Abstract
The role of protein isoaspartyl methyltransferase (PIMT) in repairing a wide assortment of damaged proteins in a host of organisms has been inferred from the affinity of the enzyme for isoaspartyl residues in a plethora of amino acid contexts. The identification of PIMT target proteins in plant seeds, where the enzyme is highly active and proteome long-lived, has been hindered by large amounts of isoaspartate-containing storage proteins. Mature seed phage display libraries circumvented this problem. Inclusion of the PIMT co-substrate, S-adenosylmethionine (AdoMet), during panning permitted PIMT to retain aged phage in greater numbers than controls lacking co-substrate or when PIMT protein binding was poisoned with S-adenosyl homocysteine. After four rounds, phage titer plateaued in AdoMet-containing pans, whereas titer declined in both controls. This strategy identified 17 in-frame PIMT target proteins, including a cupin-family protein similar to those identified previously using on-blot methylation. All recovered phage had at least one susceptible Asp or Asn residue. Five targets were recovered independently. Two in-frame targets were produced in Escherichia coli as recombinant proteins and shown by on-blot methylation to acquire isoAsp, becoming a PIMT target. Both gained isoAsp rapidly in solution upon thermal insult. Mutant analysis of plants deficient in any of three in-frame PIMT targets resulted in demonstrable phenotypes. An over-representation of clones encoding proteins involved in protein production suggests that the translational apparatus comprises a subgroup for which PIMT-mediated repair is vital for orthodox seed longevity. Impaired PIMT activity would hinder protein function in these targets, possibly resulting in poor seed performance.
Keywords: Plant, Protein Deamidation, Protein Methylation, Protein-Protein Interactions, S-Adenosylmethionine (SAM), Seed
Introduction
Spontaneous conversion of l-Asn or l-Asp residues in proteins to the unusual, uncoded amino acid, l-isoAsp, occurs at varying rates depending on primary, secondary, and higher order protein structure (1) as well as cellular environment (2). This conversion can be detrimental to protein function and/or solubility (2–5) or in rare instances necessary (6) or as an intermediate step that effects an Asn → isoAsp → Asp conversion that optimizes protein function post-translationally (7, 8). Protein l-isoaspartyl methyltransferase (PIMT)4 is an intracellular enzyme present in most life forms. It recognizes and modifies isoaspartyl residues in proteins and peptides (9, 10). Complete (Asp → isoAsp → Asp) or partial (Asn → isoAsp → Asp) repair is effected by PIMT through an iterative process of isoAsp methylation at the α-carbon atom, reformation through the loss of methanol of an unstable succinimide, and subsequent hydrolysis of one of two carbonyls, producing either isoAsp or Asp. IsoAsp formation results in another round of PIMT action (supplemental Fig. 1).
In plants, PIMT activity is primarily localized in seed tissues during the late stages of embryogenesis during and after maturation desiccation, suggesting a role in rescuing the functionally active conformation of the generally long-lived (11) orthodox seed (12) proteome upon imbibition. Considering the fact that embryos and at least some cell layers of the endosperm (if present) remain viable for extended periods of time in the dry state (13), repairing the accumulated damage to proteins upon imbibition may be important for longevity (14–17). PIMT activity may also be instrumental in maximizing fitness during abiotic stress, to which seedlings are particularly susceptible (18).
In Arabidopsis (A. thaliana L. Heynh), two PIMT genes have been identified (19–21). Transcription of the PIMT2 is particularly complex (22, 23), providing PIMT2 variants to organelles as well as the cytoplasm.
A few proteins containing l-isoaspartyl residues have been identified as PIMT substrates in cells and tissues in non-plant species, including calmodulin (10, 24) and tubulin (25, 26). The use of post-extraction (27) or post-separation (28) labeling of isoAsp-containing proteins has identified a number of PIMT targets in the mouse brain and some seed storage proteins in the Arabidopsis seed (23). In the latter study, a recombinant, human PIMT (rHsPIMT) with a dramatically lower Km for AdoMet than either Arabidopsis enzyme was used in on-blot methylation (OBM) assays to identify the seed target protein. OBM requires protein extraction from the seed and heat treatment, either of which can cause isoAsp formation through heat shock (29), oxidation (30), or other damage (31). Although using OBM on seed proteins has resulted in the identification of some seed storage proteins as isoAsp-containing proteins (23), their abundance and susceptibility to damage (32) has made identification of additional PIMT-target proteins from seeds difficult. Additionally, it was desirable to avoid isoAsp production due to chemical/thermal insult during protein extraction.
In this work we have taken an alternative approach that allows the use of the plant enzyme to identify plant target proteins and does not require protein extraction. Additionally, this approach reduced the number of seed storage protein targets that can overshadow other substrates of PIMT. Here, we constructed a phage display cDNA library for biopanning of isoAsp-containing polypeptides. The use of radiolabeled AdoMet could be avoided, and AdoMet in concentrations sufficient for the Km of the plant PIMT could be used. Potential target proteins would be produced by the phage within the bacterial host, and a natural lysis would occur after which the phage could be rapidly recovered by centrifugation at neutral pH in PEG solution. A normalized phage-display cDNA library from an amalgam of mRNA from mature, dehydrated, and germinating seeds would circumvent the problem of storage protein abundance because mRNA for storage proteins are less abundant in the mature, dehydrated seed than during mid-to-late development (33) and are decimated after imbibition (34). The paucity of storage protein mRNA in the libraries focuses the phage display on PIMT protein targets produced from stored or de novo-synthesized mRNA present during germination.
Studies of PIMT from a variety of species have determined that the enzyme works by an ordered sequential mechanism in which it must first bind AdoMet before attaining a conformation allowing protein substrate binding (35, 36). Thereafter, PIMT (serendipitously fulfilling one of the important criteria essential for successful phage display) has a low turnover rate for substrate release (37, 38). The objective of the current work was to identify PIMT target proteins from phage display cDNA libraries normalized for transcripts present in quiescent and germinating Arabidopsis seeds.
EXPERIMENTAL PROCEDURES
Feasibility and Optimization
Experimental procedures associated with establishing the feasibility of biopanning using phage display to acquire rAtPIMT1 target proteins can be found in the supplemental material.
Screening the Libraries by Biopanning
The biopanning protocol employed aliquots of amplified libraries that had been recovered from LB liquid lysates (from a multiplicity of infection (m.o.i.) of 0.001) and clarified by centrifugation (8000 × g, 10 min), and the supernatant was stored at 4 °C for 2 weeks. Three replications of three 500-μl aliquots were prepared for biopanning by adding a 0.2 volume of 5× modified TBS-Tween (mTBST) to each of the 9 aliquots. One set of three aliquots received 0.01 volume of 10 mm S-adenosylmethionine, one received 0.01 volume of 10 mm S-adenosyl homocysteine (heated until dissolved just before dilution), and one received 0.01 volume of double distilled H2O. The final library aliquots were in 1) 1× mTBST, 2) 100 μm S-adenosylmethionine (re-purified from Sigma (39)) in 1× mTBST, or 3) 100 μm S-adenosyl homocysteine in 1× mTBST. The amended libraries were added to multiple ELISA plate wells (100 μl·well−1) to which 1 μg·μl−1 rAtPIMT1 in 0.7 m urea had been previously introduced, washed in mTBS without urea, blocked using BR (Novagen Inc., La Jolla, CA), and washed with mTBST. The libraries and rAtPIMT1 in the wells of the plate were incubated at 25 °C for 30 min. The wells were then washed 10 times with 500 μl of mTBST per wash. After the last wash, 150-μl aliquots of Escherichia coli (BLT5403, A600 = 0.7) in LB were added to each well, and the plate was sealed with adhesive film, incubated for 1 h at 37 °C to allow infection, recovered, and when statistics on phage titer and/or insert presence per biopanning round were not to be collected, the cells from wells containing the same library amendment were combined. A portion of the recovered cells was plated at low density to determine the titer and the presence of insert (see below). The remainder were added to, and amplified in, 37 ml of BLT5403 cells growing at 37 °C with shaking. Incubation continued for ∼3 h until visible lysis occurred. Amplified phage were centrifuged (8000 × g, 10 min), recovered in the supernatant of spent media, titered, and stored at 4 °C to age until the next biopanning round.
PCR and Sequence Analysis of Phage Recovered during Biopanning
After four rounds of biopanning, an aliquot of the final infected bacteria was plated at low density and titered, and some of the isolated plaque was PCR-amplified and size-fractionated as in previous biopanning rounds. This entailed scraping the top agarose with the isolated plaque with a sample loop and swirling the loop in 100 μl of 1.0 mm EDTA in an Eppendorf tube. The tube was capped and heated to 65 °C for 10 min before being cooled and centrifuged at 14,000 × g for 3 min. The supernatant (2 μl) was used as a DNA template with T7Select-UP and -DOWN primers (supplemental Table S1) designed to amplify over the multicloning site between the vector arms in 50 μl of PCR reactions performed as described by the manufacturer (T7Select 10–3 cloning kit; Novagen).
