Abstract
The enzyme Qβ replicase is an RNA-dependent RNA polymerase, which plays a central role in infection by the simple single-stranded RNA virus bacteriophage Qβ. This enzyme has been used in a number of applications because of its unique activity in amplifying RNA from an RNA template. Determination of the thermal stability of Qβ replicase is important to gain an understanding of its function and potential applications, but data reported to date have been contradictory. Here, we provide evidence that these previous inconsistencies were due to the heterogeneous forms of the replicase with different stabilities. We purified two forms of replicase expressed in Escherichia coli, which differed in their thermal stability but showed identical RNA replication activity. Furthermore, we found that the replicase undergoes conversion between these forms due to oxidation, and the Cys-533 residue in the catalytic β subunit and Cys-82 residue in the EF-Tu subunit of the replicase are essential prerequisites for this conversion to occur. These results strongly suggest that the thermal stable replicase contains the intersubunit disulfide bond between these cysteines. The established strategies for isolating and purifying a thermally stable replicase should increase the usefulness of Qβ replicase in various applications, and the data regarding thermal stability obtained in this study may yield insight into the precise mechanism of infection by bacteriophage Qβ.
Keywords: Bacteriophage, Enzyme Inactivation, Enzyme Mutation, Enzyme Purification, Kinetics, RNA Polymerase, Bacteriophage Qβ, Qβ Replicase, Stability
Introduction
Bacteriophage Qβ is one of the simplest single-stranded RNA viruses and its mechanism of infection is studied as a model of RNA virus infection (1–9). As Qβ replicase plays a central role in infection, there has been a great deal of interest in its function for several decades (10-17). Qβ replicase is an RNA-dependent RNA replicase that uses single-stranded RNA as a template to synthesize a complementary strand of RNA. In the presence of excess Qβ replicase, a template RNA, such as the genomic RNA of phage Qβ, is synthesized autocatalytically (18, 19). Because of this unique autocatalytic synthesis activity of target RNA, Qβ replicase has been used in several applications, such as the in vitro demonstration of Darwinian evolution (20), a biological example of a hypercycle (21), studies on the origin of life (22), as a component of an artificial cell model (23–26), a method for detection of RNA recombination (27) or RNA viruses (28), an RNA amplification method (29, 30), RNA sequencing (31), and as a means of introducing mutations (32).
Thermal stability is an important property for understanding the function of replicase during infection, and also for various other applications. Although the replicase has been studied for several decades, data reported to date regarding its thermal stability have been contradictory. The first report published describing its thermal stability reported that 50% of replication activity was lost by incubation at 37 °C for 10 min (33), whereas RNA replication continued for 3 h at 35 °C in another study (18). In our previous report, we also reported that the replicase was highly stable at 37 °C, with retention of more than 70% activity after 2 h of incubation (34). The present study was performed to provide a consistent explanation for these contradictory reports and to determine the thermal stability of Qβ replicase.
EXPERIMENTAL PROCEDURES
Purification of Replicase
The standard method for purification of Qβ replicase was essentially as described previously (17). Escherichia coli BL21(DE3) harboring the plasmids pET-Tu-Ts and pBAD33-rep(NB), or other plasmids of cysteine mutants were grown at 30 °C in LB media supplemented with 50 μg/ml ampicillin, 20 μg/ml chloramphenicol, and 1.0 mm lactose until the optical density at 660 nm (OD660) was 1.0, then incubated for an additional 3 h with 0.2% arabinose. For the single-chain replicase constructs, E. coli XL1-blue harboring the plasmid pBAD33Ts-Tu-β-3 or cysteine mutants were grown in LB media essentially as described above except that lactose and ampicillin were omitted from the media. The collected cell pellet was disrupted using a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan) in buffer A (50 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 5 mm 2-mercaptoethanol, 1 mm EDTA, 500 mm NaCl). The cell lysate used for the replication assay in Fig. 6 was prepared using a sonicator instead of the Multi-Beads Shocker. The lysate was subjected to ammonium sulfate precipitation (0.39 g/ml ammonium sulfate) and suspended in buffer B (buffer A with 100 mm NaCl instead of 500 mm NaCl). After desalting through a PD10 column (GE Healthcare, Piscataway, NJ), the samples were applied to a HiTrapTM Q HP column (GE Healthcare) and eluted with a gradient of NaCl (100-400 mm) in buffer B. The fractions containing the heterotrimer were collected and further desalted with another PD10 column, applied to a HiTrapTM SP HP column (GE Healthcare) and eluted with a gradient of NaCl (100-400 mm). Fractions containing the heterotrimer were collected and used as the standard replicase. Plasmid pBAD33-rep(NB), a derivative of pBAD33rep (35), encodes the β subunit of Qβ replicase, and has NdeI and BglII restriction sites at the N and C termini of the β subunit, respectively.
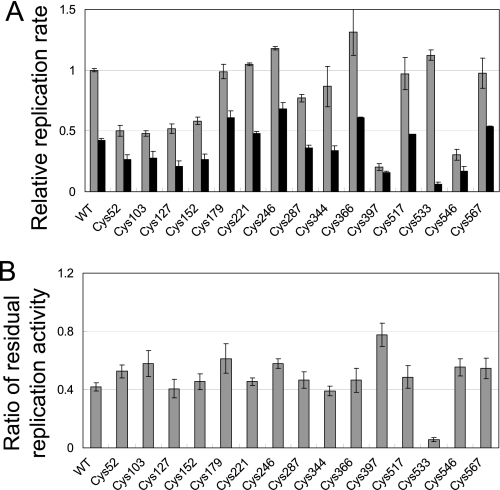
FIGURE 6.
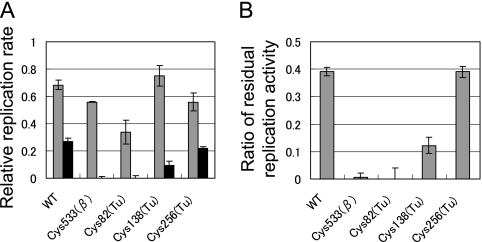
Thermal stability of cysteine mutants of the β subunit. A, replication activity of each cysteine mutant of the β subunit in the cell lysates. Replication rates were measured before (gray bar) and after (black bar) incubation of the lysate at 37 °C for 20 min without addition of template RNA and NTPs. Replication rates relative to that of the wild-type before incubation were plotted. The proportions of residual replicase activity after the incubation are shown in B.
