Abstract
Plants utilize ethylene as a hormone to regulate multiple developmental processes and to coordinate responses to biotic and abiotic stress. In Arabidopsis thaliana, a small family of five receptor proteins typified by ETR1 mediates ethylene perception. Our previous work suggested that copper ions likely play a role in ethylene binding. An independent study indicated that the ran1 mutants, which display ethylene-like responses to the ethylene antagonist trans-cyclooctene, have mutations in the RAN1 copper-transporting P-type ATPase, once again linking copper ions to the ethylene-response pathway. The results presented herein indicate that genetically engineered Saccharomyces cerevisiae expressing ETR1 but lacking the RAN1 homolog Ccc2p (Δccc2) lacks ethylene-binding activity. Ethylene-binding activity was restored when copper ions were added to the Δccc2 mutants, showing that it is the delivery of copper that is important. Additionally, transformation of the Δccc2 mutant yeast with RAN1 rescued ethylene-binding activity. Analysis of plants carrying loss-of-function mutations in ran1 showed that they lacked ethylene-binding activity, whereas seedlings carrying weak alleles of ran1 had normal ethylene-binding activity but were hypersensitive to copper-chelating agents. Altogether, the results show an essential role for RAN1 in the biogenesis of the ethylene receptors and copper homeostasis in Arabidopsis seedlings. Furthermore, the results indicate cross-talk between the ethylene-response pathway and copper homeostasis in Arabidopsis seedling development.
Keywords: Copper, Metalloproteins, Plant, Receptors, Signal Transduction, Ethylene Signaling, Silver
Introduction
Ethylene is a plant hormone that influences many aspects of development and responses to the environment, including the timing of seed germination, seedling morphology, leaf expansion, fruit ripening, senescence and abscission of fruits and flower parts, and responses to pathogens (1, 2). Various studies have identified a number of components in the ethylene-signaling pathway and led to increasingly refined models for signal transduction (3, 4). In these models, responses to ethylene are mediated by a family of receptors.
The model plant Arabidopsis thaliana contains five receptor isoforms (ETR1 (ethylene response 1), ERS1 (ethylene response sensor 1), ETR2, EIN4 (ethylene-insensitive 4), and ERS2) (5–8). These proteins contain several membrane-spanning α-helices at the N-terminal region, followed by a cytoplasmic C-terminal region containing a GAF (cGMP phosphodiesterase/adenyl cyclase/FhlA) motif, a domain similar to bacterial histidine kinases and, in some cases, a receiver domain. The receptors have homology to bacterial two-component receptors that transduce signals via the autophosphorylation of a His residue in the kinase domain, followed by the transfer of phosphate to a conserved Asp residue in the receiver domain of a response regulator protein (9). Although some of the receptors are capable of His kinase activity (10, 11) and ethylene may inhibit this activity in ETR1 (12), kinase activity is not needed for signaling (13–15). The N terminus of these receptors contains the novel ligand-binding domain (16–18), and a number of studies have now identified amino acid residues in the three α-helices of ETR1 that are important for ethylene binding and signal transduction (16, 19–21).
We have shown previously that the addition of copper to extracts of yeast cells expressing the Arabidopsis ETR1 receptor enhances ethylene-binding activity and that copper co-purifies with ETR1 (16, 22). The addition of copper to intact yeast cells expressing ETR1 has no effect on ethylene-binding activity, suggesting that yeast cells maintain internal concentrations of copper at high enough levels to support biogenesis of ETR1. These results support earlier speculations about the requirement for a transition metal cofactor for ethylene binding (23–26). Of many transition metals tested, only silver and gold ions can substitute as a cofactor for ethylene binding to ETR1 (16, 22). Interestingly, the etr1-1 mutant fails to coordinate copper and is unable to bind ethylene (16, 19). This requirement for copper is likely to be a general feature of all ethylene receptors in plants (17).
Additional support for copper as the transition metal in the receptors comes from genetic studies on the ran1 (response to antagonist 1) mutants (27–30). Two partial loss-of-function ran1 alleles (ran1-1 and ran1-2) have been identified from a mutant screen (28). Both contain point mutations predicted to reduce function of RAN1. The ran1-1 allele contains a T497I substitution in the phosphatase domain, whereas the ran1-2 allele contains a G173E substitution, which might affect the metal-binding capacity of RAN1 (28). These mutants have normal responses to ethylene. However, exposure of either ran1-1 or ran1-2 mutants to the ethylene-response antagonist trans-cyclooctene causes phenotypes similar to those caused by ethylene, whereas treatment of wild-type plants with trans-cyclooctene inhibits ethylene signaling. Support that this is related to copper comes from the observation that the addition of copper ions to ran1-1 and ran1-2 plants partially suppresses the ran1 phenotype in plants containing these weak alleles (28). Two stronger alleles of RAN1 have also been identified. The ran1-3 allele contains a G759R substitution in the predicted ATPase domain, which results in a nonfunctional protein (29), whereas ran1-4 has a T-DNA insertion within the second intron that is predicted to disrupt translation of RAN1 (30). Both ran1-3 and ran1-4 are null alleles that result in phenotypes similar to loss-of-function receptor mutants (27, 29, 30). Therefore, it is possible that copper is needed for both ethylene binding and functional receptors. RAN1 encodes a protein similar to the copper-transporting CPx class of P-type ATPases such as Ccc2 protein from Saccharomyces cerevisiae and human Menkes/Wilson proteins (31–36). One function of these ATPases is to deliver copper to copper-containing enzymes in extracytoplasmic compartments. Genetic analyses showed that RAN1 acts upstream of the ethylene receptors, prompting the hypothesis that it delivers copper to the ethylene receptors (28, 29).
