Abstract
YchF is a subfamily of the Obg family in the TRAFAC class of P-loop GTPases. The wide distribution of YchF homologues in both eukarya and bacteria suggests that they are descendents of an ancient protein, yet their physiological roles remain unclear. Using the OsYchF1-OsGAP1 pair from rice as the prototype, we provide evidence for the regulation of GTPase/ATPase activities and RNA binding capacity of a plant YchF (OsYchF1) by its regulatory protein (OsGAP1). The effects of OsGAP1 on the subcellular localization/cycling and physiological functions of OsYchF1 are also discussed. The finding that OsYchF1 and OsGAP1 are involved in plant defense response might shed light on the functional roles of YchF homologues in plants. This work suggests that during evolution, an ancestral P-loop GTPase/ATPase may acquire new regulation and function(s) by the evolution of a lineage-specific regulatory protein.
Keywords: ATPases, Electron Microscopy (EM), G Proteins, Plant, Signal Transduction, Obg, Rice
Introduction
The most common protein fold in the three domains of life is the phosphate binding loop (P-loop)-containing NTPases, comprising 11–18% of total proteins annotated from genomic data (1). P-loop GTPases are thought to be a monophyletic superclass within the P-loop NTPase fold (2), which further evolved into different groups of important regulatory proteins (3).
The activities and modes of actions of P-loop GTPases are controlled by regulatory proteins, such as GTPase-activating proteins and guanine nucleotide exchange factors (4). In general, a GTPase charged with GTP is active and exhibits greater affinities toward its target(s) (3).
YchF is a subfamily of the Obg family in the TRAFAC class of P-loop GTPases (2, 5). YchF proteins contain a G domain in the N-terminal for GTP binding (6) and a typical TGS (ThrRS, GTPase, and SpoT) domain in the C terminus that may associate with the translation machinery (7).
The G domain is universal among all “molecular switch GTPases” (8) and consists of five motifs (6): the G1/Walker A motif (the P-loop that helps to position the triphosphate moiety), the G2 and G3/Walker B motifs (coordinating a Mg2+ ion for nucleotide biding and hydrolysis), the G4 motif (conferring specificity in nucleotide binding), and the G5 motif (supporting guanine base recognition). One unique feature of the YchF subfamily is the absence of the canonical sequence (NKXD) in the G4 motif, resulting in the loss of substrate specificity toward GTP (6, 9).
Crystal structure of the YchF protein of Hemophilus influenzae revealed a crablike three-domain structure that may act as the binding site for nucleic acids. The loopy structure of RNA may better fit into the nucleic acid binding domain (9). YchF proteins were also shown to associate with ribosomes/polysomes (10). However, no direct experimental evidence verifying RNA binding by YchF proteins has been reported.
YchF is probably an ancient protein involved in fundamental life processes, based on the wide distribution of YchF proteins in eukarya and bacteria (2). However, the physiological roles of this group of GTPases are still not clearly understood. We previously identified an YchF homologue from rice (OsYchF1), which is a target of the regulatory protein OsGAP1 (11). OsGAP1 binds to OsYchF1 and activates its GTPase activity, whereas overexpression of OsGAP1 could enhance plant defense response (11).
In this study, using the OsYchF1 and OsGAP1 pair as the prototype, we provide evidence to demonstrate the regulation of a plant YchF protein by its regulatory protein. The effects of OsGAP1 on the subcellular localization and physiological functions of its target OsYchF1 are also implicated.
EXPERIMENTAL PROCEDURES
DNA and RNA Manipulation
DNA sequencing, RNA extraction, reverse transcription, and PCR (regular and real time) were performed according to a previous report (11). The relative gene expression was calculated using the 2−ΔΔCT method (12) and normalized with the Arabidopsis thaliana UBQ10 gene (13). Detailed primer information for the cloning and production of recombinant constructs is given in supplemental Table S1.
Phylogenetic Analysis and Sequence Alignment
A multiple sequence alignment of full-length protein sequences (supplemental Fig. S1) was performed using the ClustalW program (14). The phylogenetic tree was built with the MEGA program (version 4) (15), using the neighbor joining method with 1000 bootstrap replicates. YchF homologues from various higher plant species were included. Homologues from other eukarya and bacteria were chosen from well characterized representatives of different phyla. Because YchF homologues were not found in archaea, we substituted with Ygr210 proteins (the closest member to the YchF subfamily in the Obg family) from archaea in the analysis.
Fusion Proteins and Antibodies
Glutathione S-transferase (GST) fusion proteins were expressed by subcloning target cDNAs into the pGEX-4T-1 vector (GE Healthcare) to form in-frame fusion proteins with the GST tag located at the N terminus. Protein expression in the Escherichia coli host (BL21) was induced by adding 0.5 mm isopropyl-β-d-thiogalactopyranoside to the growth medium and incubating at 30 °C overnight. The proteins were then purified using the GST SpinTrapTM purification module (catalog no. 27-4570-03, GE Healthcare).
The primary antibodies for detecting OsGAP1 (11) and OsYchF1 (this work) were raised from rabbits and mice, respectively. Anti-GST antibodies and gold-labeled secondary antibodies were from Sigma-Aldrich and Electron Microscopy Sciences (Hatfield, PA), respectively.