The thermally stable replicase (S-form) was prepared by incubating the standard replicase at 37 °C for 30 min to inactivate all unstable replicases, followed by further purification by gel-filtration chromatography (Superdex 200 column; GE Healthcare) with a buffer containing 50 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 10 μm DTT, 1 mm EDTA, and 250 mm NaCl after removal of the insoluble fraction by centrifugation. The fractions containing the heterotrimer were collected and used as the S-form replicase. The thermally unstable replicase (U-form) was prepared by dialysis of the standard replicase with a buffer containing 50 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 10 mm DTT, 1 mm EDTA, and 250 mm NaCl. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis with 2-mercaptoethanol indicated that both the U- and S-forms were homogeneous heterotrimers (supplemental Fig. S1).
RNA Replication Assay
The standard reaction mixture for the RNA replication assay contained 125 mm Tris-HCl, pH 7.8, 5 mm MgCl2, 0.01% (w/v) BSA, 1.25 mm each NTP, 0.4× ROX dye (Invitrogen, Carlsbad, CA), SYBR Green II (Invitrogen), 50 nm template RNA (s130), and 100 nm replicase unless otherwise indicated. Incubation with replicase was performed before the addition of template RNA and NTPs, and therefore the concentrations of other components were ∼1.2-fold higher than in the standard reaction mixture. Most of the replication reactions were performed at 37 °C. The reactions shown in Figs. 3B, 5B, and supplemental Fig. S2B were performed at 25 °C. Fluorescence was measured on an Mx3005P QPCR System (Agilent Technologies, Santa Clara, CA) with ROX used as an internal reference. The fluorescence of SYBR Green II was converted into RNA concentration using the known concentration of the template RNA as a standard. The replication rate was determined by linear regression of the time course data for RNA replication shown in Fig. 1A. The template RNAs were prepared as described previously (34).
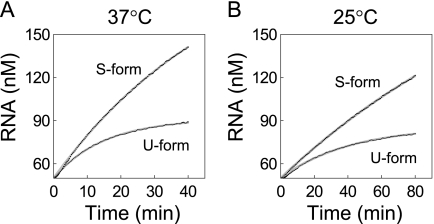
FIGURE 3.
Kinetics of RNA replication in the linear phase. U-form and S-form replicase (12.5 nm) were incubated with excess amounts of template RNA (s130, 50 nm) at 37 °C (A) or 25 °C (B). The data were fitted to Equation 2 to estimate kinact values in the working state.
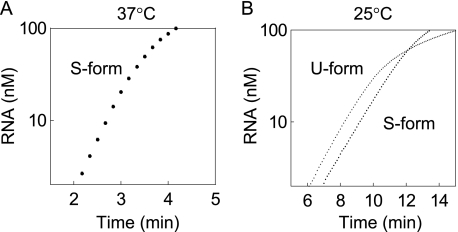
FIGURE 5.
Kinetics of RNA replication in the exponential phase. The S- and U-form replicase (112.5 nm) were incubated with template RNA (s130, 0.1 nm) at 37 °C (A) or 25 °C (B). As the replicase was excessive compared with template RNA, the RNA amplified exponentially. Replication by the U-form at 37 °C was not detected.
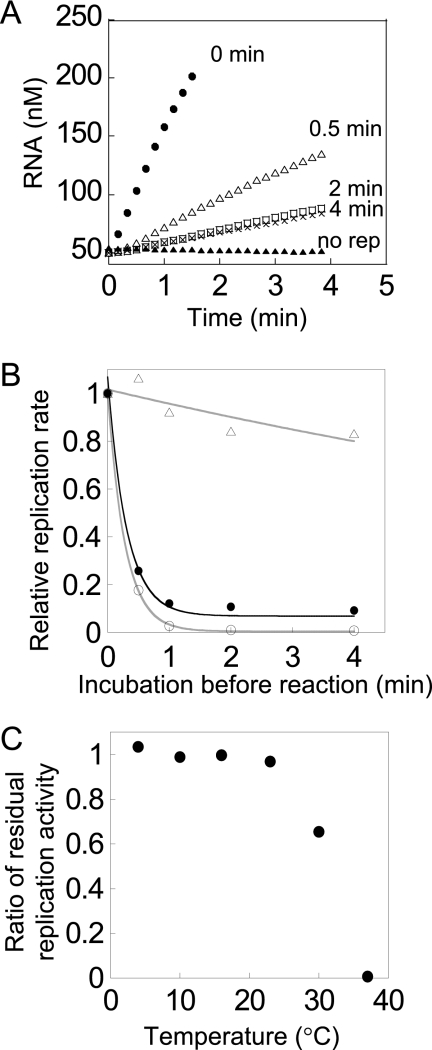
FIGURE 1.
Thermal stability of the replicase. A, time course of RNA replication by replicase purified by the standard method and the effects of incubation before the reaction. The replicase (200 nm) was incubated at 37 °C in the absence of RNA and NTPs for the indicated times (0, 0.5, 2, and 4 min) before the replication assay, and then RNA replication was measured at 37 °C after the addition of 50 nm template RNA (s130) and NTPs. The label “no rep” indicates the results of a control experiment without replicase. B, effects of preincubation on residual replication activity. Each form of replicase was incubated at 37 °C in the absence of template RNA and NTPs before the replication assay for the indicated times. The RNA replication reaction was measured as described in Fig. 1A, and the replication rates were determined by linear regression analysis. The replicase purified by the standard method is indicated by the filled circles. Replicase prepared by dialyzing the standard replicase with 10 mm DTT (U-form) is indicated by circles. Replicase prepared by heat inactivation of the standard replicase followed by gel-filtration chromatography (S-form) is indicated by triangles. The data for the S- and U-forms were fitted to a single exponential decay curve as eq. 1 (gray lines). C, effects of preincubation temperature on residual activity. U-form replicase was incubated at different temperatures before the replication reaction was carried out for 2 min in the absence of RNA and NTPs, and then RNA replication rates of the replicase were measured as described in Fig. 1A.