However, the specific role of RAN1 in the ethylene-response pathway is not clear because ran1-1 and ran1-2 mutants show normal physiology in the presence and absence of applied ethylene, and ran1-3 and ran1-4 mutations show ethylene-independent growth defects. To determine the molecular role of RAN1 in the ethylene-response pathway and to test the hypothesis that it delivers copper to the ethylene receptors, we examined whether RAN1 plays a role in the biogenesis of ethylene receptors. Expression of ETR1 in S. cerevisiae mutants lacking the Ccc2 protein, a homolog of RAN1, showed that the Ccc2 protein and RAN1 can mediate the delivery of copper ions to ETR1 to generate ethylene-binding activity. Seedlings carrying the weak alleles ran1-1 and ran1-2 displayed the ethylene triple response in the presence of copper chelators, phenocopying the stronger alleles ran1-3 and ran1-4. Additionally, ethylene binding to Arabidopsis seedlings was absent in the ran1-3 and ran1-4 mutants. Taken together, our results indicate that RAN1 is essential for the biogenesis of ethylene receptors and that this is crucial for normal growth and development.
EXPERIMENTAL PROCEDURES
The Arabidopsis ran1 mutants used in this study have been described previously (28–30) and were obtained from Joseph Ecker and Ed Himmelblau. The Δccc2 yeast cells (37) were kindly provided by Daniel Yuan. The ethylene biosynthesis inhibitor l-α-(2-aminoethoxyvinyl)glycine (AVG)4 was a gift from Rohm and Haas (Philadelphia, PA). Lysophosphatidylcholine was from Sigma. 14C2H4 and [3H]C2H4 were obtained from American Radiolabeled Chemicals (St. Louis, MO).
Plant Growth and Ethylene Treatment Conditions
Arabidopsis seedlings were grown on 0.8% (w/v) agar plates containing half-strength Murashige and Skoog agar medium (38) with 10 μm AVG to inhibit ethylene biosynthesis and no added sucrose (referred to as ½MSNS-AVG). Seeds were stratified for 4 days at 4 °C, exposed to white fluorescent lights for 12 h, and grown in darkness for 4 days. For ethylene dose-response assays, the seedlings were grown for 4 days in the indicated concentrations of ethylene or ethylene-free air. Ethylene concentrations were determined by gas chromatography using a Carboxen 1000 (45/60 mesh column, Supelco, Bellefonte, PA) with ethylene as a calibration standard (20). To test the effect of chelators and silver on plant growth, sterile solutions of the indicated compound were prepared and added at the indicated concentrations to the sterilized ½MSNS-AVG agar prior to solidification.
Seedling Ethylene Binding Assays
For ethylene binding assays on plants, 100 seeds were grown for 4 days on sterilized filter paper placed on top of ½MSNS-AVG agar plates. The filters with seedlings were transferred to sealed chambers and assayed for [3H]C2H4 binding according to the methods of Sisler (39). Saturable ethylene binding was determined by comparing seedlings treated with [3H]C2H4 (0.1 μl/liter) alone or with [3H]C2H4 (0.1 μl/liter) in the presence of excess nonradioactive ethylene (100 μl/liter). The dry weight of plants was determined by drying the filters for 1 week at 65°.
DNA Constructs, Yeast Strains, and Growth Conditions
ETR1 was expressed in S. cerevisiae using a construct containing a PCR-amplification product of ETR1 cDNA ligated into the pYcDE2 vector as described (19). RAN1/HA, ran1-1/HA, and ran1-2/HA fusion constructs contain the RAN1 cDNA sequences fused to three copies of the HA epitope tag in the pYES3 vector (28). All of the yeast expression constructs were propagated in Escherichia coli (strain DH5α), purified by alkaline lysis extraction, and introduced into yeast using the lithium acetate method (40). The role of the Ccc2 protein in the biogenesis of ETR1 in heterologous S. cerevisiae was examined by introducing the pYcDE-ETR1 construct in the yeast 2908 strain 7 (Δccc2) and its parental strain 2908 strain 6 (Ccc2) (37). The functional complementation of the ethylene-binding activity of ETR1 expressed in Δccc2 yeast (Δccc2-ETR1 strain) by RAN1, ran1-1, and ran1-2 was examined by introducing individual constructs into Δccc2-ETR1. All S. cerevisiae cells were grown in synthetic dextrose medium lacking appropriate amino acids to select cells containing the corresponding expression vectors.
Ethylene Binding Assays in Intact Yeast Cells and Yeast Membranes
Intact yeast cells were harvested and assayed for binding of 14C2H4 using methods described previously (19, 39). To determine ethylene binding in yeast membranes, the cells were then disrupted, and membranes were isolated as described previously (16). Membranes were rapidly frozen in liquid nitrogen and stored at −80 °C until used. Saturable ethylene binding in isolated yeast membranes expressing full-length ETR1 protein was determined using the methods of Sisler (39) as modified by others (16). Saturable ethylene binding was determined by comparing yeast cells treated with 14C2H4 (0.1 μl/liter) alone or with 14C2H4 (0.1 μl/liter) in the presence of excess 12C2H4 (100 μl/liter). The levels of ETR1 were examined by Western blotting using anti-ETR1 antibody HRR (41).
The effect of the copper-chelating agents on ethylene binding was determined using diethyldithiocarbamate, bathocuproinedisulfonic acid, or neocuproine. Yeast membrane preparations were solubilized with l-α-lysophosphatidylcholine (5 mg/ml) as described previously (16, 42) and incubated at 37 °C with the indicated concentrations of chelators for 30 min. The chelator complex was separated from the proteins by gel filtration in 10-ml Sephadex G-25 columns pre-equilibrated with solubilization buffer and spun for 2 min at 500 × g. Visual inspection confirmed that the chelator-copper complex was retained in the column. Ethylene binding assays were then carried out as described above in the presence or absence of 300 μm CuSO4.
RESULTS
Copper and Reconstitution of Ethylene Binding in Exogenously Expressed ETR1 Receptor
We have shown previously that copper is associated with the ETR1 receptor (16) and that of numerous transition metals tested, only copper and the other Group 11 transition metals (silver and gold) can act as cofactors for ethylene binding to ETR1 exogenously expressed in yeast (16, 22). To more completely characterize the requirement for a copper transporter in receptor biogenesis, the ETR1 ethylene receptor was expressed in yeast, and ethylene binding was assessed. This approach has previously yielded many details about ethylene binding to the receptors (16–22, 42).