Nucleotide Binding Assays
Nucleotide binding assays were performed as described (16) using non-fusion OsYchF1 proteins (see below). 31 μm Mant-GTP2 (catalog no. M12415, Invitrogen) and about 6 μm OsYchF1 were used in each 160-μl reaction. For competition with the Mant-GTP, a final concentration of 31 μm of either GTP or ATP was added to the previous reaction mixture containing 31 μm Mant-GTP and 6 μm OsYchF1 protein.
GTPase/ATPase Activity Assays
The GTPase/ATPase activities were examined by monitoring the decrease in lanthanide luminescence of Tb(III)-norfloxacin due to strong quenching by Pi during GTP/ATP hydrolysis, following the procedures in a previous report (17). The concentrations of charged GTP and charged ATP were estimated by HPLC. Initial velocity was calculated right after the reference signal was stabilized, and khyd was determined as described (17).
After protein purification, the GST tags of GST-OsYchF1 and GST-OsGAP1 were cleaved by thrombin protease (catalog no. 27-0846-01, GE Healthcare) and removed by passing the digested samples through GSTrapTM FF columns (17-5130-02, GE Healthcare). Thrombin was then removed, using HiTrap Benzamidine FF (high sub) columns (17-5143-02, GE Healthcare). The resultant non-fusion OsYchF1 and OsGAP1 were then used in this assay.
The amount of OsYchF1 used in the reaction was quantified as described (17). The assays were recorded in 96-well microplates, with each well having a final reaction volume of 100 μl. The concentrations of GTP-bound OsYchF1 and ATP-bound OsYchF1 in the assays were 13.4 and 13.0 μm, respectively (within the range tested in the method reference, 6–24 μm). To assess the GTPase activation capacity of OsGAP1, 20 μm OsGAP1 was added.
The data were recorded in a NunclonTM Delta Surface 96-well plate (catalog no. 136101, Nunc, Roskilde, Denmark) and acquired with the Safire microplate reader (catalog no. F129013, Tecan, Männedorf, Switzerland). Enzyme assays were performed in triplicate by applying the time-gated detection at a time lag of 60 μs after the excitation pulse and a signal integration time of 60 μs (17).
Identification of the RNA Target of OsYchF1
About 200 ng of purified GST-OsYchF1 (or GST as the negative control) was mixed with 1 μg of total RNA extracted from rice in 0.5 ml of RNA binding buffer (10 mm Tris (pH 7.5), 1.5 mm MgCl2, 250 mm KCl, 0.5 mm DTT, 2 μg/ml leupeptin, 10 units of RNasin inhibitor) and incubated for 30 min at room temperature. 30 μl of Protein A-agarose (catalog no. P9376-1ML, Sigma-Aldrich) presoaked with anti-GST antibodies was added to pull down the protein-RNA complex by a further incubation of 30 min at room temperature. After washing with the same buffer five times, the sample mixture was boiled in 1% SDS, extracted with phenol/chloroform/isoamylalcohol (25:24:1; v/v/v), and precipitated with ethanol (18, 19). Reverse transcription with the SuperScript II system (catalog no. 18064-071, Invitrogen) and PCR amplification were performed using a mixture of arbitrary primers (detailed in supplemental Table S1). Three discrete bands were detected by running the resulting PCR products on agarose gel and were excised and cloned into pBluescript KS II (+) for sequencing and in vitro transcription.
Binding Assays of 26 S RNA
Digoxigenin (DIG)-labeled or unlabeled RNAs were synthesized via in vitro transcription using the RiboMAX large scale RNA production system T7 (catalog no. P1300, Promega). In the pull-down assays, about 1 μg of DIG-labeled RNA was mixed with 200 ng of GST-OsYchF1 in 0.5 ml of the same RNA binding buffer described above. The same amount (200 ng) of GST and GST-OsGAP1 was employed instead of GST-OsYchF1 as negative controls. For the competition experiment, the DIG-labeled 26 S RNA was mixed with different amounts of unlabeled 26 S RNA and then incubated for 30 min at room temperature before 20 μl of Protein A-agarose presoaked with anti-GST antibodies was added to pull down the protein-RNA complex as described above.
To show the inhibitory effect of OsGAP1 on the binding of 26 S RNA to OsYchF1, different amounts (0, 50, 100, or 200 ng) of GST-OsGAP1 or GST (negative control) were mixed with 200 ng of GST-OsYchF1 and 1 μg of DIG-labeled RNA in 0.5 ml of RNA binding buffer. After incubating for 30 min at room temperature, 20 μl of Protein A-agarose presoaked with anti-GST antibodies was added to start the pull-down reactions as described above.
Pull-down products were then blotted onto nylon membrane via a slot blot apparatus. DIG-labeled RNA was detected by anti-DIG antibodies with CSPD as the substrate (Roche Applied Science). The membrane was then stripped by heating with stripping buffer (Amersham Biosciences ECL Advance Western blotting detection kit, catalog no. RPN2135, GE Healthcare) at 50 °C with occasional agitation, blocked in 2% skim milk blocking solution, and incubated with anti-GST antibodies to verify the success of protein pull-down in each reaction.