Determination of kinact
As the RNA replication velocity at the linear phase is known to be correlated with replicase concentration, which decreases at the inactivation rate, kinact, we determined the RNA replication velocity at time t in Equation 1,
where Vintial was the initial replication velocity. We fitted the results shown in Fig. 1B to Equation 1 and estimated the kinact value in the free state. To estimate the kinact value of replicase in the working state, we integrated Equation 1 to obtain the template RNA concentration at t in Equation 2,
where RNAintial and RNAfinal were the initial RNA and final amplified RNA concentration, respectively. The results shown in Fig. 3 were fitted to Equation 2 to obtain kinact in the working state.
Replication with Crude Extract
To assay replication activity in the crude extract, we modified the method reported previously (36). The reaction mixture contained 50 mm Tris-HCl, pH 7.8, 10 mm magnesium acetate, 1 mm EDTA, 1 mm phosphoenolpyruvate, 0.1 mm DTT, 2.5 μg/ml rifampicin, 7.5% (v/v) glycerol, 5 μg/ml pyruvate kinase (Wako, Osaka, Japan), 0.05 units/μl DNaseI (Takara Bio Inc., Kyoto, Japan), 1.25 mm each NTP, 0.4× ROX, SYBR Green II, 50 nm template RNA (s222), and 1/20 volume of cell lysate. Detection of specific fluorescence from template RNA was calculated by subtracting the fluorescence of the reaction mixture from that of the reaction mixture without template RNA. ROX fluorescence was used as an internal reference. The fluorescence of SYBR Green II was converted into RNA concentration using the known concentration of the template RNA as a standard. The replication rate was determined by linear regression of the time course data for RNA replication. The lysate was incubated for 20 min at 37 °C before the reaction was performed.
Construction of Cysteine Mutants
Each cysteine was mutated to serine by site-directed mutagenesis with Phusion DNA polymerase (Finnzymes, Espoo, Finland) using pBAD33rep(NB) as the template. Substituting each cysteine residue at positions 52, 103, 127, 152, 179, 221, 246, 287, 344, 366, 397, 517, and 567 to serine residues was carried out using primers Cys52(+) and (−), Cys103(+) and (−), Cys127(+) and (−), Cys152(+) and (−), Cys179(+) and (−), Cys221(+) and (−), Cys246(+) and (−), Cys287(+) and (−), Cys344(+) and (−), Cys366(+) and (−), Cys397(+) and (−), Cys517(+) and (−), and Cys567(+) and (−), respectively. For construction of Cys533(β) and Cys546(β) mutants, an In-FusionTM PCR cloning kit (Clontech, Palo Alto, CA) was used in accordance with the manufacturer's instructions. Briefly, pBAD-33rep(NB) was amplified by PCR with Phusion DNA polymerase to prepare five different PCR products using primers FW1-snaBI and RV1-ser, FW1-snaBI and RV1-cys, FW2-ser and RV2-hindIII, FW2-cys and RV2-hindIII, and vectorF and vectorR, to yield fragments 1, 2, 3, 4, and 5, respectively. Mixtures of fragments 1, 4, and 5, or 2, 3, and 5 were subjected to In-FusionTM reaction to yield Cys533(β) and Cys546(β) mutants, respectively. Construction of all mutants was confirmed by DNA sequencing. The number of cysteine residues was consistent with the replicase data in the GenBankTM database (ACY07237.1). The sequences of the primers used are listed in supplemental Table S1.
Mutants of single-chain replicase were prepared by introducing the corresponding cysteine to serine mutation into the plasmid pBAD33Ts-Tu-β-3 (35). Briefly, for the Cys533(β) mutation, the XhoI−HindIII fragment-digested from pBAD33rep(NB) with the Cys533(β) mutation was replaced with the corresponding fragment of pBAD33Ts-Tu-β-3. For EF-Tu mutants, the NdeI−BlpI fragment digested from pET-TuTs with cysteine to serine mutations was replaced with the corresponding fragment of pBAD33Ts-Tu-β-3. The pET-TuTs with cysteine mutations were constructed by site-directed mutagenesis using pET-TuTs as a template. Substituting each cysteine residue at positions 82, 138, and 256 to serine residues was carried out using primers Cys82(+) and (−), Cys138(+) and (−), Cys256(+) and (−), respectively.
Non-reducing SDS-PAGE and Western Blotting
4-Acetoamido-4-maleimidylstilbene-2, 2-disulfonic acid (AMS)2 was purchased from Invitrogen. AMS treatment and nonreducing SDS-PAGE were performed according to the method reported previously (37). Monoclonal antibodies to β subunit, EF-Tu, and EF-Ts were kindly provided by Dr. Tsukada of Osaka University.
RESULTS
Replicase Exhibits Heterogeneity in Thermal Stability
We studied the mechanisms of Qβ replicase (17, 34, 35, 38), during which we found that the replicase purified by the method used, denoted as the “standard method,” contained replicase with different thermal stabilities. Briefly, our standard method consists of ammonium sulfate precipitation and ion exchange chromatography. When we incubated the purified replicase at 37 °C for 2 min before mixing with template RNA and NTPs, the activity of replicase (replication rate) was decreased to about 10%, while prolonged incubation (4 min) did not further decrease the activity (Fig. 1, A and B). These results suggested that the purified replicase preparation contained about 90% thermally unstable enzyme, which was inactivated immediately at 37 °C, with the remaining 10% of the enzyme was thermally stable. The proportion of thermally stable replicase varied among batches of purification lots at 22% ± 14% (n = 4), even when the same purification method was employed. There have been no previous reports regarding the heterogeneity of thermal stability. Therefore, the inconsistency of previous reports regarding thermal stability of the replicase may be explained by the observed heterogeneity. To verify this suggestion, it was necessary to purify and characterize each form of replicase independently.