Aliquots of the PCR reactions were size-fractioned through 1% (w/v) agarose gels, and the remainder of those reactions providing amplicons greater than 100 bp were purified away from the primers using spun columns (QiaQuick PCR purification kit; Qiagen Inc.). These amplicons were then cycle-sequenced, aligned with Arabidopsis cDNA sequences in WU-BLAST using The Arabidopsis Information Resource (40), and the gene producing the cDNA was identified. Each sequence contained a portion of the vector arms and linker associated with the Arabidopsis cDNA from which it was possible to determine 1) whether the cDNA was ligated in-frame or not and, from this information, 2) the deduced amino acid sequence displayed on the phage coat.
Assessment of Amino Acid Frequencies and Contexts from In-frame Hits
The frequencies with which individual amino acids occurred within each of the in-frame proteins recovered during phage display was analyzed to ascertain if proteins particularly rich in any one (or subgroup of) amino acid(s) were more prone to selection. The amino acid sequence of the Arabidopsis protein or fragment thereof attached to the phage coat was subjected to non-parametric analysis (χ2, Proc Freq (41)) for significant deviations from its expected frequency of occurrence based on the deduced codon usage for 80,395 protein coding regions of the species (Kazusa DNA Research Institute (42)).
Independent in-frame protein sequences recovered by phage display were examined using a nine-amino acid moving window centered on any Asp or Asn residue not immediately followed by P- (a P1′ proline makes Asp or Asn resistant to isoAsp formation (54)). These nonamers were depicted in separate WebLogos (43) for Asp or Asn, which were examined for the occurrence of amino acids in any position from P4-P4′ that were over represented. A control data set was comprised of the same number of consecutive nonamers for both Asp and Asn from a series of proteins commencing at a randomly selected point in the Arabidopsis genome (At1g30000-At1g30100, excluding At1g30030, which is a transposable element; The Arabidopsis Information Resource (40)).
Additionally, the entire Arabidopsis proteome (UniProt data base version 15.3, 31,638 protein sequences) was analyzed by an in-house-developed perl script to determine the frequency of occurrence of individual amino acids in the proteome and also the frequency with which Asn or Asp residues were surrounded by all other amino acids in positions P4-P4′.
Recombinant Protein Production, Purification, and On-blot Methylation of Two rAtPIMT1 Target Proteins
The coding sequence of two in-frame, AtPIMT1 substrates was amplified and cloned into pET23b (Novagen) by introducing NdeI and XhoI (At4g26050, plant intracellular Ras group-related leucine-rich repeat protein 8 (PIRL8) (47)) or BspHI and EcoRI (At5g62190, plant RNA helicase 75 (PRH75) (44)) sites into the amplicon 5′- and 3′-termini, respectively (supplemental Table S1). Recombinant protein production was induced in the E. coli strain BL21(DE3)RIL (Stratagene, La Jolla, CA), solubility was determined, and proteins were purified using nickel columns (Hi-Trap, GE Healthcare) and FPLC. Pure fractions (determined by SDS-PAGE of aliquots) were combined, protein concentration was determined (Ref. 45, as modified in the DC Protein Assay, Bio-Rad), and two identical SDS-PAGE fractionations of recombinant proteins and commercial, prestained molecular weight markers (Bio-Rad) were simultaneously prepared. One gel was stained with Coomassie Brilliant Blue, and the other was transferred overnight to PVDF. After transfer, the PVDF membrane was blocked at room temperature (30 min in 0.2 mg·ml−1 BSA, 10 mm MES, pH 6.2). Recombinant and marker proteins were subjected to OBM (28) as in Dinkins et al. (23).
Assessment of the Rapidity of Isoaspartate Accumulation in Two AtPIMT1 Protein Targets
Purified PRH75 and PIRL8 recombinant proteins were quantified (45), and the concentration was determined using the polypeptide molecular masses of 75,000 and 42,309 daltons, respectively. Exactly 330 pmol of each protein was diluted to a final volume of 66 μl in 50 mm Tris-HCl, 500 mm NaCl, pH 7.4. These samples were put into thin-walled PCR tubes and floated in 37 °C water along with the same molar concentrations of hen egg ovalbumin (Sigma grade V) and bovine serum albumin (Sigma). Four tubes for each substrate (replicates) were retrieved and analyzed for isoaspartate after 0, 8, 14, 21, and 28 days. At each time point, three 20-μl aliquots (triplicates) were taken from each of the incubated tubes (each aliquot containing 100 pmol of polypeptide) and reacted with a large amount of human recombinant PIMT (based on an activity of 20 pmol/min) for an extended time (120 min) to ensure, insofar as possible, nearly stoichiometric methylation of all l-isoaspartyl (d-aspartyl) residues. The number of methyl esters formed from [14C]AdoMet in the triplicate samples was then quantified, and the results were averaged. The average from the four replicates was determined as well as the S.E.
Acquisition and Assessment of Homozygous T-DNA Insertional Mutant Seeds for Some rAtPIMT1 Targets
Only WT and heterozygotes for the T-DNA insert were recovered for the PRH75 putative mutant (SALK_016729), whereas none of the plants from seeds designated as SALK_040581.53.40.x carried a T-DNA insert in PRH75. Being unable to test whether the homozygous prh75 seeds were altered from WT, this line was reserved for future studies of embryo lethality.5 A single T-DNA-interrupted mutant for a lysine-glutamate-aspartate-rich protein (similar-to-KED from tobacco (46) (Table 1, At1G56660, SAIL_163_D09)), two T-DNA- and one transposon-interrupted mutant for the Arabidopsis late embryogenesis abundant 4 protein gene (seed maturation protein 1, At3G12960 (SAIL_1184_D08, WiscDsLox297300_22J and CSHL_ET11624), and two independent T-DNA insertional mutants of PIRL8 (At4G26050 (47) and SALK_095144) were identified (through PCR and Southern blot on pools of genomic DNA for pirl8-1 (47)) or on the SIGnAL website (83), and seeds acquired from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH or Cold Spring Harbor Laboratories, Cold Spring Harbor, NY). Homozygotes for these alleles were sought using PCR with gene-specific primers (supplemental Table S1) on either side of the insertion site (SIGnAL iSECT tool) and outward-facing primers to the T-DNA/transposon as well as with antibiotic resistance for those lines expressing this trait. Template DNA was acquired from leaf disks using a kit (Extract-N-Amp, Sigma) and amplified using RedTAQ (Sigma). Amplicons were isolated and sequenced to identify the exact insertion sites.
TABLE 1.
Seventeen in-frame rAtPIMT1 protein targets identified using phage display
Seed library (SL) hits, shown in bold type, contained significantly greater numbers of Asx than expected based on codon usage for the species in the peptide displayed on the phage coat. The amino acid deviating most from expected is indicated. TAIR, the Arabidopsis Information Resource; FLC, Flowering locus C; FLM, Flowering locus M.