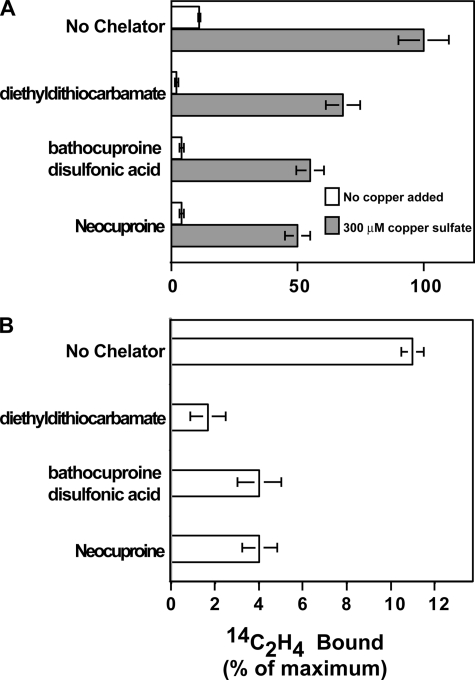
To provide further evidence that copper is the cofactor present in ETR1, we tested the effect of the copper chelators diethyldithiocarbamate, bathocuproinedisulfonic acid, and neocuproine on ethylene binding in membranes isolated from yeast. In the absence of chelators or added copper, a low level of ethylene binding was observed (Fig. 1). The addition of 300 μm CuSO4 enhanced ethylene-binding activity by ∼9-fold (Fig. 1A). The addition of any of these copper chelators at a concentration of 5 mm in the absence of added CuSO4 resulted in a reduction in ethylene binding (Fig. 1B). In the absence of chelator and added copper, ethylene binding was 11% of maximal binding. The addition of 5 mm diethyldithiocarbamate reduced this by ∼85%, whereas the addition of either 5 mm bathocuproinedisulfonic acid or neocuproine reduced this by ∼65%. The subsequent readdition of 300 μm CuSO4 restored ethylene binding. Restoration of ethylene binding ranged from ∼50 to 75% of the binding seen in the absence of chelator. These reduced levels of ethylene binding could be due to the presence of residual levels of chelator, reduced levels of ETR1 protein, or both. Nonetheless, these results indicate that the effect of these copper chelators is likely due to the removal of copper from ETR1.
FIGURE 1.
Copper chelators and ethylene-binding activity in ETR1 expressed in yeast. A, yeast membranes isolated from yeast cells expressing ETR1 were analyzed for 14C2H4 bound in the presence of various copper chelators in the presence or absence of 300 μm copper sulfate. The copper chelators diethyldithiocarbamate, bathocuproinedisulfonic acid, and neocuproine were tested at a concentration of 5 mm. B, data in the absence of added copper from A are shown at an expanded scale. Saturable ethylene binding to ETR1 was determined as described under “Experimental Procedures” and normalized to the amount of radioactivity in the presence of copper with no chelator added. The mean ± S.D. is shown.
ETR1 Ethylene-binding Activity When Expressed in Δccc2 Yeast
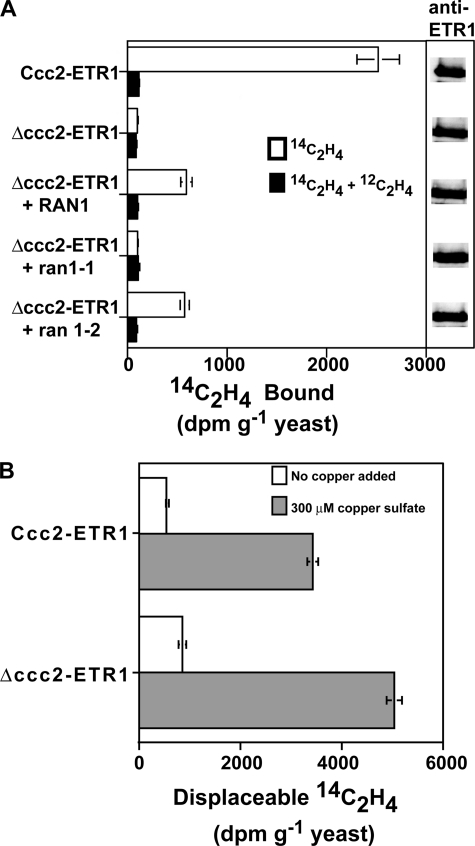
The RAN1 copper transporter has been linked to ethylene sensing in Arabidopsis and is presumed to be important in the delivery of copper to the receptors (27–30). However, this has not been directly shown to be the case. The yeast Ccc2 protein is a homolog of RAN1 that transports copper into the trans-Golgi network in S. cerevisiae (37). To investigate whether the Ccc2 protein plays a role in the generation of ethylene-binding sites in S. cerevisiae, we expressed ETR1 in mutant yeast cells lacking Ccc2 (Δccc2-ETR1) and Ccc2 control yeast cells (Ccc2-ETR1). When ethylene binding was measured in intact yeast, significant levels of ethylene binding were detected in Ccc2-ETR1 yeast (Fig. 2A). In contrast, Δccc2-ETR1 mutants lacked detectable ethylene-binding activity, pointing to a central role for Ccc2p in the biogenesis of ETR1 in yeast. Similar levels of immunolabeled ETR1 were detected in both lines, indicating that this difference is not due to different levels of ETR1 protein. To test the involvement of copper in this Ccc2-dependent binding, we isolated membranes from Ccc2-ETR1 yeast and Δccc2-ETR1 mutants and added 300 μm CuSO4 to these extracts. In the absence of added copper, low levels of ethylene binding were observed in membranes isolated from either Ccc2-ETR1 or Δccc2-ETR1 yeast, whereas the addition of CuSO4 reconstituted the ethylene-binding activity in both (Fig. 2B). The Δccc2-ETR1 yeast membranes had slightly higher binding activity. This might represent slightly higher levels of ETR1 protein in these extracts. The addition of CdSO4, CoSO4, MnSO4, NiSO4, or ZnSO4 did not restore ethylene-binding activity to these samples (data not shown). These results suggest that the copper-transporting activity of the Ccc2 protein is essential for the biogenesis of ethylene-binding sites in ETR1 exogenously expressed in S. cerevisiae.
FIGURE 2.