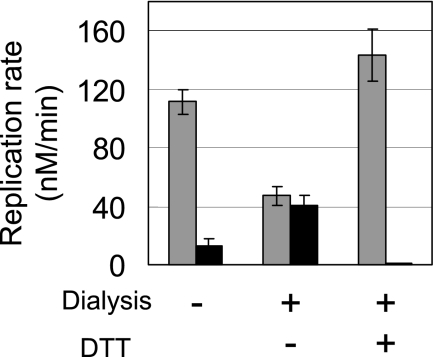
Therefore, we established strategies to prepare each form of replicase separately. The thermally stable replicase was prepared by incubating the replicase purified by the standard method at 37 °C for 10 min to inactivate all unstable replicases, followed by further purification by size exclusion chromatography. The resultant replicase retained 80% of its activity after incubation at 37 °C for 30 min (Fig. 1B and supplemental Fig. S2). We designated this stable form of replicase as the S-form. The thermally unstable replicase was prepared by dialyzing the purified replicase according to the standard method in the presence of 10 mm DTT. Dialysis with a high concentration of DTT was found to convert the thermally stable replicase into a thermally unstable form (Fig. 2). Simple addition of DTT to the thermally stable replicase also produced the thermally unstable replicase at a rate of 0.016/h (supplemental Fig. S5). The thermally unstable replicase was almost completely inactivated by incubation at 37 °C for 2 min (Fig. 1B). This unstable replicase was designated as the U-form. The replicase was a heterotrimer composed of a phage-derived β subunit and host-derived translation factors, EF-Tu and EF-Ts (11). SDS-PAGE analysis with 2-mercaptoethanol showed that both the U- and S-forms were homogeneous heterotrimers (supplemental Fig. S1).
FIGURE 2.
Effects of dialysis in the presence of DTT on the thermal stability of replicase. The replicase purified by the standard method was subjected to dialysis with or without 10 mm DTT in a buffer composed of 50 mm Tris-HCl, pH 7.8, 5 mm MgCl2, 1 mm EDTA, and 250 mm NaCl. RNA replication rates before (gray bar) and after (black bar) the incubation at 37 °C for 2 min in the absence of template RNA and NTPs were plotted.
Thermal Stability of the Two Forms of Replicase
We characterized the thermal stability of each form quantitatively. Assuming that the inactivation rate was proportional to replicase concentration, we fitted the data from Fig. 1B to a single exponential curve (Equation 1) to obtain the inactivation rate constant, kinact; the kinact of the S-form was about 18-fold lower than that of the U-form (Table 1, free state). The inactivation rate constant was dependent on the incubation temperature; the kinact values at 25 °C of the S- and U-forms were smaller than those at 37 °C, but the kinact of the S-form was less than that of the U-form. A broader range of temperature dependence for inactivation of the U-form is shown in Fig. 1C, where the U-form was incubated for 2 min at the indicated temperature before activity measurements. A substantial fraction of replicase was inactivated at temperatures higher than 30 °C.
TABLE 1.
Inactivation rate constants (kinact) of the U- and S-form replicases
|
kinact |
||||
|---|---|---|---|---|
| Free statea |
Working stateb |
|||
| 37 °C | 25 °C | 37 °C | 25 °C | |
| min | min | min | min | |
| S-form | 0.009 ± 0.002 | 0.002 ± 0.002 | 0.019 ± 0.0003 | 0.0059 ± 0.0001 |
| U-form | 3.6 ± 0.1 | 0.095 ± 0.006 | 0.066 ± 0.0004 | 0.024 ± 0.0003 |
a kinact in the free state shows the inactivation rate constant in the absence of template RNA and NTPs. The values were obtained by curve fitting the data shown in Fig. 1B (U-form at 37 °C) and supplemental Figs. S2A (S-form at 37 °C) and S2B (both forms at 25 °C).
b kinact in the working state shows the inactivation rate constant in the presence of template RNA and NTPs. The values were obtained by curve fitting the data shown in Fig. 3.
We had measured the thermal stability of replicases in the absence of template RNA and NTPs, which we designated the “free state.” We next analyzed the thermal stability during the replication reaction in the presence of template RNA and NTPs. Under these conditions, the replicase was bound to the RNA template, and therefore this was designated as the “working state.” Inactivation of the replicase in the working state was measured by the decrease in replication rate in the linear phase (34), where template RNA was in excess relative to the replicase. In the linear phase, most of the active replicase was bound to the RNA, and thus the replication rate was proportional to active replicase concentration (19). The RNA amplification continued linearly up to 2 min at 37 °C for both S- and U-forms (Fig. 3A), indicating that the replicase in the working state remained active during this time. This result suggested that U-form replicase in the working state is more stable than that in the free state, where the U-form was inactivated completely after 2 min at 37 °C (Fig. 1B). To confirm this result quantitatively, we estimated the kinact value by fitting the data in Fig. 3 with Equation 2. The kinact value of the U-form in the working state was ∼50-fold smaller than that in the free state at 37 °C (Table 1).
It is also notable that unlike the U-form, the S-form was less stable in the working state than in the free state at both 37 °C and 25 °C. This can be explained by the inactivation effect of GTP present during incubation in the working state. We showed previously that GTP, and probably small fractions of GDP present in the GTP solution, inactivated the replicase with a kinact of 0.0128/min (34), close to the value in this study at 37 °C in the working state (0.019/min).
Stabilization Factors of the U-Form
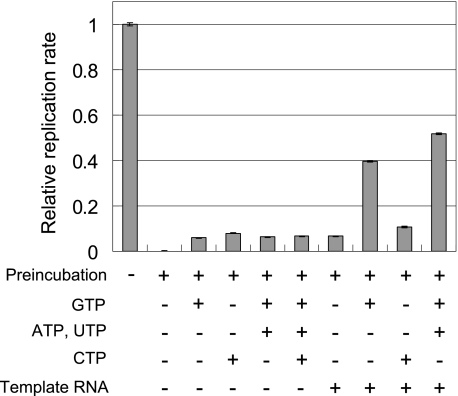
There are several possible reasons for the increased stability of the U-form in the working state, such as the presence of template RNA, NTPs, or both. To distinguish between these possibilities, we incubated the U-form replicase in reaction mixtures of various compositions, and measured residual replicase activity (Fig. 4). Incubation with template RNA or NTPs alone resulted in a small increase in residual replicase activity. In contrast, incubation with both template RNA and GTP significantly increased the residual replicase activity, and addition of ATP and UTP resulted in a further increase. Replicase is known to form an initiation complex in the presence of template RNA and GTP, and further addition of ATP and UTP in this case induced formation of an elongation complex (39). The observed stabilization with template RNA, GTP, ATP, and UTP indicated that the U-form replicase in the initiation or elongation complex was more stable than the free U-form. We also examined the effects of other factors on stability, such as ionic strength, magnesium ions, reducing agents, glycerol, and the subunits of replicase: EF-Tu, EF-Ts, ribosomal S1 protein, and Hfq protein. None of these factors showed any stabilization effect (data not shown).