| In-frame hits |
Protein identity | Subcellular residence/authority | |
|---|---|---|---|
| Library | Locus identifier | ||
| SL_ TC224 (Asp) | AT5G02050 | Mitochondrial glycoprotein family protein/MAM33 | Plastid, mitochondrial matrix/TAIR |
| SL_TC241 | AT2G33590 | Cinnamoyl-CoA reductase family protein; first step of the phenylpropanoid pathway specifically dedicated to the monolignol biosynthetic branch | Unknown/TAIR |
| SL_TC276 | AT1G56660 | Similar to KED: similar to unknown protein. (46). KED in this manuscript | Unknown/TAIR |
| SL_ TC291 | AT3G46440 | UDP-xylose synthase 5 (UXS5) | Plastid/TAIR |
| SL_ TC305 (Asp) | AT3G48690 | A. thaliana carboxyesterase 12, ATCXE12 (77) | Cytoplasm/TAIR |
| SL_ TC293 | AT2G01100 | Similar to CAX interacting protein 4 (CXIP4), a nuclear H+/Ca+ antiporter activating protein | Unknown/TAIR |
| SL_TC344 | AT4G26050; AY849578 | PIRL8 (47) | Unknown/TAIR |
| SL_ TC312 | AT2G06210 | VIP6, ELF8 ELF8 (early flowering 8). Encodes a yeast CTR9 homolog that is involved in the control of flowering time by elevating FLC expression to a level that creates the vernalization-responsive, winter-annual habit. Yeast CTR9 is a component of a five-member PAF1 complex that associates with RNA polymerase II and is thought to regulate gene expression by recruiting SET1 (a histone 3 Lys-4 (H3-K4) methyl transferase) to the initially transcribed (5′) regions of target chromatin. Mutants display reduced H3-K4 methylation in both FLC and FLM chromatin | Unknown membrane component/TAIR |
| SL_ TC331 | AT2G28450 | Zinc finger CCCH-type family protein: zinc finger, CX8CX5CX3H type; S-adenosyl-l-methionine-dependent methyltransferases | Unknown/TAIR |
| SL_ TC321 | AT3G46230 | ATHSP17.4 (A. thaliana) class I heat shock protein (78, 79) | Unknown/TAIR |
| SL_ TC335 | AT3G12960 | Similar to seed maturation protein (G, tomentella): a LEA 4 protein (57). SMP1 in this manuscript | Unknown/TAIR |
| SL_ TC338 | AT5G51150 | Similar to unknown protein (A. thaliana) | Plastid/TAIR |
| SL_ TC341 | AT3G22640 | Cupin family protein (23, 80) | Cell wall/(81) |
| SL_ TC295 | AT3G51270 | ATP binding/protein serine/threonine kinase. The N-terminal domain is found in RIO2 kinases and is similar in shape to the winged helix (wHTH) domain. This domain permits DNA binding, and in yeast, the RIO2 proteins are implicated in cell cycle progression, DNA-repair, and ribosomal maturation | Unknown/TAIR |
| SL_ TC237 (Asp and Asn) | AT5G62190 | PRH75 | Nucleolus, nucleus/(44); TAIR |
| SL_ TC333 (Asp) | AT5G55920 | Oligocellula 2, nucleolar protein, putative 16 S rRNA m5C967 methyltransferase. Yeast Nop2p is a probable RNA m(5)C methyltransferase essential for processing and maturation of 27 S pre-rRNA and large ribosomal subunit biogenesis (82) | Nucleolus/TAIR |
| SL_ TC349 | AT3G22230 | 60 S ribosomal protein L27 (RPL27B) | Cytoplasm/TAIR |
For those lines for which seeds from homozygous mutants were acquired, both wild type and mutant seeds were sown at the same time, and plants were grown on the same shelf under the same conditions and harvested on the same day. These seeds were then either sown directly (pirl8 alleles and respective WTs) or kept under the same conditions inducing dry after-ripening for at least 3 months until testing (all genotypes). Germination tests were conducted on at least 50 seeds per replication, 4 replications per experiment, sown on 2 layers of filter paper (Whatman No. 1, 55-mm diameter, Fisher, placed in the bottom of Petri dishes (60 × 15 mm, Falcon, BD Biosciences) moistened with 2 ml of double distilled H2O write and sealed with Parafilm (Fisher). All experiments were performed on at least two separate occasions and on at least two separate generations. When imbibed seeds were exposed to 40 °C, the Petri dishes were placed in a plastic bag with water-soaked paper towels, and the bag was sealed. Four Hygrothemochron iButtons (DS1923, Embedded Data Systems, Lawrenceburg, KY) in the bag were tasked to simultaneously log temperature and humidity every 10 min for the duration of the supraoptimal temperature. This monitored the uniformity of the stressful conditions within the bag (supplemental Fig. 3).
Based on the information on gene expression under environmental perturbation (The Bio-Array Resource for Plant Functional Genomics (BAR) website) and on co-expression with the genes of interest (GeneCAT website) whose proteins were rAtPIMT1 targets, mutant seeds were tested for rapidity in completing germination and final percentage germination relative to respective wild types on water at 25 °C. Seeds were also tested at sub (4 °C)- and supra-optimal (35 °C; similar to Tamura et al. (48)) temperatures. Separate analyses were performed on seeds that were or were not moist-chilled and with or without light during the germination test in the following assays. Seeds were tested at 25 °C after 4 days at 40 °C either after 3 months of dry after-ripening (ked, smp1, and pirl8 mutants) or immediately after harvest (pirl8 mutants).
Seeds were tested at 25 °C on an ABA or Paclobutrazol concentration series (49). Seeds were also tested on a salt concentration series (0–250 mm NaCl) and on 200 mm H2O2. Finally, the pirl8 mutant and respective wild type seeds were tested on glucose (0–6% w/v), sorbitol (0–6% w/v), insulin (1 or 5 nm), wortmannin (1 or 5 nm), or α-ketoglutarate (1, 10, 100 μm) concentration series. After testing, seeds failing to complete germination were moist-chilled 3 days at 4 °C and then placed at 25 °C for 1 week to assess seed viability.
Statistical Assessment of the Data for Optimizing the Biopan and for Mutant Seed Germination Attributes Was Performed with the Statistical Analysis System (SAS for Windows Version 9.1, 2002)
In each experiment analysis of variance was performed on the main effect, and if significantly different, Tukey's mean separation test was performed to distinguish among significantly deviating means. In those instances where more than one main effect was tested (e.g. urea concentration in which rAtPIMT was introduced into the wells and urea concentration in the subsequent wash buffer) interactions between the main effects were tested, and if significant, each level of one main effect was tested over the range of the other separately.
RESULTS
Library Characteristics
The primary library I titer was 1.6 × 107 pfu·ml−1, and that of primary library II was 2 × 107 pfu·ml−1. After amplification, the titers of library I and II were 8.15 × 109 pfu·ml−1 and 3.6 × 1010 pfu·ml−1, respectively.
The rAtPIMT1 from Arabidopsis was the most active of the various AtPIMT isoforms from Arabidopsis (22). However, its Km for ovalbumin was between 1.2 and 5 mm (20) for VYP(isoD)HA ∼28 μm and for AdoMet, 6 μm (20). Several rAtPIMT2 isoforms have an even greater Km for VYP(isoD)HA. Based on these results, the rAtPIMT1 was chosen to perform biopanning to capture isoAsp-containing proteins expressed from a seed library on phage coats by their interaction with AdoMet-charged rAtPIMT1 bound to a solid support.
Optimization of the rAtPIMT Biopan
Preparatory to biopanning, it was necessary to determine whether this technique would work with the plant enzyme attached to solid support and, if so, to optimize the protocol.
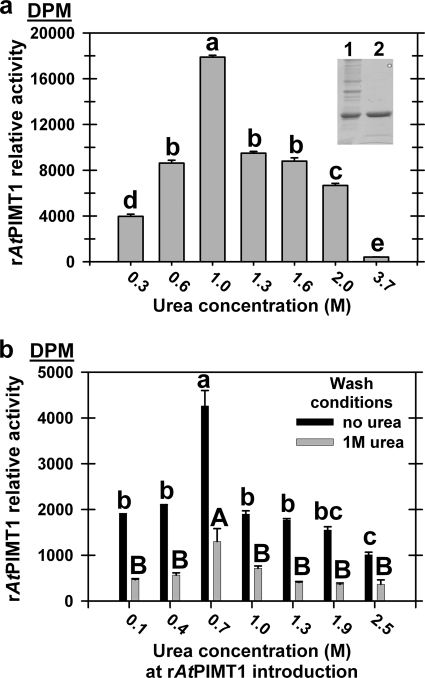
The rAtPIMT1 was most expeditiously obtained in the quantity and purity required for biopanning from inclusion bodies (see the inset Fig. 1a, lane 2) before nickel column purification. This necessitated solubilizing and purifying the protein in urea, a technique that allows recovery of considerable activity (21). rAtPIMT1 relative activity (in a range of urea concentrations) was greatest in liquid assays in 1 m urea (Fig. 1a). However, this was not the urea concentration permitting maximum active rAtPIMT1 attachment to the microtiter plate wells. In a combinatorial experiment, rAtPIMT1 maximum activity was retained by introducing the protein into the wells in 0.7 m urea and then eliminating urea from the mTBS buffer used to wash the wells to remove unbound rAtPIMT (Fig. 1b).
FIGURE 1.
Urea concentration and relative rAtPIMT1 activity in solution or when bound to microtiter plates. a, recombinant A. thaliana PIMT1 (rAtPIMT1), recovered from inclusion bodies, was solubilized in 6 m urea, purified over a nickel column, and dialyzed to remove imidazole (see the inset) First lane, rAtPIMT1 from inclusion bodies; second lane, purified, dialyzed rAtPIMT1. The enzyme was assayed in different concentrations of the chaotrope. The rAtPIMT1 (1 μl of 0.05 μg·μl−1) was most active in 1 m urea. Different lowercase letters above the bars indicate significantly different relative activities in different urea concentrations when compared using Scheffe's multiple pairwise comparison. b, 100 μl of rAtPIMT1 (0.01 μg·μl−1) was introduced into ELISA plate wells in a variety of urea concentrations (0.1–2.5 m). The wells were then washed with × mTBS containing no or 1 m urea to eliminate unbound rAtPIMT1, and the bound enzyme was assayed in reaction mix with no or 1 m urea. Greatest bound rAtPIMT1 relative activity was obtained when the enzyme was introduced into the wells in 0.7 m urea and washed with 1× mTBS buffer without urea before assay. Significant deviation was determined using Scheffe's test after an analysis of variance. In b, rAtPIMT1 activity was greater regardless of urea concentration used for rAtPIMT1 introduction, when the wash was without urea. Lowercase letters over bars indicate significantly deviating means among urea concentrations used for rAtPIMT1 introduction when no urea was included in the wash. Uppercase letters indicate the same for rAtPIMT1 activity when 1 m urea was used in the wash buffer. (500 mm Tris).
