Moreover we found that the thermal unfolding of this oligo in the presence of monovalent cations is still not complete at temperatures as high as 105 °C, reflecting the enhanced stability conferred to G-quadruplex structures by K+ and Na+. Finally we showed that the G10-loop, contrary to the T10-loop, is capable of binding the porphyrin TmPyP4 with the same stoichiometry and dissociation constants comparable with those of the c-MYC oligo, thus behaving as expected for a G-quadruplex forming oligo.
G-quadruplex forming regions are frequent in the human genome and, interestingly, they are particularly abundant at gene promoter regions, with roughly 47% of all gene promoters having putative G-quadruplex forming regions (26, 28). A large number of these regions have been investigated and shown to form fully folded quadruplexes at least in vitro (34). In particular, it is interesting to note that these regions are frequently associated to oncogene promoters, including those of c-Myc, k-ras, bcl2, and c-kit genes, whereas a markedly reduced frequency at tumor suppressor promoters was observed (34). Moreover telomeres, which are made of the repetitive sequence TTAGGG, were also shown to form G-quadruplexes both in vitro and in vivo, these structures being detrimental to telomerase function (35). These findings suggested that drugs capable of stabilizing G-quadruplexes might have a therapeutic value in cancer treatment both because of telomerase inhibition or oncogene suppression (27). As a proof of concept, it was shown that the NHEIII1 region at the c-MYC promoter suppresses c-MYC gene transcription when adopting a G-quadruplex structure and that the porphyrin TmPyP4, which stabilizes the G-quadruplex structure, is able to down-regulate this gene in vivo and to inhibit tumor growth in xenograft tumor models (36). The role of G-quadruplexes in gene regulation may, however, differ in other cases and be associated to gene activation, possibly through protein binding (28). This might be the case of the SOD2 promoter, because we showed a G-quadruplex arrangement for the G10-loop region.
Given their importance in processes such as gene transcription and regulation and telomere elongation control, it is not surprising that a considerable number of proteins have been reported to interact with G-quadruplexes, most of them associated to telomere function (28). These include proteins that only bind G-quadruplexes, proteins that promote G-quadruplex formation, helicases that unwind G-quadruplexes, proteins that destabilize G-quadruplexes, and nucleases that are specific for G-quadruplexes (37). We have shown here that NPM1-C70, besides binding G-quadruplex forming regions, is also capable of promoting G-quadruplex formation in vitro, a property that may be relevant for NPM1 function in the cell. Having established that G10-loop folds as a G-quadruplex, and because this particular sequence is recognized by NPM1 in vivo and contributes to SOD2 gene activation (24), it is tempting to speculate that there might be other G-quadruplex forming regions in the genome that are recognized and stabilized by NPM1.
This will be an important subject for future investigations. In fact, we know that mutations of NPM1 associated with acute myeloid leukemia map to the C-terminal G-quadruplex binding domain, destabilize the protein, and drive its aberrant translocation to the cytosol (both wild-type and mutant) (17). Therefore, a significant amount of NPM1 DNA binding activity occurring in the nucleus is likely to be lost in acute myeloid leukemia blasts and this may contribute to driving transformation through the loss of uncharacterized gene regulation activities.
Our studies also suggest that further work is needed to understand the structure and function of the NPM1 C-terminal domain. In fact whereas we showed that NPM1-C70 is the functional domain for G-quadruplex binding, only the structure of NPM1-C53 is available to date (19). This observation led us to focus our attention on the sequence differentiating these two constructs (see Fig. 1A). We mutated all five lysines contained in this sequence and found that single alanine substitutions cannot explain the marked loss of function of NPM1-C53 with respect to NPM1-C70. However, when mutating both Lys229 and Lys230 to alanine, we obtained dissociation constants comparable with those of NPM1-C53 when testing both the G10-loop and c-MYC oligos, suggesting that these two residues cooperate in recognizing some presently unknown structural feature within the G-quadruplex arrangement.