FIGURE 4.
Stabilizing factors of the U-form replicase during preincubation. Replication rates were measured following incubation of the U-form replicase with the indicated components, concentrations of which were the same as in the standard reaction mixture. Relative replication rates with and without preincubation are plotted. The error bars indicate the standard deviation of the measurements.
S- and U-Forms Exhibit Similar Replication Activity
We compared kinetic parameters other than thermal stability between the S- and U-forms. In previous studies, the kinetic parameters, Km and kcat, were estimated based on replication reactions performed with excess replicase compared with template RNA at 37 °C (19, 34). Here we performed similar experiments using S- and U-forms of the replicase. Our results indicated exponential amplification of the S-form but not the U-form at 37 °C (Fig. 5A). The lack of detectable replication by the U-form can be explained by the immediate inactivation of the U-form in the free state at 37 °C. We then measured replication at 25 °C, where the kinact values of both forms were significantly lowered (Table 1), and observed exponential amplification by both S- and U-forms (Fig. 5B). The replication rate constant, kobs, was measured for two template RNAs, s130 and s222, at various concentrations of the replicase (supplemental Fig. S3), and we estimated Km and kcat values at 25 °C (Table 2). There were no significant differences in the values of these parameters between the U- and S-forms. These results indicated that while thermal stability was different, the S-form and U-form showed similar replication activity.
TABLE 2.
Km and kcat values of the U- and S-form replicases at 25 °C
| s130 RNA |
s222 RNA |
|||
|---|---|---|---|---|
| kcat | Kma | kcat | Kma | |
| min | nm | min | nm | |
| S-form | 0.68 ± 0.002 | 2.8 ± 0.9 | 0.25 ± 0.001 | 1.7 ± 2.8 |
| U-form | 0.70 ± 0.005 | 5.2 ± 1.5 | 0.29 ± 0.001 | 1.3 ± 1.1 |
a To estimate the Km value, we assumed that the active ratio of the replicase was 100%.
Cys533(β) of the β Subunit Is Responsible for Replicase Stability
We next examined the chemical differences between the S- and U-forms of the replicase. As conversion of the S-form to the U-form was dependent on the reducing reagent DTT (Fig. 2), oxidation was considered to play a role in S-form formation. Cysteine residues are well-known oxidative targets in proteins; these residues undergo oxidation to form disulfide bonds or may be modified by several chemical groups. The replicase used in the present study was a heterotrimer, consisting of a β subunit, EF-Tu, and EF-Ts. First we focused on the β subunit, which is the catalytic subunit and contains 15 cysteines. We constructed cysteine mutants in which each cysteine residue was mutated to serine, and examined the proportion of the S-form in enzyme preparations.
Each cysteine mutant was expressed in E. coli, cell lysates were prepared, and the thermal stability of replication activity in the cell lysate was determined. We measured the replication activity of the lysates before and after incubation at 37 °C for 20 min in the absence of the template RNA and NTPs (Fig. 6A), and calculated the proportion of residual activity after incubation (Fig. 6B). For the wild-type and 14 of the mutants (Cys52, 103, 127, 152, 179, 221, 246, 287, 344, 366, 397, 517, 546, 567), ∼40% of the activity remained after incubation, whereas the residual activity was significantly decreased in the Cys533(β) mutant. This result was further confirmed using replicase purified by the standard method (supplemental Fig. S6). These results indicated that the cysteine 533 residue is responsible for S-form formation.
Nonreducing SDS-PAGE of the S-Form after Blocking Free Thiols Demonstrated a High Molecular Weight Protein Consisting of the β Subunit and EF-Tu
We have identified only one cysteine residue (Cys533(β)) responsible for S-form formation, rather than two in the β subunit. Therefore, if the S-form has a disulfide bond, an intersubunit bond is likely to be present. To examine this possibility, S- and U-forms of the replicase were subjected to SDS-PAGE under various conditions.
We took into account the possibility that alterations in disulfide bonds may have occurred during sample preparation. Disulfide bond exchange is known to occur for certain proteins during manipulation for nonreducing SDS-PAGE unless free thiols are blocked (40, 41). We first precipitated the replicases with TCA, which repressed the exchange reaction by lowering the pH, and then modified free thiols with AMS. The modified S- and U-form replicase were subjected to non-reducing SDS-PAGE (Fig. 7A).
FIGURE 7.
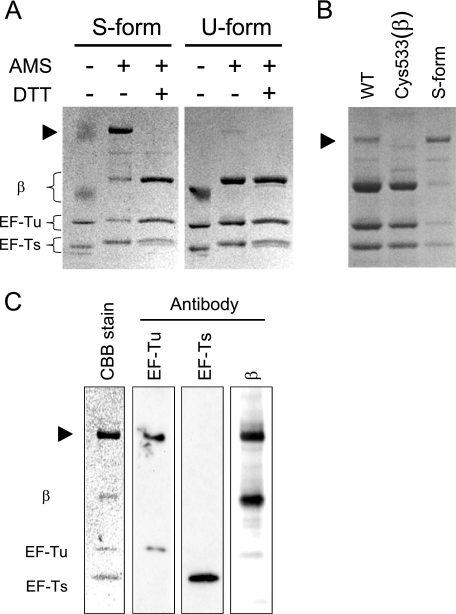
Nonreducing SDS-PAGE after blocking of free thiols. A, S- and U-forms of replicase were precipitated with 5% TCA and treated with the oxidative agent, AMS, to modify free thiols. This procedure is known to inhibit disulfide bond exchange. The replicases were then subjected to SDS-PAGE (5%-15%) without any reducing agent. In the AMS− lane, the TCA-precipitated replicase before AMS treatment was applied. In the DTT+ lanes, the applied replicase was treated with DTT (10 mm) for 5 min at 37 °C after modification with AMS. The black arrowhead indicates the high molecular weight proteins. As AMS has a molecular weight of 0.5 kDa, the bands showed a shift in migration following AMS treatment. B, wild-type and Cys533(β) mutant replicase purified by standard methods were treated with AMS and subjected to nonreducing SDS-PAGE. The S-form replicase was applied again for comparison. C, Western blotting analysis. S-form replicase subjected to nonreducing SDS-PAGE was analyzed by Western blotting using monoclonal antibodies specific for β, EF-Tu, and EF-Ts subunits.