Ccc2 and RAN1 copper transporters and saturable ethylene binding to ETR1 expressed in yeast. A, intact yeast cells expressing the ETR1 receptor in the Ccc2 (Ccc2-ETR1) or Δccc2 (Δccc2-ETR1) background were analyzed for [14C]ethylene binding. The effects of transforming Δccc2-ETR1 yeast with RAN1, ran1-1, and ran1-2 on ETR1 ethylene binding are shown. Ethylene binding using [14C]ethylene is compared between samples treated with [14C]ethylene (0.1 μl/liter) and identical samples treated with [14C]ethylene (0.1 μl/liter) plus [12C]ethylene (100 μl/liter). Equal amounts of yeast used in the binding assays were analyzed on Western blots probed with anti-ETR1 antibodies. B, displaceable ethylene binding in the presence and absence of 300 μm copper sulfate is shown for membranes isolated from Ccc2-ETR1 and Δccc2-ETR1 yeast. Displaceable binding was calculated by subtracting the amount of [14C]ethylene (0.1 μl/liter) bound in the presence of excess [12C]ethylene (100 μl/liter) from amount of [14C]ethylene (0.1 μl/liter) bound in the absence of added [12C]ethylene. In both panels, the mean ± S.D. for disintegrations/min/g of yeast is shown.
RAN1 Reconstitutes Ethylene-binding Activity of ETR1 in Δccc2 Yeast
Previous studies showed that expression of Arabidopsis RAN1 can rescue copper-dependent high affinity iron uptake in Δccc2 mutants (28). This demonstrated that RAN1 is a functional homolog of the Ccc2 copper transporter. To determine whether RAN1 can also rescue the ethylene-binding activity of Δccc2-ETR1 yeast, we introduced constructs for the expression of the RAN1, ran1-1, or ran1-2 proteins into Δccc2-ETR1 yeast. Expression of either RAN1 or ran1-2 increased the ethylene-binding activity of Δccc2-ETR1 cells to ∼30% of the levels seen in Ccc2-ETR1 cells (Fig. 2A). In contrast, expression of ran1-1 did not restore the ethylene-binding activity to Δccc2-ETR1 cells (Fig. 2A). The difference in ethylene binding was not due to different levels of expression of ETR1 because similar levels of immunolabeled ETR1 were detected in samples from all cell lines (Fig. 2A). It is possible that the differences were due to different expression levels of RAN1, ran1-1, and ran1-2 in the yeast cells, but these results are consistent with previous studies indicating that RAN1 and ran1-2, but not ran1-1, can efficiently rescue high affinity iron uptake in Δccc2 mutants (28). This provides further support for the idea that RAN1 plays a function in the biogenesis of ETR1 ethylene-binding activity.
Ethylene Binding in Arabidopsis ran1 Mutants
The results above provide evidence that RAN1 may play a role in the delivery of copper to the ETR1 ethylene receptor and the biogenesis of the ethylene receptors in planta. Previous studies have shown that mutant Arabidopsis seedlings carrying the strong allele ran1-3 or ran1-4 display a constitutive activation of ethylene responses similar to the phenotype of mutants lacking multiple ethylene receptor isoforms (27, 29, 30). In contrast, the ran1-1 and ran1-2 mutants show no obvious alteration in responses to ethylene but have an altered response to the ethylene antagonist trans-cyclooctene (28). The various phenotypes of ran1 mutants suggest the possibility that these mutations cause a defect in ethylene receptor function because copper is not delivered to the receptor properly. To investigate whether ran1 mutants have a defect in ethylene perception, we assessed ethylene binding in seedlings homozygous for each of the ran1 alleles.
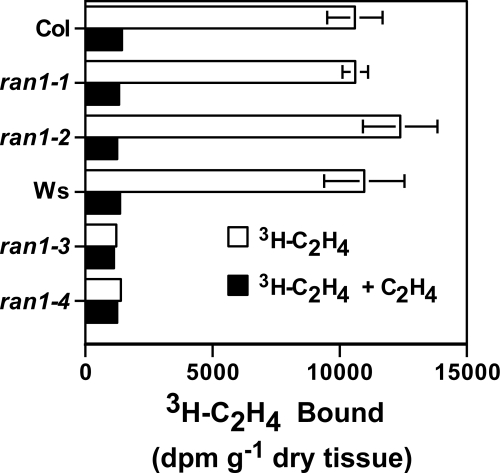
Ethylene binding assays using radiolabeled ethylene showed that the levels of ethylene binding in the weak ran1-1 and ran1-2 mutants were indistinguishable from ethylene binding in Columbia control seedlings (Fig. 3). In contrast, there was no detectable binding in the ran1-3 and ran1-4 loss-of-function mutant seedlings, whereas the wild-type Wassilewskija controls had high levels of ethylene binding (Fig. 3). Previous work has shown that ran1 mutants have wild-type levels of ETR1 protein (43). Thus, loss of ethylene binding is not due to loss of the ETR1 receptor. These results show that eliminating the RAN1 protein leads to severe deficits in ethylene binding and are consistent with earlier studies indicating that a functional RAN1 is required for ethylene responses in plants.
FIGURE 3.
Ethylene binding in Arabidopsis ran1 mutants. [3H]Ethylene binding to receptors in wild-type seedlings and the four ran1 mutants was analyzed. Analysis was performed on 100 seedlings that were first allowed to grow for 4 days. Saturable ethylene binding is shown as the difference in [3H]ethylene between samples treated with [3H]ethylene (0.1 μl/liter) and identical samples treated with [3H]ethylene (0.1 μl/liter) plus nonradioactive ethylene (100 μl/liter). The mean ± S.D. for disintegrations/min/g of plant (dry weight) is shown. Col, Columbia; Ws, Wassilewskija.
Hypersensitivity of Arabidopsis ran1-1 and ran1-2 Mutants to Copper-chelating Agents
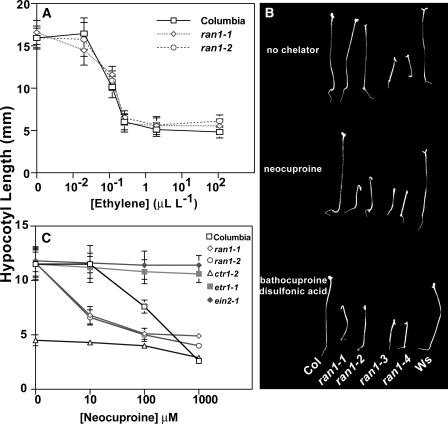
It has been hypothesized that the ran1-1 and ran1-2 mutations alter the copper-transporting activity of RAN1, resulting in a reduction of copper delivery to the ethylene receptors (28). However, consistent with previous work (28), neither ran1-1 nor ran1-2 seedlings showed an altered growth inhibition response upon application of ethylene over a wide concentration range (Fig. 4A).