A variety of substances were used to block the wells once rAtPIMT1 had been introduced. These resulted in similar PIMT activity when assayed in the well by the addition of peptide substrate (data not shown). However, considerably more phage was retained in wells blocked with the commercial blocking reagent (BR) sold with the phage display kit than with the other three substances assayed (Fig. 2a). This may have been beneficial due to BR not competing with phage for PIMT1 binding (as did BSA and nonfat milk protein in liquid assays when each blocking reagent was added as rAtPIMT1 substrate; data not shown) or detrimental due to BR indiscriminately binding phage, artificially inflating titer. To distinguish between these two possibilities, equal plaque-forming units were introduced into ELISA wells that had or had not (buffer only) been coated with rAtPIMT1 before blocking with BR. Subsequently, the background titer could be lowered to insubstantial amounts in the non-rAtPIMT1-containing wells by frequent washing with mTBST, whereas phage titer in wells coated with rAtPIMT1 before blocking with BR was significantly greater after extended washing (Fig. 2b).
FIGURE 2.
Blocking reagent and washing buffer optimization. a, different blocking solutions influence phage titer. PVP, polyvinylpyrolidine. The company product achieved high titers either by preserving rAtPIMT1 activity or by binding phage indiscriminately. Different lowercase letters over the bars indicate significantly deviating mean plaque forming units retained in the wells coated with different blocking agents. b, these alternatives were tested by washing wells to which rAtPIMT1 or no protein (buffer only) had been bound before blocking with BR. Effect of the number of washing steps before phage elution on phage titer during the biopanning is shown. BR preserves rAtPIMT1 activity rather than indiscriminately binding phage. Different lowercase letters above bars (within a washing regime) indicate significantly deviating means dependent on the presence of rAtPIMT1. c, shown is SDS-amended mTBST washing buffer and rAtPIMT1 activity. The inclusion of even small percentages of SDS in the mTBST washing buffer detrimentally influenced the rAtPIMT1 activity. Significantly deviating means identified using Scheffe's test after an analysis of variance.
Boiling rAtPIMT1 or exposing it to low pH before washing and introduction of aged phage resulted in significantly lowered phage titers relative to the un-boiled or un-acidified control (data not shown). Efforts to eliminate indiscriminate phage binding to the ELISA plate wells, blocking reagent, and/or rAtPIMT1 using either low concentrations of SDS in the TBST wash buffer (Fig. 2c) or by performing negative selection before biopanning (data not shown) were unfruitful. Greatest phage titer was consistently obtained by introducing the E. coli directly into the wells after the last stringency wash (data not shown).
Phage Titer Increased Only When rAtPIMT1 Was Active
After each round of biopanning, an aliquot of the cells introduced into each well for infection by the bound phage was titered (Fig. 3a). During the biopan, the phage libraries were introduced into the wells as controls (without AdoMet or AdoHcy) or in the presence of either AdoMet (a co-substrate of PIMT) or AdoHcy (a potent inhibitor of PIMT activity; negative control). Titer increased with successive rounds of biopanning only when AdoMet was present (Fig. 3a). In fact, titer decreased from the original by the final round of biopanning when nothing or AdoHcy were added to the phage during panning (Fig. 3a).
FIGURE 3.
a, shown is the effect of AdoMet (SAM) and AdoHcy (SAH) on phage titer from successive rounds of biopanning. None, neither AdoMet nor AdoHcy was added to the phage before introduction into the microtiter plate wells. S-Adenosylmethionine, a PIMT co-substrate, was added to the phage library to 100 μm just before biopanning. S-Adenosyl homocysteine, a PIMT inhibitor, was added to the phage library to 100 μm just before biopanning. Significantly deviating means among the three treatments were identified using Scheffe's test within each of the four rounds and are represented by different lowercase letters over each bar. b, shown is the effect of AdoMet and AdoHcy on the retention of phage with inserts in successive rounds of biopanning. c, shown is a graphical depiction of the 111 hits retrieved from the biopans. Out-of-frame (O.O.F.) and in-frame (I.F.) hits have been divided into those clones that were recovered only once (or the same clone (identical sequence) obtained multiple times; shades of red-purple), and those hits that are represented by at least two different clones (different lengths and/or portions of the same cDNA; shades of blue and green). All clones, after four rounds of biopanning, had at least one Asp or Asn residue. d, shown is clone identity for those recovered only once or for those recovered more than once but from the same biopan. e, clone identity for those recovered from different biopans and/or those that were different in sequence length and/or position in the cDNA and/or those that were from different libraries is shown. Different clones of At5g66400 recovered from library II are depicted in blue text. In all instances, the numbers in the pie slice represent the number of clones recovered and sequenced in that category.
PCR amplification of isolated plaque from plates used to ascertain titer revealed considerably more plaque from the AdoMet-containing biopan housing inserts relative to either the control- or AdoHcy-containing biopans in the second, third (Fig. 3b), and fourth (not shown) rounds. After the third round, the size of the amplicons for an experiment (successive rounds of phage aging and introduction to rAtPIMT1 in the presence of AdoMet) tended to consolidate around a few representative plaques, which were usually identical cDNAs (Fig. 3b and data not shown). Hence, in the gel image depicted for AdoMet-containing biopans after round 3, 6 of 18 amplicons were of the same size and sequence (lanes 5, 6, 7, 9, 16, 17).
Identification of rAtPIMT1 Target Proteins Displayed on Phage
From all biopans, 111 Arabidopsis sequences were read from phage recovered from wells containing AdoMet. All of these C-terminal additions to the phage coat, regardless of whether they were ligated in-frame or not, resulted in at least one Asp/Asn residue for which isoAsp conversion was possible (i.e. not followed by proline; Fig. 3c). There were 80 sequences recovered that translated proteins out-of-frame and 31 in-frame (Fig. 3c). All in-frame proteins had more than one susceptible Asp/Asn residue (Fig. 3c). For any one biopanning experiment, some clones when sequenced were identical to others recovered in the same biopan (Fig. 3, c and d). There was no means of determining whether multiple recoveries within an experiment were due to a single phage being retained and later multiplying (despite no effort to amplify phage before plating to ascertain titer) or whether multiple independent but identical phages were retained. On the other hand, the same sequence recovered from the other library within the same or different experiments or from the same library in different experiments could be counted as independent recoveries of the same clone (Fig. 3e). The gene loci are provided for those clones recovered multiple times regardless of the frame. The identity of all in-frame hits is provided in Table 1, and their sequences are in supplemental Table 2.
The Amino Acid Sequence of Recovered, In-frame Proteins Was Atypical
Sixty-five percent of the in-frame proteins had amino acid frequencies significantly deviating from those expected based on amino acid frequencies derived from the codon usage for this species and assuming a random chance of any amino acid occupying a position (i.e. Leu should be most abundant with a probability of occupying any position of 0.0935; Trp should be least abundant with a 0.0125 probability (42)). However, only three of these proteins had greater than expected numbers of Asp, and one had greater frequencies of both Asp and Asn (Table 1, bold text). Nevertheless, 9 of 20 had more Glu, and 7 of 20 had more Lys than expected.