The resulting U-form exhibited three bands corresponding to the β subunit, EF-Tu, and EF-Ts. In contrast, the S-form treated with AMS showed another major band above the β subunit, as well as a significant decrease in intensity of the band corresponding to the β subunit. Similar results were also obtained with another reagent that modifies thiols, N-ethylmaleimide (NEM) (data not shown). This high molecular weight protein was also observed even in the sample before AMS treatment as an indistinct smeared band (Fig. 7A, lane 1, upper band), indicating that protein appearance was not an artifact of AMS treatment.
The appearance of this high molecular weight protein, observed in the wild-type but not in Cys533(β) mutant (Fig. 7B), was sensitive to treatment with DTT (Fig. 7A). To identify the subunits constituting the high molecular weight protein, Western blotting analysis was performed using antibodies against EF-Tu, EF-Ts, and β subunit, respectively. The results clearly showed that the high molecular weight protein consisted of a β subunit and EF-Tu (Fig. 7C). Taken together, these findings strongly suggest that cysteine 533 of the β subunit is connected to EF-Tu via a disulfide bond in the S-form replicase.
Cys82(Tu) of EF-Tu Is Responsible for Replicase Stability and High Molecular Weight Protein Formation
To identify the partner of the Cys533(β) disulfide bond, we constructed mutants of EF-Tu in which each cysteine residue was mutated to serine. Here, we used fusion replicase, in which the β subunit, EF-Tu, and EF-Ts were connected as a single polypeptide. Use of the fusion protein allowed us to obtain a mutant replicase containing each mutant EF-Tu without affecting the original function of EF-Tu in the host cell, and reduced the possibility of integration of host-derived EF-Tu into the replicase. We measured the replication activities of the lysates from each mutant before and after incubation at 37 °C for 20 min in the absence of the template RNA and NTPs (Fig. 8A), and calculated the proportion of residual activity after incubation (Fig. 8B). The wild-type fusion replicase and Cys533(β) mutant showed similar results to the heterotrimeric replicases (Fig. 6, A and B); the activity remained after preincubation at a certain level for the wild-type, whereas the activity was significantly decreased for the Cys533(β) mutant. EF-Tu has three cysteine residues, Cys82, -138, and -256. For Cys138(Tu) and Cys 256(Tu), significant levels of activity remained after the incubation. In contrast, the residual activity decreased for the Cys82(Tu) mutant to the same level as in the Cys533(β) mutant. These results indicated that Cys82(Tu) is required for formation of the S-form and suggested that Cys82(Tu) is the disulfide bond partner with Cys533(β).
FIGURE 8.
Thermal stability of the fusion replicase containing cysteine mutants of EF-Tu. A, replication activity of the fusion replicase with each cysteine mutant of EF-Tu in the cell lysates. Replication rates were measured before (gray bars) and after (black bars) incubation of the lysate at 37 °C for 20 min without addition of template RNA and NTPs. Replication rates relative to that of the wild-type before incubation are plotted. The proportions of residual replicase activity after incubation are shown in B. Error bars indicate the standard deviation from duplicate experiments.
DISCUSSION
The results of the present study indicated that a replicase purified by the standard method contained at least two forms of the enzyme with different thermal stabilities but essentially identical Km and kcat values. We also demonstrated that oxidation of the replicase was responsible for the differences between the two forms. In addition, our findings supported the suggestion that the β subunit and EF-Tu are connected by a disulfide bond between Cys533(β) and Cys82(Tu) in the thermally stable replicase.
Previous Inconsistencies Can Be Explained by the Heterogeneity of Thermal Stability
Previous reports on the thermal stability of Qβ replicase were contradictory. We carefully examined the purification methods and thermal stability, and our results indicated that the previous inconsistencies could be explained by the heterogeneity of the replicase. The previous report of 50% inactivation by Eoyang et al. (33) can be explained by their replicase containing 50% S- and 50% U-form replicase. Haruna et al. (18) reported continuous replication for more than 3 h, which can be explained by the presence of the S-form replicase. In our previous study, we incubated the replicase before analysis to remove the U-form replicase (34). Therefore, the detailed kinetic analysis of the replicase that we reported previously was that of the S-form. Although purification of the replicase in previous studies was mostly carried out in the presence of reducing agents, conversion from the S- to the U-form was very slow and required dialysis for more than 1 day with high concentrations of a reducing agent (10 mm DTT; data not shown). Such procedures were not performed in previous purification methodologies (10, 33, 35, 36, 42, 43, 44), resulting in a mixture of the S- and U-forms in preparations. With the exception of thermal stability, there were no other significant differences between the S- and U-forms of the replicase. Therefore, previous knowledge regarding the characteristics of the replicase other than thermal stability would still be valid even though heterogeneous replicase preparations were used.
Chemical Differences between the S- and U-Forms of the Replicase
The results of the present study indicated that oxidation was responsible for the difference between the S- and the U-form, and a disulfide bridge between Cys533(β) and Cys82(Tu) was important for S-form formation. The disulfide bond is known to stabilize protein conformation (45) by destabilizing the unfolded state of the protein.