What then is the structure of NPM1-C70 and how are these two lysines positioned with respect to each other? We have previously underlined that all disorder prediction algorithms indicate this protein construct to resemble the structure of NPM1-C53, with an additional natively unfolded tail at the N terminus. However, we have shown here that (i) NPM1-C70 does not acquire additional secondary structure upon DNA binding, (ii) its stability is considerably higher than that of NPM1-C53, and (iii) that structure compactness is not increased by the presence of salts. These findings contrast the predictions and suggest instead that NPM1-C70 is folded as a compact domain, with the region comprised between amino acids 225 and 241 contributing to shape a G-quadruplex-binding site whose structure is presently unknown.
Therefore structural studies of NPM1-C70 and its complexes with relevant G-quadruplex forming oligos are much in need, also because there is a paucity of structural information concerning the mechanism of G-quadruplex recognition by their interacting proteins (27). Moreover the C-terminal domain of NPM1 is considered as a possible drug target for the treatment of acute myeloid leukemia with cytoplasmic NPM1 (13, 21). In principle, any molecule that targets this domain could stabilize a native-like state even in the mutated protein and prevent its association with the nuclear export system or translocation. Under this light, the structural analysis of NPM1 binding to nucleic acids would help in the rational design of such molecules.
Acknowledgment
We thank Benedetta Mattei for valuable advice in the design and execution of surface plasmon resonance experiments.
This work was supported by grants from the Associazione Italiana Ricerca sul Cancro (AIRC) (to M. B. and B. F.), Research Programs D.1 2009-2010 “Kinetic Study of Nucleophosmin by Surface Plasmon Resonance” and D.2.2 2008-2009 “SPR of Biological Macromolecules” from the Università Cattolica del Sacro Cuore (to A. A.) and grants from the University of Chieti “G. D'Annunzio” (to L. F.).
- NPM1
- nucleophosmin
- SOD
- superoxide dismutase.
REFERENCES
- 1.Yung B. Y., Busch H., Chan P. K. (1985) Biochim. Biophys. Acta 826, 167–173 [DOI] [PubMed] [Google Scholar]
- 2.Feuerstein N., Mond J. J. (1987) J. Biol. Chem. 262, 11389–11397 [PubMed] [Google Scholar]
- 3.Wang D., Baumann A., Szebeni A., Olson M. O. (1994) J. Biol. Chem. 269, 30994–30998 [PubMed] [Google Scholar]
- 4.Herrera J. E., Savkur R., Olson M. O. (1995) Nucleic Acids. Res. 23, 3974–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo E., Marine J. C., Danovi D., Falini B., Pelicci P. G. (2002) Nat. Cell Biol. 4, 529–533 [DOI] [PubMed] [Google Scholar]
- 6.Korgaonkar C., Hagen J., Tompkins V., Frazier A. A., Allamargot C., Quelle F. W., Quelle D. E. (2005) Mol. Cell. Biol. 25, 1258–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertwistle D., Sugimoto M., Sherr C. J. (2004) Mol. Cell. Biol. 24, 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W., Budhu A., Forgues M., Wang X. W. (2005) Nat. Cell Biol. 7, 823–830 [DOI] [PubMed] [Google Scholar]
- 9.Grisendi S., Bernardi R., Rossi M., Cheng K., Khandker L., Manova K., Pandolfi P. P. (2005) Nature 437, 147–153 [DOI] [PubMed] [Google Scholar]
- 10.Borer R. A., Lehner C. F., Eppenberger H. M., Nigg E. A. (1989) Cell 56, 379–390 [DOI] [PubMed] [Google Scholar]
- 11.Szebeni A., Olson M. O. (1999) Protein Sci. 8, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grisendi S., Mecucci C., Falini B., Pandolfi P. P. (2006) Nat. Rev. Cancer 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 13.Falini B., Sportoletti P., Martelli M. P. (2009) Curr. Opin. Oncol. 21, 573–581 [DOI] [PubMed] [Google Scholar]