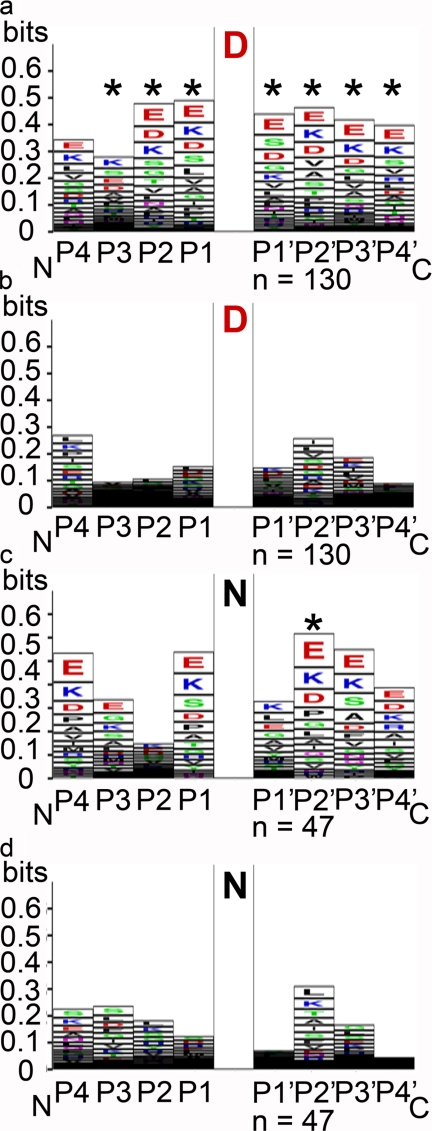
When a nine-amino acid window was examined centered on Asx, amino acid frequencies were significantly different from the expected (based on codon usage for A. thaliana and assuming a random chance of any amino acid occupying a position within 4 of Asx) for all positions except the P4 position surrounding Asp. Only the amino acid frequencies in the P2′ position in Asn-centered nonapeptides was significantly different from the expected (Fig. 4c). Examining the in-frame hits for amino acid-nearest neighbors in a nonapeptide window centered on Asx, Glu is the most frequent amino acid in positions P2′–P4′ followed by Lys, except for P4′ in Asn-containing proteins, where it is preceded in abundance by Asp (Fig. 4, a and c, supplemental Table 2). For positions P4–P1, Lys and Glu were also the most, or among the most prominent amino acids in all but position P2 for Asn-containing proteins (Fig. 4, a and c). In addition to Glu, Asp, and Lys, positions P3-P1 and P1′ and P4′ contained Ser in high frequencies for Asp-containing proteins and P1 in Asn-containing proteins, whereas no discernable trend in amino acid frequency was present at P1′ in Asn-containing proteins (Fig. 4c). Based on an analysis of the entire A. thaliana proteome, amino acids Glu and Lys are not more likely than expected due to a random chance to be located within four positions of Asn; in fact, the opposite is more probable (supplementary Table S3A). Although the same is true for Lys within two positions either side of Asp, Glu is more likely to be located within four positions of Asp (supplementary Table S3B).
FIGURE 4.
A representation of the frequency of neighboring amino acids at positions from non-prime 4 to prime 4 on either side of the Asp (D) (A and B) or Asn (N) residues (c and d) from the in-frame hits (a and c) and from proteins (b and d) along a randomly chosen segment of the A. thaliana genome. An asterisk above a bar indicates a significantly greater abundance of one/some amino acids relative to that expected based on codon usage for the species. The n under the graphs represents the number of Asp or Asn amino acid positions evaluated.
Recombinant rAtPIMT1 Target Proteins Are Susceptible to IsoAsp Formation and PIMT Methylation
The two rAtPIMT1 targets generated as recombinant proteins (PIRL8, At4g26050; PRH75, At5g62190: Figs. 5, a–c) were methyl-accepting proteins from the rHsPIMT in on-blot methylation assays (Fig. 5d). Of the molecular weight markers also included on this blot, carbonic anhydrase was a strong methyl acceptor in this assay (Fig. 5d). Carbonic anhydrase has been identified previously as a protein susceptible to isoAsp formation (50).
FIGURE 5.
Two full-length PIMT substrates, protein fragments of which were identified through biopanning, were cloned into pET23 and expressed in E. coli BL21(DE3)RIL. a, the Coomassie-stained PRH75 protein gel includes molecular weight (M), a blank, lysate from uninduced cells at harvest (1), insoluble (2)and soluble protein (3) from induced cells after harvest and centrifugation, a nickel column wash after introduction of soluble protein (4), a post-wash before imidazole (5), elution (2 ml of 1 m imidazole) (6), blank, and protein post-dialysis (7). b, The PIRL8 recombinant protein was recovered from lysed cells as an inclusion body and solubilized in 6 m urea, and Ni-NTA column-purified. The Coomassie-stained gel includes lysate from uninduced cells (1), molecular weight markers (M), lysate from IPTG-induced cells (2), the insoluble pellet from the lysate after centrifugation (3), the soluble proteins post-centrifugation (4), protein from the inclusion body after urea solubilization and Ni-NTA column purification, undialyzed in 6 m urea and imidazole (5), purified inclusion body in 6 m urea dialyzed to remove imidazole (6). In all lanes, for both purifications, 10 μl of the lysate/eluate/dialyzed sample was mixed with 10 μl of SDS-containing loading dye, boiled, and loaded. c, both purified proteins were electrophoresed using SDS-PAGE and Coomassie-stained or assayed for isoaspartyl formation through on-blot methylation (fluorograph depicted here) and liquid assay after incubation at 37 °C (Table 2) using human recombinant PIMT (d). kDa, kDa of the Bio-Rad-prestained molecular weight markers.
The rate with which these proteins generated isoaspartate was explored by incubating them at 37 °C. Initially, neither protein contained considerable isoaspartate (Table 2). At 37 °C PRH75 accumulated some isoaspartate, approaching that of BSA, which was aged under the same conditions, although BSA commenced the assay with considerably more isoaspartate (Table 2). PIRL8 accumulated similar amounts of isoaspartate as ovalbumin, although, like PRH75, it commenced the assay with many fewer isoaspartyl residues than ovalbumin (Table 2). During the course of the assay, the relative rate of isoaspartate accumulation for both PRH75 and PIRL8 was considerably greater than that of ovalbumin, a protein known to be susceptible to isoaspartyl formation (Ref. 51, Table 2).
TABLE 2.
Isoaspartyl accumulation in ovalbumin, bovine serum albumin, and the PIMT1 target proteins, recombinant PRH75 and PIRL8
ME, methyl equivalents
| Substrate Identity | 37 °C | pmol of ME/pmol of substrate | Relative increase in isoaspartate |
|---|---|---|---|
| Days | Mean ± S.D. | ||
| PRH75 | 0 | 0.0031 ± 0.0003 | 0 |
| 8 | 0.0176 ± 0.0004 | 5.7 | |
| 14 | 0.0356 ± 0.001 | 11.6 | |
| 21 | 0.0317 ± 0.0011 | 10.3 | |
| 28 | 0.0397 ± 0.0009 | 12.9 | |
| PIRL8 | 0 | 0.0018 ± 0.0003 | 0 |
| 8 | 0.0183 ± 0.0009 | 10.2 | |
| 14 | 0.0350 ± 0.0005 | 19.5 | |
| 21 | 0.0620 ± 0.0007 | 34.5 | |
| 28 | 0.0924 ± 0.0012 | 51.4 | |
| Ovalbumin | 0 | 0.0442 ± 0.0011 | 0 |
| 8 | 0.0589 ± 0.0003 | 1.3 | |
| 14 | 0.0715 ± 0.0011 | 1.6 | |
| 21 | 0.0907 ± 0.0010 | 2.1 | |
| 28 | 0.1330 ± 0.0015 | 3.0 | |
| BSA | 0 | 0.0162 ± 0.0008 | 0 |
| 8 | 0.0259 ± 0.0018 | 1.6 | |
| 14 | 0.0303 ± 0.0004 | 1.9 | |
| 21 | 0.0395 ± 0.0004 | 2.4 |
Absence of rAtPIMT1 Target Proteins Impacts Seed Attributes
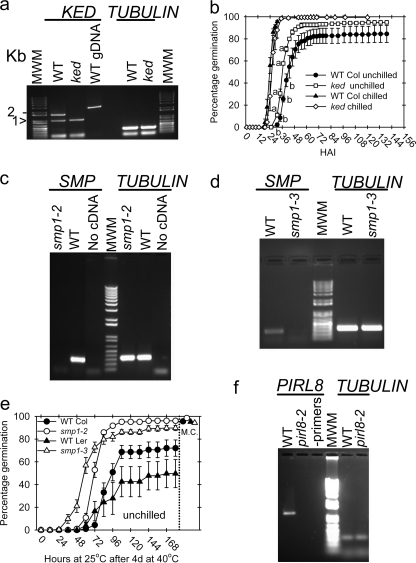
Insertional mutants of some of the genes encoding rAtPIMT1 target proteins were acquired. Attempts to isolate homozygotes of plant RNA helicase 75 (prh75) failed, suggesting lethality at some stage.5 For three other genes, homozygotes were analyzed for physiological consequences of the genetic lesions. Freshly harvested seeds with a T-DNA insertion in a lysine/glutamate/aspartate-rich protein (ked), a strong knockdown (Fig. 6a), completed germination demonstrably faster than WT (selected from the same batch of seeds from ABRC) if not moist-chilled (Fig. 6b). Three insertional mutants for the seed maturation protein 1 (SMP1) gene were analyzed, but the insertion in the WiscDsLox297300_22J line was in the 3′-UTR and did not diminish the amount of mRNA of the SMP1. The other two alleles were strong knockdowns (Figs. 6, c and d), and seeds from these lines completed germination faster than WT when they were placed at 25 °C after heat shock for 4 days at 40 °C (Fig. 6e) as did the ked mutant (data not shown). Both plant intracellular Ras group-related leucine-rich repeat protein 8 (pirl8-1 and pirl8–2) mutants were strong knockdowns (see pirl8-2 in Fig. 6f and Ref 47), but no consistent phenotype could be demonstrated for these mutants.