FIGURE 4.
Effects of copper chelators on the ran1-1 and ran1-2 mutants. A, the ethylene dose responses on hypocotyl length in wild-type (Columbia (Col)) and ran1 mutants are shown. Etiolated Arabidopsis seedlings were treated with the indicated concentrations of ethylene. B, the effects of 10 μm neocuproine and 100 μm bathocuproinedisulfonic acid on etiolated seedlings are shown. For comparison, seedlings not treated with a copper chelator are shown. The ran1 mutants are compared with their wild-type controls (Columbia for ran1-1 and ran1-2 and Wassilewskija (Ws) for ran1-3 and ran1-4). C, the dose response for neocuproine on the growth of hypocotyls of etiolated seedlings is shown. In all panels, seedlings were grown in the dark for 4 days under the conditions indicated. In A and C, data represent the mean ± S.D. for hypocotyl length.
To determine whether the ran1-1 and ran1-2 mutations have subtle effects on copper homeostasis in Arabidopsis, we examined the growth of ran1-1 and ran1-2 etiolated seedlings on plates containing either bathocuproinedisulfonic acid or neocuproine to chelate copper. In this way, we hoped to determine whether the mutants have altered copper responses that are masked at higher copper levels. Neither 10 μm neocuproine nor 100 μm bathocuproinedisulfonic acid had a measurable effect on growth of wild-type seedlings (Fig. 4B). However, both ran1-1 and ran1-2 seedlings grew ∼50% slower than wild-type seedlings under these conditions and had enhanced apical hook curvature (Fig. 4B). These responses are similar to the responses seen when etiolated Arabidopsis seedlings are treated with ethylene. Neither ran1-3 nor ran1-4 mutants responded to treatment with these chelators.
Dose-response measurements with neocuproine revealed that the half-maximal growth inhibition response for wild-type seedlings occurred at ∼100 μm neocuproine, whereas the application of <10 μm neocuproine caused half-maximal growth inhibition in the ran1-1 and ran 1–2 seedlings (Fig. 4C). Ethylene-insensitive mutants such as etr1-1 and ein2-1 showed little or no response to neocuproine up to a concentration of 1 mm. Similarly, the constitutive response mutant ctr1-2 showed only a minor decrease in length upon treatment with 1 mm neocuproine (Fig. 4C). These results suggest that the ran1-1 and ran1-2 mutants have altered copper homeostasis and that the ethylene-like responses observed in ran1-1 and ran1-2 grown in the presence of copper chelators occur via signaling through the known ethylene-signaling pathway.
Reduced Inhibition of Ethylene Responses by Silver in ran1-1 and ran1-2 Mutants
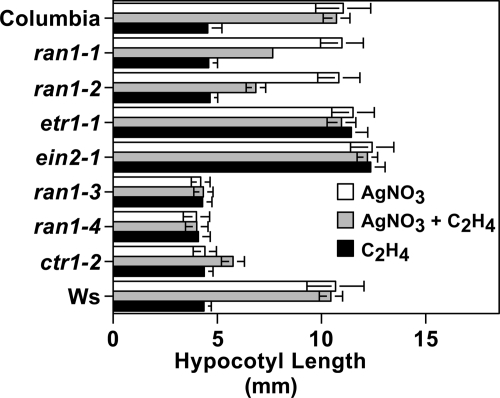
Silver ions act as a potent inhibitor of ethylene responses in plants (24). Interestingly, silver ions can act as a cofactor for ethylene binding in the ETR1 receptor (16, 22). Because of this, silver has been presumed to work by displacing the native metal cofactor in the ethylene-binding site to interfere with signal transduction. The constitutive ethylene response of ran1-3 seedlings is not affected by silver ions, despite the fact that genetic analysis showed that the ethylene receptors are epistatic to RAN1 (28, 29). Because our results indicate that RAN1 delivers copper to the ethylene receptors and a similar protein, CopB from Enterococcus hirae, can transport both copper and silver ions (44), we hypothesized that RAN1 is required for the delivery of silver to the receptors. To test this hypothesis, we examined ethylene responses of ran1 mutant seedlings grown in medium containing 10 μm silver nitrate. Columbia and Wassilewskija control seedlings had significant growth inhibition in the presence of ethylene; 10 μm silver nitrate blocked this response entirely in the control seedlings (Fig. 5). Similarly, ran1-1 and ran1-2 mutants responded to ethylene in the absence of silver. However, unlike wild-type seedlings, the addition of silver to ran1-1 and ran1-2 mutants only partially blocked responses to ethylene (Fig. 5). In the presence of silver, the application of ethylene inhibited growth by ∼50% in these mutants. The ran1-3 and ran1-4 mutants were unresponsive to both silver and ethylene (Fig. 5). These results are consistent with a role for RAN1 in the delivery of silver to the ethylene receptors.
FIGURE 5.
Effects of silver on the response to ethylene in ran1 mutants. Seedlings were grown in the dark for 4 days in the presence of 10 μm silver nitrate, 10 μm silver nitrate plus 100 μl/liter ethylene, or 100 μl/liter ethylene with no added silver. Data represent the mean ± S.D. for hypocotyl length. Ws, Wassilewskija.
DISCUSSION
Prior work demonstrated that expression of Arabidopsis ETR1 generates membrane-associated ethylene-binding sites in S. cerevisiae (19). The results presented here expand upon these earlier results to show that the Ccc2 copper transporter, a homolog of RAN1, is required for the biogenesis of ETR1 expressed heterologously in S. cerevisiae. Similarly, RAN1 in Arabidopsis is required for the development of normally functioning receptors in planta.