- 14.Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A., Bigerna B., Pacini R., Pucciarini A., Liso A., Vignetti M., Fazi P., Meani N., Pettirossi V., Saglio G., Mandelli F., Lo-Coco F., Pelicci P. G., Martelli M. F.GIMEMA Acute Leukemia Working Party (2005) N. Engl. J. Med. 352, 254–266 [DOI] [PubMed] [Google Scholar]
- 15.Falini B., Nicoletti I., Martelli M. F., Mecucci C. (2007) Blood 109, 874–885 [DOI] [PubMed] [Google Scholar]
- 16.Garzon R., Garofalo M., Martelli M. P., Briesewitz R., Wang L., Fernandez-Cymering C., Volinia S., Liu C. G., Schnittger S., Haferlach T., Liso A., Diverio D., Mancini M., Meloni G., Foa R., Martelli M. F., Mecucci C., Croce C. M., Falini B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falini B., Bolli N., Liso A., Martelli M. P., Mannucci R., Pileri S., Nicoletti I. (2009) Leukemia 23, 1731–1743 [DOI] [PubMed] [Google Scholar]
- 18.Falini B., Bolli N., Shan J., Martelli M. P., Liso A., Pucciarini A., Bigerna B., Pasqualucci L., Mannucci R., Rosati R., Gorello P., Diverio D., Roti G., Tiacci E., Cazzaniga G., Biondi A., Schnittger S., Haferlach T., Hiddemann W., Martelli M. F., Gu W., Mecucci C., Nicoletti I. (2006) Blood 107, 4514–4523 [DOI] [PubMed] [Google Scholar]
- 19.Grummitt C. G., Townsley F. M., Johnson C. M., Warren A. J., Bycroft M. (2008) J. Biol. Chem. 283, 23326–23332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scaloni F., Gianni S., Federici L., Falini B., Brunori M. (2009) FASEB J. 23, 2360–2365 [DOI] [PubMed] [Google Scholar]
- 21.Scaloni F., Federici L., Brunori M., Gianni S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5447–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dumbar T. S., Gentry G. A., Olson M. O. (1989) Biochemistry 28, 9495–9501 [DOI] [PubMed] [Google Scholar]
- 23.Hingorani K., Szebeni A., Olson M. O. (2000) J. Biol. Chem. 275, 24451–24457 [DOI] [PubMed] [Google Scholar]
- 24.Xu Y., Fang F., Dhar S. K., St. Clair W. H., Kasarskis E. J., St. Clair D. (2007) J. Biol. Chem. 282, 15981–15994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santoro M. M., Bolen D. W. (1988) Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 26.Huppert J. L., Balasubramanian S. (2005) Nucleic Acids Res. 33, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidle S. (2009) Curr. Opin. Struct. Biol. 19, 239–250 [DOI] [PubMed] [Google Scholar]
- 28.Huppert J. L. (2008) Chem. Soc. Rev. 37, 1375–1384 [DOI] [PubMed] [Google Scholar]
- 29.González V., Hurley L. H. (2010) Annu. Rev. Pharmacol. Toxicol. 50, 111–129 [DOI] [PubMed] [Google Scholar]
- 30.González V., Guo K., Hurley L., Sun D. (2009) J. Biol. Chem. 284, 23622–23635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freyer M. W., Buscaglia R., Kaplan K., Cashman D., Hurley L. H., Lewis E. A. (2007) Biophys. J. 92, 2007–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei C., Jia G., Yuan J., Feng Z., Li C. (2006) Biochemistry 45, 6681–6691 [DOI] [PubMed] [Google Scholar]
- 33.Wei C., Wang J., Zhang M. (2010) Biophys. Chem. 148, 51–55 [DOI] [PubMed] [Google Scholar]
- 34.Qin Y., Hurley L. H. (2008) Biochimie 90, 1149–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipps H. J., Rhodes D. (2009) Trends Cell Biol. 19, 414–422 [DOI] [PubMed] [Google Scholar]
- 36.Grand C. L., Han H., Muñoz R. M., Weitman S., Von Hoff D. D., Hurley L. H., Bearss D. J. (2002) Mol. Cancer Ther. 1, 565–573 [PubMed] [Google Scholar]
- 37.Fry M. (2007) Front. Biosci. 12, 4336–4351 [DOI] [PubMed] [Google Scholar]