FIGURE 6.
Homozygous insertional mutant lines for three of the genes whose proteins were identified as being PIMT1 targets in phage display were recovered as homozygotes. a, one insertional mutant in the KED gene was determined to be a severe knockdown. The chevron points to a nonspecific amplicon from cDNA of both wild type and the ked mutant. b, seeds of this mutant, along with wild type seeds, were tested at 25 °C on water in constant light without moist chilling. c and d, three independent mutants of an Arabidopsis LEA 4 protein, similar to seed maturation protein1 (SMP1) from Glycine tomentella were obtained. One of these, smp1-1 located in the 3′-UTR, did not reduce SMP1 transcript and showed no phenotype in the screens used on the seeds (data not shown). The other two insertions resulted in severe knockdowns (smp1-2 and smp1-3). e, mutant and wild type seeds were placed on water-saturated filter paper at 40 °C in light in a sealed plastic bag with soaked paper towels for 4 days before being placed at 25 °C in light to complete germination. The smp1 mutants completed germination to a greater percentage than WT without moist chilling. f, two independent insertional mutants of the PIRL8 were identified (pirl8-2 depicted) and determined to be severe knockdowns. After heat shock to induce secondary dormancy and after the seeds had completed germination at 25 °C to the extent possible, seeds were moist-chilled for 3 days and placed at 25 °C to complete germination to distinguish between dead and dormant seeds (final germination percentage is depicted to the right of the vertical dashed line over M.C. in e). When possible, primers were designed to span an intron. For each germination time point, an analysis of variance was conducted between mutant and wild type to distinguish between significantly deviating means. This is depicted in b where different lowercase letters distinguish those time points where the ked mutant seeds averaged a greater percentage germination than their WT. For the other mutant/wild type combination significance is obvious.
DISCUSSION
Exploration of the recombinant seed proteome using phage display libraries and rAtPIMT1 as bait identified 17 in-frame target proteins in an optimized biopan. The salient feature of the optimized biopan was that the phage titer increased only when the PIMT co-substrate AdoMet was included in the assays. Additionally, after four rounds of biopanning, the majority of recovered phage contained a cDNA insert that was not the case for unsupplemented or AdoHcy-poisoned negative controls. None of the recovered targets were without susceptible Asp or Asn residues, which (despite the lenient recognition requirements for PIMT binding) were situated in amino acid contexts that overrepresented Lys and, for Asn-containing proteins, Glu residues. Two of the in-frame targets were susceptible to isoAsp formation in vitro and were methylated by PIMT. The elimination of any of three rAtPIMT1 targets through insertional mutagenesis resulted in demonstrable phenotypes.
Phage Display and Biopanning Successfully Identified rAtPIMT1 Target Proteins
AtPIMT1 target proteins were identified by biopanning normalized seed, phage-display libraries. Previous results using OBM with a recombinant human PIMT found some seed storage proteins to be strong methyl acceptors (23). However, due to their abundance in seeds (33), these protein species decrease the sensitivity of the OBM technique to other, less abundant targets. Additionally, when OBM was performed using the plant PIMT recombinant enzymes, the high Km values of the plant enzymes for protein substrate and AdoMet prevented the successful employment of this technique (23). Although the use of rHsPIMT (with a lower Km for both protein substrate and AdoMet) was a necessary expedient for examination of the seed proteome for proteins containing isoAsp using OBM, it was desirable to examine the seed proteome for plant PIMT target proteins using a plant PIMT. Seed storage protein mRNA abundance peaks during seed development declines drastically by the time the seed enters maturation desiccation (33) and is greatly diminished after imbibition (34). Hence, using phage display libraries from mRNA extracted from mature, dehydrated seeds and seeds during germination but before the completion of this event permitted scrutiny of the mature seed proteome and that arising post-imbibition during seed germination. This minimized the interference presented by the seed storage proteins, although phage-displaying peptide from a protein in the same family as the storage proteins identified previously (23) was recovered in-frame in the biopan (Table 1). Due to its nonradioactive nature and the limited surface area within the microtiter plate wells, phage display also permitted greater AdoMet concentrations to be added to the reactions than was financially possible using OBM. This alleviated the constraint caused by the high Km of the plant PIMT for AdoMet. Finally, the protein moieties exposed on the phage coat have not been subjected to chemical insult experienced in extraction procedures using detergent or other chaotropes. Many of the plant proteins on the phage coat are only fragments of the native proteins and, due to this and other reasons, are possibly in an incorrect secondary structure because of improper folding. They also may not be able to form tertiary or higher order structures in which they may be involved in vivo. It is possible that the curtailed protein on the phage coat places Asx residues in abnormal secondary structures that greatly facilitates their conversion to isoAsp, particularly regions of a protein that are not usually solvent-exposed in the intact protein. Nevertheless these arguments can also be levied against OBM where the proteome has been extracted into buffers, denatured, and fractionated using SDS-PAGE and adhered to a solid support.
The success keeping rAtPIMT1 active when bound to the microtiter plate wells and defining an optimized biopanning strategy for use with the phage display libraries allowed the exploitation of the small well volumes to increase the concentration of AdoMet present with the recombinant plant enzyme and libraries. The fact that phage titers increased only in the presence of AdoMet suggests that, at this concentration of AdoMet, biopanning permitted detection of protein fragments in which at least one isoAsp residue was present. Additionally, the convergence of clones retained by the rAtPIMT1 in the presence of AdoMet to a few identical species during successive rounds of biopanning (Fig. 3b) is a hallmark of success in this endeavor (52), suggesting that these clones were particularly susceptible to isoAsp formation, out-competing others for available rAtPIMT1. This fact also necessitated re-commencement of the biopan with the naïve libraries after a (several) target had been recovered in the fourth biopanning round. Selection of protein fragments by rAtPIMT1 in the presence of AdoMet was not simply a matter of numerical probability, i.e. proteins that contained a large number of Asx residues, automatically inflating the possibility of having one convert to isoAsp, as only 4 such proteins of the 17 in frame hits were preferentially retained by rAtPIMT1. Rather, some biochemical basis (resulting from primary sequence context) for the destabilization of Asx residues in these proteins is probable.
It is remarkable that there is a high frequency of Asn and Asp residues that are surrounded by charged, hydrophilic amino acids (Lys, Glu, and Asp) in the in-frame rAtPIMT1 substrates. The former two amino acids belong to those contributing most to a disordered structure (53), which in some instances enhances flexibility, which in turn promotes isoAsp formation (1). Examining deamidation rates of synthetic peptides, Robinson and Robinson (54) determined that paired acidic-basic nearest-neighbors decelerated Asn deamidation considerably, whereas basic-acidic nearest-neighbors also retarded deamidation, although this effect was not as pronounced. However, there was not a single Asn in this context (somewhat stabilized) in the in-frame hits and only 10 Asp residues (significantly, 8 of the 10 occurring in one protein; similar-to-KED, AT1G56660; Table 1, supplemental Table 2). Perhaps more important to the current study is that both Glu and, especially, Lys have been demonstrated to accelerate Asn deamidation rates when in the P2′-P4′ position, whereas the P1′ position is where Lys is thought to be incapable of accelerating Asn deamidation (55). In mouse histone H2B, a protein known to undergo constant conversion to isoAsp at Asp25 (4), Lys is in the P2, P1, P2′, and P3′ positions (56). Note that in the in-frame hits, the frequency of Lys in P1 and P2′-P4′ positions is quite high (Figs. 4, a and c), whereas it is less in the P1′ position (Fig. 4, a and c).
Both Ser and Gly enhance the ability of peptides to form isoAsp when they occur in the P1′ position of Asx (1) as well as N-terminal to these amino acids (55), the influence of Ser (at least) lessening as it becomes more distal to Asx (55). A trend, although not obvious for Asn (Fig. 4c), can be seen of increasing Ser abundance from P4 to P1 in Asp-containing in-frame hits (Fig. 4a). Additionally, the proteins assessed in this fashion from the out-of-frame hits (data not shown) and those from randomly chosen proteins (At1g30100-At1g31000; Fig. 4, b and d) upon examination of the entire A. thaliana proteome (supplemental Table S3, a and b) showed no such amino acid bias surrounding Asx. The subcellular location of the various in-frame hits (Table 1) included the mitochondria, plastid, nucleus, and cytoplasm, all of which have some variant of PIMT-1 and/or -2 resident (23) as well as the cell wall, which is without PIMT protection.
Following the lead from a demonstration of the importance of AtPIMT1 for both seed longevity and vigor (17), an important question was whether any of the proteins identified using phage display were demonstrably crucial for either seed trait. Indeed, insertional mutagenesis of the four genes examined in this manner altered the seed developmental or germination phenotype relative to that of the WT for all but pirl8. Two prh75 mutants examined were embryo lethal.5 Mutations in the phosphorylated seed maturation protein 1 (57) resulted in faster completion of germination than wild type when seeds were first subjected to 4 days of 40 °C in the light before 25 °C. The same phenotype was recorded for the ked mutant. The ked mutant also resulted in faster than WT completion of germination at 25 °C without prior heat shock or moist chilling (Fig. 6b). Only the pirl8 mutants were similar to their respective WTs in all aspects examined.