Depending on the sample, the band derived from the inter-subunit disulfide was weak or absent in samples untreated with AMS (Fig. 7A, lane 1 upper band, and supplemental Fig. S1, far right lane). On the other hand, the intersubunit disulfide bond was clearly detected when the samples were labeled with AMS (Fig. 7). For some proteins, chemically blocking the free thiols was required to prevent the disulfide exchange reaction detection of true disulfide bonds (40, 41). This may also be the case for the replicase, which may explain why the intersubunit disulfide bond was not detected previously in the replicase despite the large number of studies of this enzyme.
Based on the recently reported three-dimensional structure of Qβ replicase (46), Cys533(β) and Cys82(Tu) are located at the interface of the two subunits. Although the distance between the two cysteines (21 Å) was larger than the length of the disulfide bond (about 2 Å), this fact does not exclude the possibility of disulfide bond formation because the x-ray structure represents only a snapshot of fluctuating structures of the protein. In addition, the structure around Cys533(β) was considered to be flexible, and therefore Cys533(β) and Cys82(Tu) may be in close proximity in solution.
Comparative Analysis of the Cys533(β) Residue
Compared with related group III phages, the Cys533(β) residue is conserved in the Qβ replicase of other strains, such as BZ1, VK HL4–9, and TW18. However, it is not conserved in the closely related replicases MX1 and M11 or in other phage groups. These observations indicate that the function of Cys533(β) in replicase stability is restricted to a narrow range of phage strains. Alternatively, the function of Cys533(β) may be compensated by other cysteine residues. For example MX1 and M11 have acquired another cysteine located nearby (Cys521), which may function in the same manner as Cy533(β) of Qβ replicase.
Biological Significance of the S- and U-Forms
The cytoplasm of E. coli cells usually represents a reducing environment. Therefore, the replicase expressed in these cells during phage infection would be in the U-form, which should be inactivated immediately in the free state. During infection by the phage, however, genomic RNA replicates exponentially at 37 °C for about 15 min (21), suggesting that the replicase is stabilized within the cell. These observations suggest that there is a mechanism that stabilizes the U-form replicase in the cell. One possibility is the binding of the replicase to the cellular RNA, which stabilizes the U-form, as shown in Fig. 4. The replicase in the cell may exist bound to cellular RNAs, such as mRNA, tRNA, and rRNA. There may also be a crowding effect where the replicase is present in a crowded environment in vivo (47), and such macromolecular crowding is known to stabilize proteins. Another possibility is the presence of an as yet unidentified stabilizing factor, which may bind or modify the replicase and thus assist in its stabilization. We found that the proportion of the S-form increased when the cell lysate of cysteine mutants was stored at 4 °C for several days, suggesting that a factor in the cell lysate converted the U-form into the S-form (supplemental Fig. S4). Further studies focusing on stability may lead to the identification of a novel factor or factors involved in the phage genome RNA replication process.
There is still insufficient evidence on which to base a conclusion regarding whether conversion of the U-form to the S-form is biologically relevant. Nevertheless, the possibility that the S-form formed in the cytoplasm may play a role in RNA replication should not be excluded. This possibility is not unrealistic because the cellular conditions when replicase is expressed are unusual; when the cell is infected by the Qβ phage, the phage genome and protein are expressed at high levels, and therefore the cells are lysed within several minutes. Oxidized protein may form under such abnormal conditions, which is supported by the observation that phage infection impairs the function of the E. coli oxidative resistance systems (48). A more attractive possibility is that the Qβ phage may regulate the replication activity by conversion between the S- and U-forms. As RNA replication and phage protein translation are mutually inhibitory (26, 49), the two reactions must be regulated temporally for efficient amplification of the phage. Some proteins related to the oxidative stress response, such as OxyR, are known to be regulated by disulfide bond formation (50). The Qβ phage may utilize the same strategy to regulate replication activity. Further studies using Cys533 mutant phage in vivo are required to verify this possibility.
Acknowledgments
We thank Naoko Miki, Kumiko Nakamura, Hitomi Komai, and Mizuki Ohzawa for technical assistance. We are grateful to Dr. Koji Tsukada of Osaka University for providing the antibodies used in this study.
This work was supported in part by the Global Centers of Excellence Program and Special Coordination Funds for Promoting Science and Technology: Yuragi Project from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- AMS
- 4-acetoamido-4-maleimidylstilbene-2, 2-disulfonic acid
- NTP
- nucleotide triphosphate.
REFERENCES
- 1.Spiegelman S., Pace N. R., Mills D. R., Levisohn R., Eikhom T. S., Taylor M. M., Peterson R. L., Bishop D. H. (1968) Cold Spring Harb. Symp. Quant. Biol. 33, 101–124 [DOI] [PubMed] [Google Scholar]
- 2.Weissmann C., Billeter M. A., Goodman H. M., Hindley J., Weber H. (1973) Annu. Rev. Biochem. 42, 303–328 [DOI] [PubMed] [Google Scholar]
- 3.Weissmann C. (1974) FEBS Lett. 40, suppl:S10–S18 [DOI] [PubMed] [Google Scholar]