Phospholipid Dot Blot
Phospholipids were extracted from rice leaves using a modified protocol (20). About 0.5 g of rice tissue was ground and extracted. The final residue was resuspended in 1 ml of chloroform/methanol (2:1; v/v). 5 μl of phospholipid crude extract was dotted on supported nitrocellulose membrane (0.2 μm; catalog no. 162-0095, Bio-Rad). The membranes were dried, blocked, incubated with GST or GST fusion proteins (0.5 μg/ml) in 2% skim milk blocking solution, and washed as described (21). The incubation and subsequent washing steps were performed in TBST (10 mm Tris, pH 8.0, 150 mm NaCl, 0.1% (v/v) Tween 20), supplemented with 200 μm CaCl2 or 2 mm EGTA (22). When studying the effect of the interactions between OsGAP1 and OsYchF1 on the association with phospholipids, GST-OsGAP1 and GST-OsYchF1 fusion proteins were detected with anti-GST and anti-OsYchF1 antibodies, respectively.
Electron Microscopy
For single labeling experiments, 8-week-old rice plants at booting stage were wounded by a clipping method (23). Leaf tissues within 3 cm from the wounding site or from non-wounded plants were collected 3 days after treatment. Embedding and electron microscopy were performed as described with some modifications (24, 25). Freshly prepared leaf discs were immediately frozen in a high pressure freezing apparatus (catalog no. EM PACT2, Leica, Hesse, Germany) and were kept at −85 °C for 16 h before being gradually warmed up to −50 °C over 5 h in the substitution medium (0.1% (w/v) uranyl acetate in dry acetone) using an automatic freeze-substitution unit (EMAFS; Leica). At −50 °C, the medium was replaced with 100% ethanol for dehydration and then infiltrated stepwise with 33% HM20 (catalog no. 14340, EMS) and 66% HM20 in 100% ethanol (1 h/step at −50 °C), followed by substitution with 100% HM20 for 16 h at −50 °C. The resulting tissue blocks were gradually warmed up to −35 °C over 4 h and polymerized under UV light for more than 48 h. Polymerized tissue blocks were ultrathin sectioned to 70-nm-thick slices for immunolabeling. After labeling with primary antibodies (1:50 in 1% PBS-BSA), the subcellular localization of targeted proteins was subsequently detected by gold-labeled secondary antibodies (1:50 in 1% PBS-BSA) against rabbits (for OsGAP1) or mice (for OsYchF1) IgGs. For poststaining, samples were incubated in 3% uranyl acetate for 10 min at room temperature, followed by treatment with lead citrate for 10 min. Between and after these two steps, samples were washed with double distilled H2O gently but thoroughly. The grids were then air-dried on filter paper until observation.
For double labeling experiments, 6-week-old plants of an OsGAP1-overexpressing rice line (11), and the untransformed parent were grown on half Murashige and Skoog salt mixture agar (catalog no. MSP01-50LT, Caisson Laboratories, North Logan, UT) in sterilized magenta boxes. The sample preparation and poststaining of the co-localization study were similar to single labeling experiments. The primary antibodies (rabbit anti-OsGAP1 and mouse anti-OsYchF1; 1:50 in 1% PBS-BSA) were mixed and added after BSA blocking. Detection was by gold-labeled secondary antibodies (goat anti-rabbit, 6-nm IgG, (catalog no. 25104, EMS) and goat anti-mouse 15-nm IgG/IgM (catalog no. 25173, EMS); 1:50 in 1% PBS-BSA).
Plant Materials, Growth Conditions, and Transgenic Plants
AtYchF1 knockdown mutant was from the Arabidopsis Biological Research Centre (stock number CS855214). Chemicals for plant growth were from Sigma-Aldrich. Potting soil for the growth of A. thaliana was from Floragard Vertriebs GmbH (Gerhard-Stalling, Germany). Rice was grown in regular field soil in a greenhouse under natural light. For constructing transgenic A. thaliana expressing OsYchF1, the coding region of OsYchF1 was subcloned into the binary vector V7 (26) and transformed (27) into Col-0.
Pathogen Inoculation
Eight-week-old A. thaliana plants for pathogen inoculation tests were first grown on Murashige and Skoog salt mixture agar plates for 2 weeks before being transferred to Floragard potting soil and cultivated in a growth chamber (22–24 °C; relative humidity 70–80%; light intensity 80–120 microeinsteins of a 16-h light/8-h dark cycle). Inoculation of Pseudomonas syringae pv. tomato DC3000 and subsequent titer determination of 5-day postinoculation samples were performed using a plate count method (28).
Statistical Analysis
Numerical data were analyzed using the Statistical Package for Social Sciences (version 12.0) (SPSS, Inc.). Mean difference was analyzed using one-way analysis of variance followed by Tukey's post hoc test. Errors of real time PCR were calculated as described previously (12).
RESULTS
OsYchF1 Is a Descendent of an Ancient YchF-type P-loop GTPase/ATPase
OsYchF1 is encoded by a single gene located on chromosome 8 of the rice genome. Its protein sequence was used to search for homologues from major lineages using the Blastp algorithm, using different subsets of the genome data base in GenBankTM. Phylogenetic analysis (Fig. 1A) confirmed that OsYchF1 is a member of the YchF subfamily. YchF proteins were found in bacteria and eukarya. Among completely sequenced genomes, YchF proteins are generally encoded by one or a few genes. However, direct YchF homologues were not found in archaea. The closest P-loop GTPase subfamily in archaea is the Ygr210 proteins (2).