Because excess copper ions restored the ethylene-binding activity of ETR1 in Δccc2 mutants, it is the copper ions transported by Ccc2, and not the Ccc2 protein itself, that are required for the ethylene-binding activity of ETR1. This restoration was specific to copper because other transition metals tested did not restore ethylene binding in ETR1. The ability of the Ccc2 protein to deliver copper ions to a heterologous protein with no similarity to any of its native proteins suggests that the delivery of copper to proteins in the secretory compartment is not strictly regulated. Expression of Arabidopsis RAN1 rescued the ethylene-binding activity of ETR1 in Δccc2 mutants, supporting earlier speculation that RAN1 can deliver copper to ethylene receptors (28, 29). The ability of RAN1 to mediate the biogenesis of ETR1 in a heterologous expression system implies that both proteins are located, at least transiently, in the same subcellular compartment. However, the subcellular localization of ETR1 and RAN1 when expressed in yeast remains to be determined. In Arabidopsis, ETR1 is an integral membrane protein located primarily in the membranes of the endoplasmic reticulum (45–52). Although the subcellular location of RAN1 in Arabidopsis is unknown, our results suggest that it is co-localized in the same subcellular compartment with the receptors at some point during the biogenesis of the receptors in plants.
Assessment of ethylene binding in ran1 mutant seedlings demonstrated that RAN1 plays an essential role in the biogenesis of ethylene receptors in Arabidopsis. Seedlings carrying the strong alleles ran1-3 and ran1-4 lacked detectable levels of ethylene binding. Thus, these mutants lack functional ethylene receptors. These mutants displayed a constitutive ethylene-response phenotype similar to mutants carrying loss-of-function alleles for multiple ethylene receptor genes. In contrast, the ethylene binding levels detected in seedlings carrying the weak alleles ran1-1 and ran1-2 were similar to those in wild-type controls. This is consistent with the normal growth of these mutants in the presence and absence of ethylene. However, in the presence of low levels of copper chelators, both ran1-1 and ran1-2 seedlings showed growth defects that had remarkable similarity to seedlings grown in the presence of ethylene. Higher concentrations of copper chelators induced similar effects on wild-type control seedlings but not etr1-1 or ein2-1 mutants, indicating that the effect of the copper chelators on seedling growth is related to the activation of the ethylene-response pathway. Additional support for a decrease in the ion-transporting activity of ran1 mutant proteins is the effect of silver upon ethylene responses in these mutants. Weak ran1 mutants showed a reduced inhibition of ethylene responses by silver, whereas the loss-of-function mutants were insensitive to silver. Previous studies have demonstrated that CopB from E. hirae can transport silver (44), supporting the idea that RAN1 plays a role in the delivery of silver to the ethylene receptors. Together, these results are consistent with a model in which RAN1 delivers copper to the ethylene receptors and is required for normally functioning ethylene receptors in Arabidopsis.
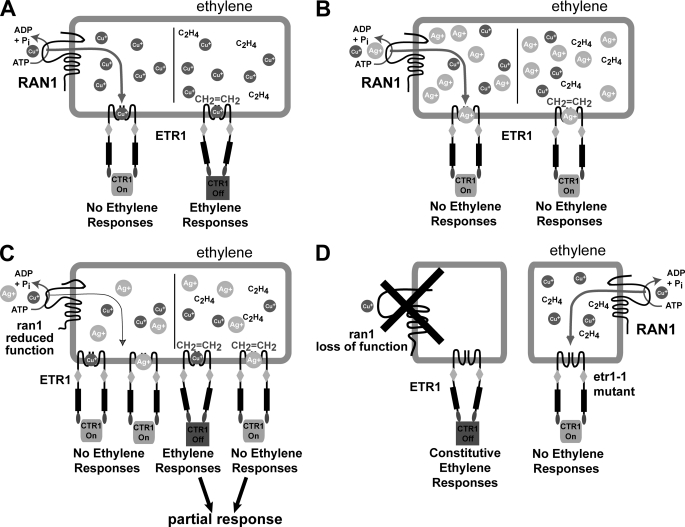
The inverse agonist model for the activation of ethylene responses is based on the constitutive activation of the ethylene-response pathway in plants carrying loss-of-function alleles for three or more ethylene receptor isoforms (7). On the basis of our results and others, we hypothesize that in the absence of ethylene, holoreceptors actively signal to keep CTR1 active, thus inhibiting the ethylene response. Ethylene binding to the copper cofactor in the ethylene-binding site causes conformational changes in the receptors that inhibit their ability to activate CTR1, leading to the derepression of the ethylene responses (Fig. 6A). On the other hand, silver-bound receptors can bind ethylene but are predicted to be incapable of changing their signaling state in response to ethylene binding (Fig. 6B) (53). Weak ran1 mutants such as ran1-1 and ran1-2 have normal responses to ethylene. However, they respond abnormally to the addition of silver. One model for this is that under these conditions, silver is not delivered as efficiently, so part of the receptor population contains silver and part contains copper (Fig. 6C). In this case, an attenuated response to ethylene might occur. It is noteworthy that strong ran1 alleles cause constitutive ethylene responses by eliminating copper delivery to the receptors, whereas the etr1-1 mutation eliminates copper binding to ETR1 but produces ethylene insensitivity (Fig. 6D). Thus, the etr1-1 mutation must not only eliminate copper binding but also produce an altered signaling state of the mutant receptors that keeps CTR1 permanently active and the ethylene-response pathway repressed. One possibility is that the etr1-1 protein is in a hyperactive signaling state that is enough to keep CTR1 active (54). Another possibility is that there is a physical interaction between the receptors so that an interaction between holoreceptors and etr1-1 aporeceptors results in the constitutive activation of CTR1 and the inhibition of ethylene responses. Indirect support for such a receptor clustering model comes from several studies (52, 55–57). In either case, it is clear that the etr1-1 protein and wild-type receptors lacking the copper cofactor are not equivalent.
FIGURE 6.