- 4.Zinder N. D. (1975) RNA Phages, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 5.van Duin J. (1988) in The Bacteriophages (Calendar R. ed) Plenum Press, New York [Google Scholar]
- 6.Jacobson A. B., Arora R., Zuker M., Priano C., Lin C. H., Mills D. R. (1998) J. Mol. Biol. 275, 589–600 [DOI] [PubMed] [Google Scholar]
- 7.Kim H., Yin J. (2004) Biotechnol. Bioeng. 88, 148–156 [DOI] [PubMed] [Google Scholar]
- 8.Tsukada K., Okazaki M., Kita H., Inokuchi Y., Urabe I., Yomo T. (2009) Biochim. Biophys. Acta 1790, 65–70 [DOI] [PubMed] [Google Scholar]
- 9.Karring H., Mathu S. G., van Duin J., Clark B. F., Kraal B., Knudsen C. R. (2004) J. Biol. Chem. 279, 1878–1884 [DOI] [PubMed] [Google Scholar]
- 10.Haruna I., Nozu K., Ohtaka Y., Spiegelman S. (1963) Proc. Natl. Acad. Sci. U.S.A. 50, 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blumenthal T., Carmichael G. G. (1979) Annu. Rev. Biochem. 48, 525–548 [DOI] [PubMed] [Google Scholar]
- 12.Biebricher C. K., Eigen M., McCaskill J. S. (1993) J. Mol. Biol. 231, 175–179 [DOI] [PubMed] [Google Scholar]
- 13.Brown D., Gold L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11558–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inokuchi Y., Kajitani M. (1997) J. Biol. Chem. 272, 15339–15345 [DOI] [PubMed] [Google Scholar]
- 15.Schuppli D., Georgijevic J., Weber H. (2000) J. Mol. Biol. 295, 149–154 [DOI] [PubMed] [Google Scholar]
- 16.Ugarov V. I., Chetverin A. B. (2008) J. Mol. Biol. 379, 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urabe H., Ichihashi N., Matsuura T., Hosoda K., Kazuta Y., Kita H., Yomo T. (2010) Biochemistry 49, 1809–1813 [DOI] [PubMed] [Google Scholar]
- 18.Haruna I., Spiegelman S. (1965) Science 150, 884–886 [DOI] [PubMed] [Google Scholar]
- 19.Biebricher C. K., Eigen M., Luce R. (1981) J. Mol. Biol. 148, 391–410 [DOI] [PubMed] [Google Scholar]
- 20.Mills D. R., Peterson R. L., Spiegelman S. (1967) Proc. Natl. Acad. Sci. U.S.A. 58, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eigen M., Biebricher C. K., Gebinoga M., Gardiner W. C. (1991) Biochemistry 30, 11005–11018 [DOI] [PubMed] [Google Scholar]
- 22.Biebricher C. K., Eigen M., Luce R. (1986) Nature 321, 89–91 [DOI] [PubMed] [Google Scholar]
- 23.Oberholzer T., Wick R., Luisi P. L., Biebricher C. K. (1995) Biochem. Biophys. Res. Commun. 207, 250–257 [DOI] [PubMed] [Google Scholar]
- 24.Szostak J. W., Bartel D. P., Luisi P. L. (2001) Nature 409, 387–390 [DOI] [PubMed] [Google Scholar]
- 25.Kita H., Matsuura T., Sunami T., Hosoda K., Ichihashi N., Tsukada K., Urabe I., Yomo T. (2008) Chem. Biochem. 9, 2403–2410 [DOI] [PubMed] [Google Scholar]
- 26.Ichihashi N., Matsuura T., Kita H., Hosoda K., Sunami T., Tsukada K., Yomo T. (2008) Chem. Biochem. 9, 3023–3028 [DOI] [PubMed] [Google Scholar]
- 27.Chetverin A. B., Chetverina H. V., Demidenko A. A., Ugarov V. I. (1997) Cell 88, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyagi S., Landegren U., Tazi M., Lizardi P. M., Kramer F. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5395–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morozov I. Y., Ugarov V. I., Chetverin A. B., Spirin A. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9325–9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abramson R. D., Myers T. W. (1993) Curr. Opin. Biotechnol. 4, 41–47 [DOI] [PubMed] [Google Scholar]
- 31.Mills D. R., Kramer F. R. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 2232–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopsidas G., Roberts A. S., Coia G., Streltsov V. A., Nuttall S. D. (2006) Immunol. Lett. 107, 163–168 [DOI] [PubMed] [Google Scholar]
- 33.Eoyang L., August J. T. (1968) Methods Enzymol. 12B, 530–540 [Google Scholar]
- 34.Hosoda K., Matsuura T., Kita H., Ichihashi N., Tsukada K., Yomo T. (2007) J. Biol. Chem. 282, 15516–15527 [DOI] [PubMed] [Google Scholar]
- 35.Kita H., Cho J., Matsuura T., Nakaishi T., Taniguchi I., Ichikawa T., Shima Y., Urabe I., Yomo T. (2006) J. Biosci. Bioeng. 101, 421–426 [DOI] [PubMed] [Google Scholar]
- 36.Kamen R. (1972) Biochim. Biophys. Acta 262, 88–100 [DOI] [PubMed] [Google Scholar]
- 37.Inaba K., Ito K. (2002) EMBO. J. 21, 2646–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakaishi T., Iio K., Yamamoto K., Urabe I., Yomo T. (2002) J. Biosci. Bioeng. 93, 322–327 [DOI] [PubMed] [Google Scholar]
- 39.Ugarov V. I., Demidenko A. A., Chetverin A. B. (2003) J. Biol. Chem. 278, 44139–44146 [DOI] [PubMed] [Google Scholar]
- 40.Kono M., Yu H., Oprian D. D. (1998) Biochemistry 37, 1302–1305 [DOI] [PubMed] [Google Scholar]
- 41.Johnson J. L., Lasagna M. D., Reinhart G. D. (2001) Protein Sci. 10, 2186–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pace N. R., Haruna I., Spiegelman S. (1968) Methods Enzymol. 12B, 540–556 [Google Scholar]
- 43.Blumenthal T. (1979) Methods Enzymol. 60, 628–638 [DOI] [PubMed] [Google Scholar]
- 44.Moody M. D., Burg J. L., DiFrancesco R., Lovern D., Stanick W., Lin-Goerke J., Mahdavi K., Wu Y., Farrell M. P. (1994) Biochemistry 33, 13836–13847 [DOI] [PubMed] [Google Scholar]
- 45.Perry L. J., Wetzel R. (1984) Science 226, 555–557 [DOI] [PubMed] [Google Scholar]
- 46.Kidmose R. T., Vasiliev N. N., Chetverin A. B., Andersen G. R., Knudsen C. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10884–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H. X., Rivas G., Minton A. P. (2008) Annu Rev. Biophys. 37, 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlsson F., Malmborg-Hager A. C., Albrekt A. S., Borrebaeck C. A. (2005) Can. J. Microbiol. 51, 29–35 [DOI] [PubMed] [Google Scholar]
- 49.Kolakofsky D., Weissmann C. (1971) Biochim. Biophys. Acta 246, 596–599 [DOI] [PubMed] [Google Scholar]
- 50.Zheng M., Aslund F., Storz G. (1998) Science 279, 1718–1721 [DOI] [PubMed] [Google Scholar]