FIGURE 1.
Phylogenetic analysis and sequence alignment of OsYchF homologues. A, a phylogenetic analysis of YchF homologues was performed for selected members from eukarya and bacteria. Those that have been previously studied are boxed. B, plant YchF homologues were aligned to show the conserved YchF domain (residues 26–386 of OsYchF1, BAD03576), including the G1–G5 motifs of the G domain, and the TGS domain (underlined). Zma, Zea mays (ACG38260); Sbi, Sorghum bicolor (XP_002445191); Gma, G. max (ACU21485); Ath, A. thaliana (AAD25745); Vvi, Vitis vinifera (XP_002263885); Can, Capsicum annuum (AAF65513); Psi, Picea sitchensis (ABK25108); Ppa, Physcomitrella patens subsp. patens (EDQ58637); Cre, Chlamydomonas reinhardtii (EDP07459); Mpu, Micromonas pusilla CCMP1545 (EEH53875); Hsa, Homo sapiens (NP_037473); Mmu, Mus musculus (BAE39543); Xtr, Xenopus tropicalis (CAJ83205); Cel, Caenorhabditis elegans (CAB07131); Dme, Drosophila melanogaster (NP_572580); Pkn, Plasmodium knowlesi strain H (CAQ38331); Rle, Rhizobium leguminosarum bv. viciae 3841 (YP_769059); Bsu, Bacillus subtilis subsp. subtilis str. 168 (NP_391972); Sau, Staphylococcus aureus subsp. aureus Mu50 (NP_370887); Xor, Xanthomonas oryzae pv. oryzae MAFF 311018 (YP_452431); Pfl, Pseudomonas fluorescens Pf0-1 (YP_350484); Eco, E. coli O157:H7 str. Sakai (NP_309735); Hin, H. influenzae Rd KW20 (NP_438555); Mtu, Mycobacterium tuberculosis H37Rv (NP_215628); Tcr, T. cruzi (EAN95922); Hsp, Halobacterium sp. NRC-1 (NP-280498); Aful, Archaeoglobus fulgidus DSM 4304 (NP_070193); Neq, Nanoarchaeum equitans Kin4-M (NP_963747); Tko, Thermococcus kodakarensis KOD1 (YP_182919); Tne: Thermoproteus neutrophilus V24Sta (YP_001794985); Afum, Aspergillus fumigatus Af293 (EAL90309); Pch, Penicillium chrysogenum Wisconsin 54-1255 (CAP93291); Pbr, Paracoccidioides brasiliensis Pb01 (EEH40843); Cgl, Candida glabrata (CAG58059); Sce, Saccharomyces cerevisiae YJM789 (EDN64640).
Within the domain eukarya, YchF proteins could be further divided into three lineages: plantae, animalia, and fungi (Fig. 1A). On the other hand, YchF homologues of protista are scattered among different lineages, indicating their polyphyletic nature (29). Within the kingdom plantae, YchF proteins in bryophyte (e.g. Physcomitrella patens subsp. patens) became an apparent outgroup of that in spermatophyte, which was further divided into two closely related lineages, monocots (e.g. Oryza sativa, Zea mays, and Sorghum bicolor) and dicots (e.g. Glycine max, A. thaliana, Vitis vinifera, and Capsicum annuum).
An alignment of the YchF members of plants revealed their strong homology (Fig. 1B). They all possess the highly conserved YchF signature (2) which includes the G domain and the YchF-type TGS domain. A more comprehensive alignment was also performed (supplemental Fig. S1) to reveal the extensive homology and conserved sequences shared by YchF proteins from bacteria and eukarya.
In contrast, the regulatory protein OsGAP1, which interacts with OsYchF1, is specific to higher plants. We performed homology searches using representative members (that have genome data deposited in GenBankTM) from Dicotyledoneae, Monocotyledoneae, Bryophyta, Chlorophyta (green algae), Cryptophyta (red algae), Cyanobacteria (blue green algae), Fungi, Mammalia, and Insecta. Homologues were only found in Monocotyledoneae and Dicotyledoneae. In Bryophyta, Chlorophyta, Cryptophyta, Cyanobacteria, Fungi, Mammalia, and Insecta, sequence similarities were confined to only the C2 domain consensus sequence. The result of a tblastn search is included in supplemental Table S2.
OsYchF1 Exhibited both GTPase and ATPase Activities That Were Activated by OsGAP1
As in other YchF homologues, the NKXD consensus sequence is absent in the G4 motif of OsYchF1. Because this may lead to the loss of substrate specificity toward GTP (6, 9), we tested both GTPase and ATPase activities of OsYchF1. By measuring the release of inorganic phosphate, we previously demonstrated the GTPase activity of OsYchF1 and its activation by OsGAP1, using GST-OsYchF1 and GST-OsGAP1 fusion proteins (11). A negative control using free GST protein showed that the GTPase activities of OsYchF1 could not be activated by GST alone (11). Because the fold activation by OsGAP1 was very low when compared with other well characterized GTPase-activating proteins (30), we further confirmed this result with a more sensitive commercial kit (supplemental Fig. S2).