Model for the function of RAN1 in ethylene receptor biogenesis and function. In this model, RAN1 and the ethylene receptors are presumed to be co-localized in the same membrane system, at least transiently, during receptor biogenesis. A, left, in the absence of ethylene, the receptors are active and function to activate CTR1. This in turn inhibits downstream signaling. Right, ethylene inhibits the receptors by binding to copper in the binding site, leading to decreased CTR1 activity and the release of inhibition to downstream components. When both RAN1 and ETR1 are expressed normally, copper is delivered to the receptor, and normal biogenesis occurs, leading to a normally functioning receptor that binds ethylene and undergoes the required conformational change to turn off. B, in the presence of silver, RAN1 delivers both silver and copper to the receptors but in such a ratio that most contain silver. In this situation, ethylene binds to the silver ion, but the binding event is not transduced through the receptor, and the receptor remains on. C, in the reduced-function ran mutants (ran1-1 and ran1-2), fewer ions are transported to the receptors. When silver is added in this situation, we predict that the population of copper-containing and silver-containing receptors is shifted, so more contain copper, and a partial response to ethylene occurs. D, left, in the ran1 loss-of-function mutants (ran1-3 and ran1-4), little or no copper is transported to the receptors and abnormal biogenesis occurs, resulting in receptors that are permanently turned off. This leads to a constitutive ethylene phenotype. Right, in contrast, the etr1-1 mutant cannot coordinate the copper ion. Therefore, even though RAN1 transports copper to the receptor, it cannot incorporate the ion. However, unlike the wild-type receptors lacking copper in ran1 loss-of-function mutants, the etr1-1 receptor is locked in the signaling state, leading to ethylene-insensitive plants.
The role of RAN1 in the biogenesis of ethylene receptors parallels the function of similar proteins in the biogenesis of cuproenzymes inside intracellular compartments. The related Ccc2 copper transporter plays an essential role in the maturation of Fet3p, a multicopper ferroxidase involved in high affinity iron uptake in S. cerevisiae (37). Menkes/Wilson proteins in humans likely perform an analogous function. For instance, tyrosinase is a copper-containing enzyme. When expressed from Menkes patients in recombinant fibroblast cell lines, it is inactive. However, coexpression of normal Menkes protein in the same cells results in an active tyrosinase (58).
The Arabidopsis genome contains two other genes encoding putative copper-transporting ATPases closely related to RAN1 (PAA1 (P-type ATPase of Arabidopsis 1) and HMA5 (heavy metal-associated 5)) (59). PAA1 functions in the delivery of copper to plastocyanin within chloroplasts, whereas HMA5 may have a mitochondrial localization and is important in copper detoxification in roots (60, 61). It is unlikely that RAN1 plays a similar role in copper resistance. Excess copper ions produce only partial rescue of ran1 mutants (28, 29), inconsistent with a role for RAN1 in copper resistance. The fact that ran1 mutant seedlings can support some limited growth and development is consistent with the possibility that one of these RAN1 homologs has overlapping functions with RAN1 at the seedling stage. However, our results clearly show that RAN1 has a prominent role in the delivery of copper to the ethylene receptors at the seedling stage. The ethylene-independent, rosette-lethal phenotype of strong ran1 mutants indicates that RAN1 function is completely essential for vegetative development at the adult stage (29). Other roles for RAN1 in plant growth remain to be established.
Acknowledgments
We express our gratitude to Daniel Yuan for providing the Ccc2 mutant yeast strains, Joseph Ecker and Ed Himmelblau for providing ran1 mutants, and members of A. B. Bleecker's laboratory for insightful discussions. We thank Shimon Meir, Sonia Philosoph-Hadas, and Dan Roberts for helpful discussions.
This work was supported by National Science Foundation Grants MCB-0918430 (to B. M. B.) and MCB-0131564 (to A. B. B.).
- AVG
- l-α-(2-aminoethoxyvinyl)glycine.
REFERENCES
- 1.Abeles F., Morgan P., Saltveit M. J. (1992) Ethylene in Plant Biology, 2nd Ed., Academic Press, San Diego, CA [Google Scholar]
- 2.Mattoo A. K., Suttle J. C. (1991) The Plant Hormone Ethylene, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 3.Hall B., Shakeel S., Schaller G. (2007) J. Plant Growth Regul. 26, 118–130 [Google Scholar]
- 4.Li H., Guo H. (2007) J. Plant Growth Regul. 26, 107–117 [Google Scholar]
- 5.Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M. (1993) Science 262, 539–544 [DOI] [PubMed] [Google Scholar]
- 6.Hua J., Meyerowitz E. M. (1998) Cell 94, 261–271 [DOI] [PubMed] [Google Scholar]
- 7.Hua J., Sakai H., Nourizadeh S., Chen Q. G., Bleecker A. B., Ecker J. R., Meyerowitz E. M. (1998) Plant Cell 10, 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakai H., Hua J., Chen Q. G., Chang C., Medrano L. J., Bleecker A. B., Meyerowitz E. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West A. H., Stock A. M. (2001) Trends Biochem. Sci. 26, 369–376 [DOI] [PubMed] [Google Scholar]
- 10.Gamble R. L., Coonfield M. L., Schaller G. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moussatche P., Klee H. J. (2004) J. Biol. Chem. 279, 48734–48741 [DOI] [PubMed] [Google Scholar]
- 12.Voet-van-Vormizeele J., Groth G. (2008) Mol. Plant 1, 380–387 [DOI] [PubMed] [Google Scholar]