Disruption of KED or SMP1 reduced either the primary and/or secondary dormancy of the seeds. After heat shock, seeds entering a period of secondary dormancy may do so to ensure repair and proper functioning of systems permitting subsequent seedling establishment before completing germination. Certainly seed dormancy has been correlated with seed longevity in storage in a number of species for a variety of reasons (58–61).
The proteins of the translational machinery are hypothesized to be important targets of PIMT (62). The abundance of proteins involved in the coordinated generation and subsequent functioning of the eukaryotic ribosome notwithstanding (63–67), almost one quarter (4 of 17) of the in-frame targets identified in this paper are integral to the ribosome (At3g22230), its function (At5g62190; PRH75), or its assembly and/or maturation (At3g51270, At5g55920).
PRH75 shares considerable homology with DED1 from Bakers' yeast (Saccharomyces cerevisiae) and the mung bean, Vigna radiata RNA helicase 1 (VrRH1). The former is thought to be involved in translation (68–71) and in mRNA storage in cytoplasmic processing bodies (P-bodies) (72). The latter was isolated as a differentially expressed cDNA, noticeably more abundant in artificially aged but viable seeds (73), relative to un-aged seeds just after the completion of germination of both groups (axis ∼0.5 cm long). PRH75 may also be engaged in the assembly of functional ribosomes and/or processing of rRNA (73), and its expression varies based on developmental state (more abundant in rapidly growing cells (44, 73). PIMT is thought to protect the proteome from one effect of aging (74) and has been recently demonstrated to do so in seeds (17). Hence, the identification of PRH75, an orthologue of which has been implicated by others to be important for the seed to survive aging, as a PIMT1 substrate is probably not serendipitous. This is particularly noteworthy because the putative function of the PRH75 orthologue in yeast is to aid translation.
Translation of stored mRNA but not production of new transcripts is known to be vital for the completion of seed germination (75, 76). Together, the stored proteome and intact, stored mRNA provide the germinating seed with two options for obtaining a particular functional enzyme system after storage in the dehydrated state and without recourse to transcription; 1) use of pre-existing protein X that is functional or can be made to be functional or 2) use of pre-existing mRNA from which to manufacture de novo, functional protein X. However, for the subset of proteins essential for translation, loss of option 1 (i.e. all of the population of a particular stored protein, crucial for translation, is non-functional and beyond repair) automatically renders option 2 (and transcription) useless. Therefore, at least some of this category of protein must be maintained functional or at least be repairable if the seed is to be able to complete germination, quite an impressive feat given the fact that under some conditions seeds have remained viable for ∼1000 years (11, 13). If the population of any one of these vital proteins of the translational machinery is beyond repair during seed storage, the consequence is death because no mRNA (stored or de novo-synthesized) can be translated without them. Additionally, de novo ribosome biogenesis has been hypothesized to be the most energetically costly process in any cell (67), constituting an investment well worth salvaging through repair if possible. Therefore, we hypothesize that the proteins of the translational machinery can be viewed as an Achilles heel of orthodox seed longevity and may be a major target of PIMT-mediated repair in organisms capable of entering a period of quiescence.
Acknowledgments
We acknowledge David Martin and Manoj Majee for excellent technical advice, assistance, and troubleshooting during the phage cDNA library construction and biopanning optimization, Venkata Krishna Doppalapudi for excellent technical assistance planting, screening, harvesting, cleaning, and sowing seeds of the various mutant lines, Dr. Lynnette Dirk for expert advice in recombinant protein production and purification using FPLC, and Dr. Sarah Villa for work defining enzyme kinetics for rAtPIMT1. This work was also supported by the Math Science and Technology Center program, Paul Laurence Dunbar High School (to A. B. D. and A. C. E.) and by the University of Kentucky eUreKa! program.
This work was supported in part by National Science Foundation Molecular and Cellular Biochemistry Grant 0449649 (to A. B. D. and S. G. C.). This work was also supported by University of Kentucky undergraduate research and creativity Grant 14-28 (to A. B. D. and T. D. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S2 and Figs. 1 and 2.
N. Nayak and A. B. Downie, unpublished data.
- PIMT
- protein isoaspartyl methyltransferase
- AdoMet
- S-adenosylmethionine
- AdoHcy
- S-adenosyl homocysteine
- OBM
- on-blot methylation
- m.o.i.
- multiplicity of infection
- rHsPIMT
- recombinant, human PIMT
- rAtPIMT
- recombinant, A. thaliana PIMT
- BR
- blocking reagent
- Asx
- Asp or Asn
- mTBST
- modified TBS-Tween
- KED
- lysine/glutamate/aspartate-rich protein.
REFERENCES
- 1.Stephenson R. C., Clarke S. (1989) J. Biol. Chem. 264, 6164–6170 [PubMed] [Google Scholar]
- 2.Kern R., Malki A., Abdallah J., Liebart J. C., Dubucs C., Yu M. H., Richarme G. (2005) J. Bacteriol. 187, 1377–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle H. A., Gee R. J., Mamula M. J. (2003) J. Immunol. 171, 2840–2847 [DOI] [PubMed] [Google Scholar]
- 4.Reissner K. J., Aswad D. W. (2003) Cell. Mol. Life Sci. 60, 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanthier J., Desrosiers R. R. (2004) Exp. Cell Res. 293, 96–105 [DOI] [PubMed] [Google Scholar]
- 6.Curnis F., Longhi R., Crippa L., Cattaneo A., Dondossola E., Bachi A., Corti A. (2006) J. Biol. Chem. 281, 36466–36476 [DOI] [PubMed] [Google Scholar]
- 7.Tolleter D., Jaquinod M., Mangavel C., Passirani C., Saulnier P., Manon S., Teyssier E., Payet N., Avelange-Macherel M. H., Macherel D. (2007) Plant Cell 19, 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bidinosti M., Ran I., Sanchez-Carbente M. R., Martineau Y., Gingras A. C., Gkogkas C., Raught B., Bramham C. R., Sossin W. S., Costa-Mattioli M., DesGroseillers L., Lacaille J. C., Sonenberg N. (2010) Mol. Cell 37, 797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan T. V., Anderson J. W., Jia Z., Waygood E. B., Clarke S. (1994) J. Biol. Chem. 269, 24586–24595 [PubMed] [Google Scholar]
- 10.Johnson B. A., Langmack E. L., Aswad D. W. (1987) J. Biol. Chem. 262, 12283–12287 [PubMed] [Google Scholar]
- 11.Shen-Miller J., Mudgett M. B., Schopf J. W., Clarke S., Berger R. (1995) Am. J. Bot. 82, 1367–1380 [Google Scholar]
- 12.Roberts E. H. (1973) Seed Science and Technology 1, 499–514 [Google Scholar]
- 13.Sallon S., Solowey E., Cohen Y., Korchinsky R., Egli M., Woodhatch I., Simchoni O., Kislev M. (2008) Science 320, 1464. [DOI] [PubMed] [Google Scholar]
- 14.Mudgett M. B., Clarke S. (1993) Biochemistry 32, 11100–11111 [DOI] [PubMed] [Google Scholar]
- 15.Mudgett M. B., Lowenson J. D., Clarke S. (1997) Plant Physiol. 115, 1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kester S. T., Geneve R. L., Houtz R. L. (1997) J. Exp. Bot. 48, 943–949 [Google Scholar]
- 17.Ogé L., Bourdais G., Bove J., Collet B., Godin B., Granier F., Boutin J. P., Job D., Jullien M., Grappin P. (2008) Plant Cell 20, 3022–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villa S. T., Xu Q., Downie A. B., Clarke S. G. (2006) Physiologia Plantarum 128, 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mudgett M. B., Clarke S. (1996) Plant Mol. Biol. 30, 723–737 [DOI] [PubMed] [Google Scholar]
- 20.Thapar N., Clarke S. (2000) Protein Expr. Purif 20, 237–251 [DOI] [PubMed] [Google Scholar]
- 21.Xu Q. (2004) Horticulture, Doctoral dissertation, University of Kentucky, Lexington, KY [Google Scholar]