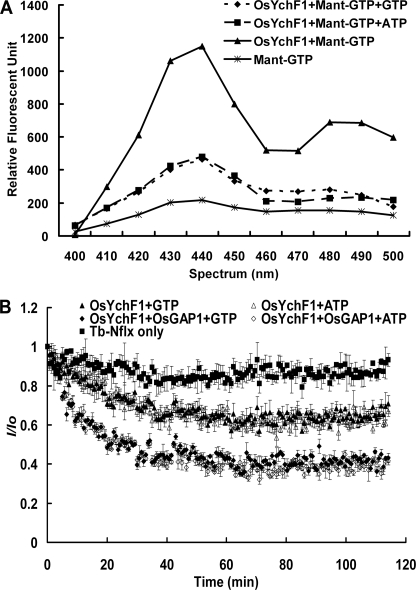
To learn more about the enzymatic properties of OsYchF1 and its regulation by OsGAP1, we obtained free OsYchF1 and OsGAP1 proteins by removing the GST tags. Binding of OsYchF1 to both GTP and ATP were shown by competition assays (Fig. 2A). The GTPase and ATPase activities were then measured by means of a luminescent terbium complex (17) (Fig. 2B). The initial velocity was calculated right after the reference signal had stabilized (after around 4–5 min), as indicated in Fig. 2B (Tb-Nflx only). The khyd was determined by dividing the absolute value of the slope of the linear range from around 4 to 10 min by the corresponding charged GTP/ATP concentration. OsYchF1 exhibited similar binding affinities and hydrolytic activities toward both GTP and ATP. The khyd (L/(min mol)) values of GTP (charging efficiency 96%) and ATP (charging efficiency 99%) were estimated to be 650 ± 94 and 603 ± 182, respectively. Comparing the initial reaction velocities of the hydrolytic curves, both GTPase and ATPase activities increased in the presence of OsGAP1, by 3.6- and 3.2-fold, respectively (Fig. 2B).
FIGURE 2.
Nucleotide binding and GTPase/ATPase activities of OsYchF1. A, fluorescent signals emitted when OsYchF1 bound to Mant-GTP (triangles). These signals decreased when the binding was challenged with unlabeled GTP (diamonds) or ATP (squares). B, the activation effect of OsGAP1 was indicated by the more rapid drop of fluorescent signals. Error bars, S.E. values of three replicates in each data point.
OsYchF1 Could Bind to the 26 S RNA, and Such Binding was Negatively Regulated by OsGAP1
The presence of the TGS domain in OsYchF1 suggests its possible interactions with RNAs (9). To examine the RNA binding ability of OsYchF1 and uncover the identity of the bound RNA, we mixed the GST-OsYchF1 fusion protein with the RNA extracted from rice leaves and performed an in vitro pull-down assay using anti-GST antibodies, followed by reverse transcription and PCR amplification using arbitrary primers. We obtained three clones that were all fragments from the 26 S rDNA (GenBankTM accession number AP008210; the positions of the three clones obtained were 5598–5806, 7438–7465, and 7445–8082). As a negative control, pull-down experiments using GST alone did not result in any successful PCR amplification.
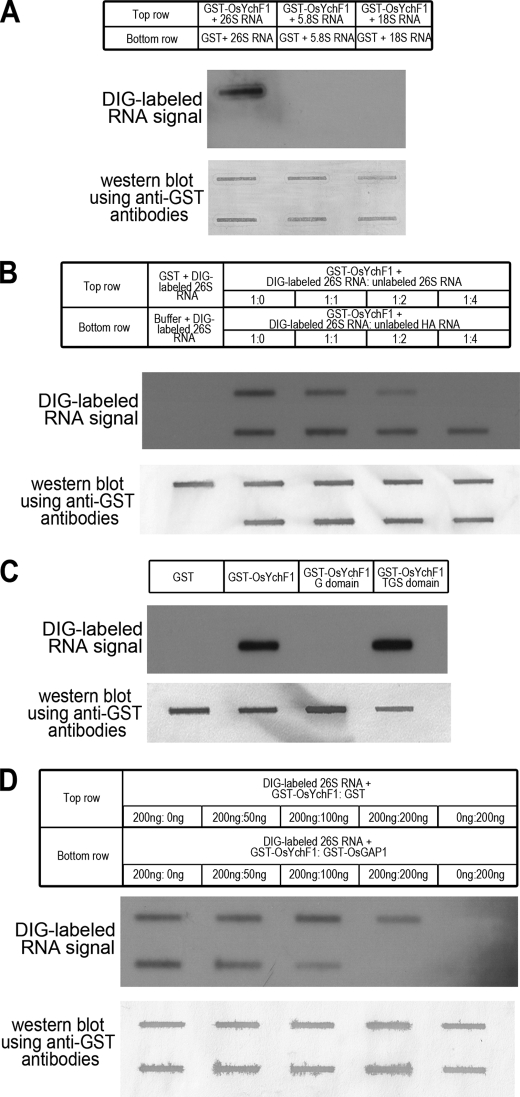
To illustrate the binding of OsYchF1 to the 26 S RNA, we produced single species of DIG-labeled 26, 5.8, and 18 S RNA by in vitro transcription. After mixing each of the RNA species with GST-OsYchF1, an in vitro pull-down assay was performed using anti-GST antibodies. The pull-down product was applied to slot blots, and the DIG-labeled RNA was detected by chemiluminescence (Fig. 3A). The results showed that OsYchF1 only bound to the 26 S RNA and not to the 5.8 or 18 S RNA.