- 13.Binder B. M., O'Malley R. C., Wang W., Moore J. M., Parks B. M., Spalding E. P., Bleecker A. B. (2004) Plant Physiol. 136, 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W., Hall A. E., O'Malley R., Bleecker A. B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu X., Schaller G. E. (2004) Plant Physiol. 136, 2961–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez F. I., Esch J. J., Hall A. E., Binder B. M., Schaller G. E., Bleecker A. B. (1999) Science 283, 996–998 [DOI] [PubMed] [Google Scholar]
- 17.O'Malley R. C., Rodriguez F. I., Esch J. J., Binder B. M., O'Donnell P., Klee H. J., Bleecker A. B. (2005) Plant J. 41, 651–659 [DOI] [PubMed] [Google Scholar]
- 18.Hall A. E., Findell J. L., Schaller G. E., Sisler E. C., Bleecker A. B. (2000) Plant Physiol. 123, 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaller G. E., Bleecker A. B. (1995) Science 270, 1809–1811 [DOI] [PubMed] [Google Scholar]
- 20.Hall A. E., Chen Q. G., Findell J. L., Schaller G. E., Bleecker A. B. (1999) Plant Physiol. 121, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Esch J. J., Shiu S. H., Agula H., Binder B. M., Chang C., Patterson S. E., Bleecker A. B. (2006) Plant Cell 18, 3429–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binder B. M., Rodriguez F. I., Bleecker A. B., Patterson S. E. (2007) FEBS Lett. 581, 5105–5109 [DOI] [PubMed] [Google Scholar]
- 23.Thompson J., Harlow R., Whitney J. (1983) J. Am Chem. Soc. 105, 3522–3527 [Google Scholar]
- 24.Beyer E. M. (1976) Plant Physiol. 58, 268–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burg S. P., Burg E. A. (1967) Plant Physiol. 42, 144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisler E. C. (1976) Tobacco Sci. 21, 43–45 [Google Scholar]
- 27.Hall A. E., Bleecker A. B. (2003) Plant Cell 15, 2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirayama T., Kieber J. J., Hirayama N., Kogan M., Guzman P., Nourizadeh S., Alonso J. M., Dailey W. P., Dancis A., Ecker J. R. (1999) Cell 97, 383–393 [DOI] [PubMed] [Google Scholar]
- 29.Woeste K. E., Kieber J. J. (2000) Plant Cell 12, 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Himelblau E., Amasino R. M. (2001) J. Plant Physiol. 158, 1317–1323 [Google Scholar]
- 31.Fu D., Beeler T. J., Dunn T. M. (1995) Yeast 11, 283–292 [DOI] [PubMed] [Google Scholar]
- 32.Himelblau E., Amasino R. M. (2000) Curr. Opin. Plant Biol. 3, 205–210 [PubMed] [Google Scholar]
- 33.Solioz M., Vulpe C. (1996) Trends Biochem. Sci. 21, 237–241 [PubMed] [Google Scholar]
- 34.Chelly J., Tümer Z., Tønnesen T., Petterson A., Ishikawa-Brush Y., Tommerup N., Horn N., Monaco A. P. (1993) Nat. Genet. 3, 14–19 [DOI] [PubMed] [Google Scholar]
- 35.Mercer J. F., Livingston J., Hall B., Paynter J. A., Begy C., Chandrasekharappa S., Lockhart P., Grimes A., Bhave M., Siemieniak D., Glover T. W. (1993) Nat. Genet. 3, 20–25 [DOI] [PubMed] [Google Scholar]
- 36.Vulpe C., Levinson B., Whitney S., Packman S., Gitschier J. (1993) Nat. Genet. 3, 7–13 [DOI] [PubMed] [Google Scholar]
- 37.Yuan D. S., Stearman R., Dancis A., Dunn T., Beeler T., Klausner R. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2632–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murashige T., Skoog F. (1962) Physiol. Plant. 15, 473–497 [Google Scholar]
- 39.Sisler E. C. (1979) Plant Physiol. 64, 538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaller G. E., Ladd A. N., Lanahan M. B., Spanbauer J. M., Bleecker A. B. (1995) J. Biol. Chem. 270, 12526–12530 [DOI] [PubMed] [Google Scholar]
- 42.Schaller G. E., Binder B. M. (2007) Methods Enzymol. 422, 270–287 [DOI] [PubMed] [Google Scholar]
- 43.Zhao X. C., Qu X., Mathews D. E., Schaller G. E. (2002) Plant Physiol. 130, 1983–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solioz M., Odermatt A. (1995) J. Biol. Chem. 270, 9217–9221 [DOI] [PubMed] [Google Scholar]
- 45.Chen Y. F., Randlett M. D., Findell J. L., Schaller G. E. (2002) J. Biol. Chem. 277, 19861–19866 [DOI] [PubMed] [Google Scholar]
- 46.Ma B., Cui M. L., Sun H. J., Takada K., Mori H., Kamada H., Ezura H. (2006) Plant Physiol. 141, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong C. H., Rivarola M., Resnick J. S., Maggin B. D., Chang C. (2008) Plant J. 53, 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao W. H., Liu J., He X. J., Mu R. L., Zhou H. L., Chen S. Y., Zhang J. S. (2007) Plant Physiol. 143, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y. F., Shakeel S. N., Bowers J., Zhao X. C., Etheridge N., Schaller G. E. (2007) J. Biol. Chem. 282, 24752–24758 [DOI] [PubMed] [Google Scholar]
- 50.Grefen C., Städele K., Růzicka K., Obrdlik P., Harter K., Horák J. (2008) Mol. Plant 1, 308–320 [DOI] [PubMed] [Google Scholar]
- 51.Zhong S., Lin Z., Grierson D. (2008) J. Exp. Bot. 59, 965–972 [DOI] [PubMed] [Google Scholar]
- 52.Chen Y. F., Gao Z., Kerris R. J., 3rd, Wang W., Binder B. M., Schaller G. E. (2010) PLoS ONE 5, e8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binder B. M. (2008) Plant Sci. 175, 8–17 [Google Scholar]
- 54.Binder B. M., Bleecker A. B. (2003) Acta Hortic. 628, 177–187 [Google Scholar]
- 55.Binder B. M., Mortimore L. A., Stepanova A. N., Ecker J. R., Bleecker A. B. (2004) Plant Physiol. 136, 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao Z., Wen C. K., Binder B. M., Chen Y. F., Chang J., Chiang Y. H., Kerris R. J., 3rd, Chang C., Schaller G. E. (2008) J. Biol. Chem. 283, 23801–23810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gamble R. L., Qu X., Schaller G. E. (2002) Plant Physiol. 128, 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petris M. J., Strausak D., Mercer J. F. B. (2000) Hum. Mol. Genet. 9, 2845–2851 [DOI] [PubMed] [Google Scholar]
- 59.Axelsen K. B., Palmgren M. G. (2001) Plant Physiol. 126, 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrés-Colás N., Sancenón V., Rodríguez-Navarro S., Mayo S., Thiele D. J., Ecker J. R., Puig S., Peñarrubia L. (2006) Plant J. 45, 225–236 [DOI] [PubMed] [Google Scholar]
- 61.Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K. K., Pilon M. (2003) Plant Cell 15, 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]