- 22.Xu Q., Belcastro M. P., Villa S. T., Dinkins R. D., Clarke S. G., Downie A. B. (2004) Plant Physiol. 136, 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinkins R. D., Majee S. M., Nayak N. R., Martin D., Xu Q., Belcastro M. P., Houtz R. L., Beach C. M., Downie A. B. (2008) Plant J. 55, 1–13 [DOI] [PubMed] [Google Scholar]
- 24.Desrosiers R. R., Romanik E. A., O'Connor C. M. (1990) J. Biol. Chem. 265, 21368–21374 [PubMed] [Google Scholar]
- 25.Najbauer J., Orpiszewski J., Aswad D. W. (1996) Biochemistry 35, 5183–5190 [DOI] [PubMed] [Google Scholar]
- 26.Lanthier J., Bouthillier A., Lapointe M., Demeule M., Béliveau R., Desrosiers R. R. (2002) J Neurochem 83, 581–591 [DOI] [PubMed] [Google Scholar]
- 27.Vigneswara V., Lowenson J. D., Powell C. D., Thakur M., Bailey K., Clarke S., Ray D. E., Carter W. G. (2006) J. Biol. Chem. 281, 32619–32629 [DOI] [PubMed] [Google Scholar]
- 28.Zhu J. X., Doyle H. A., Mamula M. J., Aswad D. W. (2006) J. Biol. Chem. 281, 33802–33813 [DOI] [PubMed] [Google Scholar]
- 29.Ladino C. A., O'Connor C. M. (1992) J. Cell. Physiol. 153, 297–304 [DOI] [PubMed] [Google Scholar]
- 30.Ingrosso D., Cimmino A., D'Angelo S., Alfinito F., Zappia V., Galletti P. (2002) Eur. J. Biochem. 269, 2032–2039 [DOI] [PubMed] [Google Scholar]
- 31.D'Angelo S., Ingrosso D., Perfetto B., Baroni A., Zappia M., Lobianco L. L., Tufano M. A., Galletti P. (2001) Free Radic. Biol. Med. 31, 1–9 [DOI] [PubMed] [Google Scholar]
- 32.Job C., Rajjou L., Lovigny Y., Belghazi M., Job D. (2005) Plant Physiol. 138, 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bewley J. D., Black M. (1994) Seed; Physiology of Development and Germination, 2nd Ed., pp. 81–83, Plenum Press Publishing Corp., New York [Google Scholar]
- 34.Hudson M. E., Bruggink T., Chang S. H., Yu W., Han B., Wang X., van der Toorn P., Zhu T. (2007) Crop Science 47, S96–S112 [Google Scholar]
- 35.Bennett E. J., Bjerregaard J., Knapp J. E., Chavous D. A., Friedman A. M., Royer W. E., Jr., O'Connor C. M. (2003) Biochemistry 42, 12844–12853 [DOI] [PubMed] [Google Scholar]
- 36.Smith C. D., Carson M., Friedman A. M., Skinner M. M., Delucas L., Chantalat L., Weise L., Shirasawa T., Chattopadhyay D. (2002) Protein Sci. 11, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowenson J. D., Clarke S. (1991) J. Biol. Chem. 266, 19396–19406 [PubMed] [Google Scholar]
- 38.Kagan R. M., Clarke S. (1995) Biochemistry 34, 10794–10806 [DOI] [PubMed] [Google Scholar]
- 39.Chirpich T. P., Zappia V., Costilow R. N., Barker H. A. (1970) J. Biol. Chem. 245, 1778–1789 [PubMed] [Google Scholar]
- 40.Swarbreck D., Wilks C., Lamesch P., Berardini T. Z., Garcia-Hernandez M., Foerster H., Li D., Meyer T., Muller R., Ploetz L., Radenbaugh A., Singh S., Swing V., Tissier C., Zhang P., Huala E. (2008) Nucleic. Acids Res. 36, D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Statistical Analysis System (1988) SAS/STAT User's Guide: Statistics, Version 5, Statistical Analysis Systems Institute., Cary, NC [Google Scholar]
- 42.Nakamura Y., Gojobori T., Ikemura T. (2000) Nucleic Acids Res. 28, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorković Z. J., Herrmann R. G., Oelmüller R. (1997) Mol. Cell. Biol. 17, 2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 46.Hara K., Yagi M., Koizumi N., Kusano T., Sano H. (2000) Plant Cell Physiol. 41, 684–691 [DOI] [PubMed] [Google Scholar]
- 47.Forsthoefel N. R., Cutler K., Port M. D., Yamamoto T., Vernon D. M. (2005) Plant Cell Physiol. 46, 913–922 [DOI] [PubMed] [Google Scholar]
- 48.Tamura N., Yoshida T., Tanaka A., Sasaki R., Bando A., Toh S., Lepiniec L., Kawakami N. (2006) Plant Cell Physiol. 47, 1081–1094 [DOI] [PubMed] [Google Scholar]
- 49.Salaita L., Kar R. K., Majee M., Downie A. B. (2005) J. Exp. Bot. 56, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 50.O'Connor C. M., Clarke S. (1984) J. Biol. Chem. 259, 2570–2578 [PubMed] [Google Scholar]
- 51.Ingrosso D., Perna A. F., D'Angelo S., Buro M. D., Malanga P., Galletti P., De Rosa M., Zappia V. (1997) J. Nutr. Biochem. 8, 535–540 [Google Scholar]
- 52.Rudgers G. W., Palzkill T. (2001) Protein Eng. 14, 487–492 [DOI] [PubMed] [Google Scholar]
- 53.Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., Obradovic Z. (2001) J. Mol. Graph. Model 19, 26–59 [DOI] [PubMed] [Google Scholar]
- 54.Robinson N. E., Robinson A. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson N. E., Robinson A. B., Merrifield R. B. (2001) J. Pept. Res. 57, 483–493 [DOI] [PubMed] [Google Scholar]
- 56.Young A. L., Carter W. G., Doyle H. A., Mamula M. J., Aswad D. W. (2001) J. Biol. Chem. 276, 37161–37165 [DOI] [PubMed] [Google Scholar]
- 57.Wolschin F., Weckwerth W. (2005) Plant Methods 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Debeaujon I., Léon-Kloosterziel K. M., Koornneef M. (2000) Plant Physiol. 122, 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clerkx E. J., Vries H. B., Ruys G. J., Groot S. P., Koornneef M. (2003) Plant Physiol. 132, 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008) Annu. Rev. Plant Biol. 59, 387–415 [DOI] [PubMed] [Google Scholar]
- 61.Owens M. K., Wallace R. B., Archer S. (1995) J. Arid. Environ. 29, 15–23 [Google Scholar]
- 62.Bidinosti M., Martineau Y., Frank F., Sonenberg N. (2010) J. Biol. Chem. 285, 19402–19408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linder P. (2006) Nucleic Acids Res. 34, 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. (2003) Gene 313, 17–42 [DOI] [PubMed] [Google Scholar]
- 65.Barakat A., Szick-Miranda K., Chang I. F., Guyot R., Blanc G., Cooke R., Delseny M., Bailey-Serres J. (2001) Plant Physiol. 127, 398–415 [PMC free article] [PubMed] [Google Scholar]
- 66.Chang I. F., Szick-Miranda K., Pan S., Bailey-Serres J. (2005) Plant Physiol. 137, 848–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henras A. K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. (2008) Cell. Mol. Life Sci. 65, 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noueiry A. O., Chen J., Ahlquist P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12985–12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. (1997) Science 275, 1468–1471 [DOI] [PubMed] [Google Scholar]
- 70.Coller J., Parker R. (2005) Cell 122, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de la Cruz J., Iost I., Kressler D., Linder P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beckham C., Hilliker A., Cziko A. M., Noueiry A., Ramaswami M., Parker R. (2008) Mol. Biol. Cell 19, 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S. C., Chung M. C., Chen C. S. (2001) Plant Mol. Biol. 47, 761–770 [DOI] [PubMed] [Google Scholar]
- 74.Clarke S. (2003) Ageing Res. Rev. 2, 263–285 [DOI] [PubMed] [Google Scholar]
- 75.Rajjou L., Lovigny Y., Groot S. P., Belghazi M., Job C., Job D. (2008) Plant Physiol. 148, 620–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rajjou L., Gallardo K., Debeaujon I., Vandekerckhove J., Job C., Job D. (2004) Plant Physiol. 134, 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gershater M. C., Cummins I., Edwards R. (2007) J. Biol. Chem. 282, 21460–21466 [DOI] [PubMed] [Google Scholar]
- 78.Wehmeyer N., Vierling E. (2000) Plant Physiol. 122, 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wehmeyer N., Hernandez L. D., Finkelstein R. R., Vierling E. (1996) Plant Physiol. 112, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chibani K., Ali-Rachedi S., Job C., Job D., Jullien M., Grappin P. (2006) Plant Physiol. 142, 1493–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Irshad M., Canut H., Borderies G., Pont-Lezica R., Jamet E. (2008) BMC Plant Biol. 8, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujikura U., Horiguchi G., Ponce M. R., Micol J. L., Tsukaya H. (2009) Plant J. 59, 499–508 [DOI] [PubMed] [Google Scholar]
- 83.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Science 301, 653–657 [DOI] [PubMed] [Google Scholar]