FIGURE 3.
Slot blot analysis showing the interaction between OsYchF1 and the 26 S RNA. DIG-labeled RNA signals (upper panels) and Western blot signals (lower panels; using anti-GST antibodies) in each part of the figure are used to illustrate the successful pull-down of RNA and proteins (free GST or GST fusion proteins), respectively. A, binding of OsYchF1 to the 26 S RNA. The binding capacities of OsYchF1 to the in vitro-transcribed 26, 5.8, and 18 S RNA were compared by monitoring the DIG label signals. Free GST was used as a negative control of the RNA binding. B, competition by unlabeled RNA. GST-OsYchF1 was mixed with DIG-labeled 26 S RNA in the presence of different ratios of unlabeled 26 S RNA or unlabeled HA RNA as competitors. Free GST and protein-free buffer (first lane from the left) were included as negative controls. C, binding of the 26 S RNA to the TGS domain. In vitro-transcribed DIG-labeled 26 S RNA was mixed with 200 ng each of free GST, full-length GST-OsYchF1, GST-OsYchF1 G domain (amino acid residues 1–301), or GST-OsYchF1 TGS domain (amino acid residues 302–394), followed by a pull-down using anti-GST antibodies. D, competition by OsGAP1. GST-OsYchF1 (200 ng) was mixed with the 26 S RNA in the presence of increasing amounts (0, 50, 100, and 200 ng) of either GST-OsGAP1 or free GST. Without OsYchF1, free GST or GST-OsGAP1 did not bind to the 26 S RNA (first lane from the right).
Subsequently, we used unlabeled RNA to challenge the binding between DIG-labeled 26 S RNA and GST-OsYchF1 (Fig. 3B). Two negative controls were added. In the absence of proteins (Buffer + DIG-labeled 26 S RNA), no protein was pulled down by anti-GST antibodies, and no RNA signal was observed. When free GST was used (GST + DIG-labeled 26 S RNA), the protein was successfully pulled down, but no RNA signal was observed, indicating that the binding of the 26 S RNA to the GST-OsYchF1 fusion protein was unrelated to the GST tag.
In the competition experiment on the binding of the 26 S RNA to GST-OsYchF1, the DIG-labeled 26 S signals diminished as the quantity of unlabeled 26 S RNA increased. Competition was observed starting at a ratio of labeled/unlabeled RNA of 1:1 (Fig. 3B, top row; see also supplemental Fig. S3). On the other hand, when an unrelated unlabeled RNA (hemagglutinin (HA)) was used as the challenger, a slight reduction of signal was only observed when the quantity of unlabeled HA RNA reached 2 or 4 times that of the labeled 26 S RNA (Fig. 3B, bottom row; see also supplemental Fig. S3). This may be due to nonspecific binding under high concentrations of the competing RNA but of a much lower affinity than the binding of OsYchF1 to the 26 S RNA.
To locate the binding domain of OsYchF1 for the 26 S RNA, GST fusion proteins were made for the G domain and the TGS domain of OsYchF1. Only the TGS domain and not the G domain could bind to the 26 S RNA (Fig. 3C).
The result of a previous yeast two-hybrid experiment (11) showed that a truncated construct covering mostly the TGS domain of OsYchF1 was sufficient to interact with OsGAP1. Consequently, we studied the effect of OsGAP1 on the binding of OsYchF1 to the 26 S RNA. As negative controls, neither the GST-OsGAP1 fusion protein nor free GST could bind to the 26 S RNA directly (Fig. 3D, first lane from the right). When GST-OsGAP1 was used as the challenger, a dosage-dependent competition was observed (Fig. 3D; see also supplemental Fig. S4). The RNA signal disappeared almost completely when 200 ng of GST-OsGAP1 was added to challenge the GST-OsYchF1 binding of labeled RNA. On the other hand, the addition of 200 ng of free GST only resulted in a slight signal reduction, probably due to nonspecific binding at a high GST concentration. These results indicated that the presence of OsGAP1 might reduce the interaction between the 26 S RNA and OsYchF1, probably via blocking the TGS domain of OsYchF1.
Relocalization of OsYchF1 and OsGAP1 from the Cytoplasm to the Plasma Membrane Was Triggered by Wounding Signals
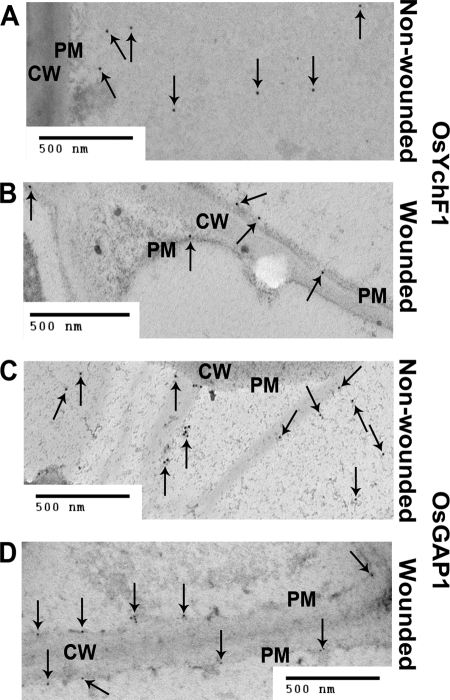
OsGAP1 is a positive factor for the plant defense response (11). It is expected to interact with the plasma membrane due to the presence of a C2 domain (22). On the other hand, OsYchF1 is predicted to be cytosolic and contains no transmembrane domain. To clarify the subcellular localization of OsYchF1 and OsGAP1, immunogold electron microscopic studies were performed using anti-OsYchF1 and anti-OsGAP1 antibodies, respectively (Fig. 4). In cells of untreated rice leaves, the majority of signals for both OsYchF1 and OsGAP1 were localized in the cytoplasm, with only a small portion on the plasma membrane (Fig. 4, A and C). However, when the rice leaves were wounded by a clipping method, there was a massive relocalization of OsYchF1 and OsGAP1 to the plasma membrane in plant cells (Fig. 4, B and D). The percentages of OsYchF1 and OsGAP1 localized on plasma membrane under different conditions were estimated by counting the total number of gold particles in different photographs (Table 1).
FIGURE 4.
Immunogold electron microscopic studies of the subcellular localizations of OsYchF1 and OsGAP1. Leaf samples of 8-week-old rice plants were collected from non-wounded (A and C) or wounded (B and D) plants 3 days after treatment by a leaf clipping method. Subcellular localization of targeted proteins was detected using anti-OsYchF1 (A and B) and anti-OsGAP1 antibodies (C and D). The arrows indicate the locations of some gold particles. CW, cell wall. PM, plasma membrane. Scale bar, 500 nm. The percentages of immunogold-labeled dots of OsYchF1 and OsGAP1 localized on plasma membrane with or without wounding were estimated using multiple photographs (Table 1).
TABLE 1.
The percentages of immunogold-labeled dots of OsGAP1 and OsYchF1 localized on the plasma membrane with or without wounding treatment
The two rice lines CBB14 (sample for Fig. 4) and SN1033 (sample for supplemental Fig. S5) were described previously (23). PM, plasma membrane. n, number of photographs counted.
| OsYchF1 |
OsGAP1 |
|||
|---|---|---|---|---|
| Total dots (n) | Percentage of dots on the PM | Total dots (n) | Percentage of dots on the PM | |
| % | % | |||
| CBB14 (non-wounded) | 73 (8) | 5 | 236 (6) | 17 |
| CBB14 (wounded) | 82 (5) | 66 | 380 (12) | 59 |
| SN1033 (non-wounded) | 145 (9) | 6 | 115 (6) | 10 |
| SN1033 (wounded) | 100 (9) | 66 | 77 (4) | 53 |
To confirm this result, we performed a similar experiment using another rice line. Consistent results were obtained (supplemental Fig. S5 and Table 1). Therefore, the wounding signal can trigger the relocalization of both OsYchF1 and OsGAP1 to the plasma membrane.
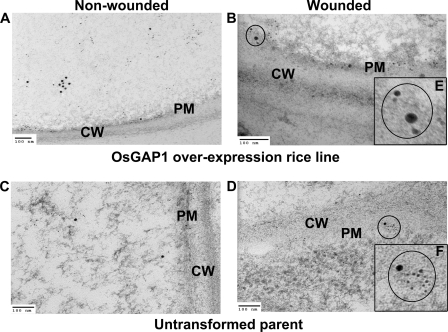
To further demonstrate the effect of OsGAP1 on the plasma membrane localization of OsYchF1 and to show the possible co-localization of OsYchF1 and OsGAP1, double labeling experiments were performed on an OsGAP1 overexpression rice line (11) and the untransformed parent (Fig. 5). Anti-mouse antibodies conjugated with large gold particles (15 nm) and anti-rabbit antibodies conjugated with small gold particles (6 nm) were used to detect OsYchF1 and OsGAP1, respectively. OsYchF1 and OsGAP1 signals could be found in close proximity (Fig. 5). Overexpression of OsGAP1 in rice increased the proportion of plasma membrane-localized OsYchF1 even without wounding (Fig. 5, A and B, and Table 2). However, wounding could give a further boost to the plasma membrane-localized OsYchF1 signals (Fig. 5, C and D, and Table 2).
FIGURE 5.
Co-localization studies of OsYchF1 and OsGAP1 on the plasma membrane via immunogold electron microscopic examination. Leaf samples of 6-week-old plants of an OsGAP1-overexpressing rice line (A and C) and the untransformed control (B and D), with (A and B) and without (C and D) wounding, were collected. E and F, enlarged views of co-localized signals. Co-localization of OsYchF1 (larger dots) and OsGAP1 (smaller dots) are circled and magnified (E and F). Double labeling of OsGAP1 and OsYchF1 was achieved by using primary antibodies (rabbit-anti-OsGAP1 and mouse-anti-OsYchF1) and secondary antibodies (goat-anti-rabbit (6-nm IgG) and goat-anti-mouse (15-nm IgG/IgM)). CW, cell wall. PM, plasma membrane. Scale bar, 100 nm. The percentages of immunogold-labeled dots of OsYchF1 localized on plasma membrane with or without wounding were estimated using multiple photographs (Table 2).
TABLE 2.
The percentages of immunogold-labeled dots of OsYchF1 localized on the plasma membrane in an OsGAP1-overexpressing rice line, with and without wounding treatment
The OsGAP1-overexpressing rice line was described previously (11). Only the OsYchF1 signals were counted. PM, plasma membrane. n, number of photographs counted.
| OsYchF1 |
||
|---|---|---|
| Total dots (n) | Percentage of dots on the PM | |
| % | ||
| OsGAP1-overexpressing rice line (without wounding) | 69 (16) | 33 |
| OsGAP1-overexpressing rice line (with wounding) | 43 (16) | 72 |
| Untransformed parent (without wounding) | 101 (17) | 2 |
| Untransformed parent (with wounding) | 64 (18) | 61 |
OsYchF1 Could Associate with Phospholipids via OsGAP1
Because amino acid sequence analysis revealed no indication that OsYchF1 by itself can associate with the plasma membrane, we hypothesized that the association of OsYchF1 with the plasma membrane is through its interaction with OsGAP1. Previous in vitro experiments showed that OsGAP1 can interact with OsYchF1 (11). We also confirmed the in vivo interaction between OsYchF1 and OsGAP1 by bimolecular fluorescence complementation experiments (supplemental Fig. S6).
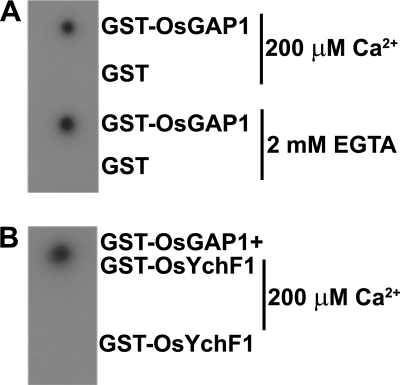
Phospholipids were extracted from rice leaves and dotted onto nitrocellulose membranes, which were then incubated with the fusion proteins, GST-OsGAP1 and GST-OsYchF1. Anti-GST and anti-OsYchF1 antibodies were used to detect GST/GST-OsGAP1 and GST-OsYchF1, respectively (Fig. 6). Protein associations with the phospholipids were not detected when GST (Fig. 6A) or GST-OsYchF1 fusion protein (Fig. 6B) was used. By contrast, GST-OsGAP1 could interact with the phospholipids (Fig. 6A). Although the interaction between the C2 domain and the plasma membrane was dependent on calcium ions in some cases (22), we did not observe any difference in the interaction between OsGAP1 and the phospholipids when either calcium ions or a divalent ion chelator (EGTA) was added (Fig. 6A). When GST-OsYchF1 was mixed with GST-OsGAP1, GST-OsYchF1 was found to be retained on the phospholipid dot blot (detected by anti-OsYchF1 antibodies) as a result of the interaction between OsYchF1 and OsGAP1 (Fig. 6B).
FIGURE 6.
Phospholipid dot blot assays. A, the membrane was incubated with GST-OsGAP1 or GST in a buffer supplemented with 200 μm CaCl2 or 2 mm EGTA, followed by detection using anti-GST antibodies. B, the membrane was incubated with GST-OsGAP1 + GST-OsYchF1 or GST-OsYchF1 only, followed by detection using anti-OsYchF1 antibodies.
Ectopically Expressing OsYchF1 and Knocking Down AtYchF1 in A. thaliana Exhibited Opposite Effects on the Resistance toward the Pathogen P. syringae pv. tomato DC3000
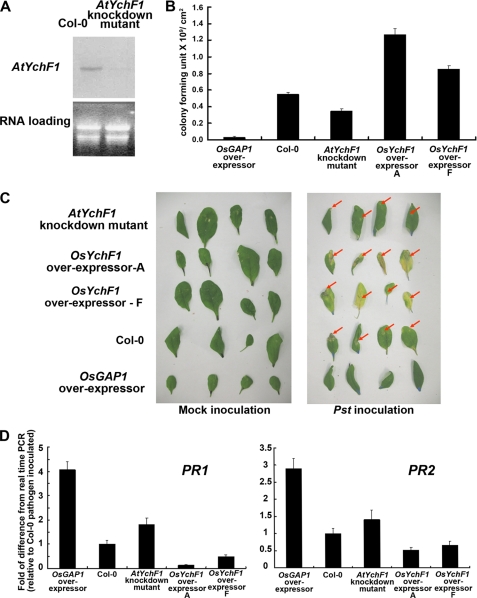
Based on the strong homology between OsYchF1 and AtYchF1 (Fig. 1) and the previous finding that the ectopic expression of OsGAP1 in transgenic A. thaliana could enhance defense response (11), we investigated the possible function of OsYchF1 using the model plant A. thaliana. An AtYchF1 knockdown mutant (with Col-2 as the wild type genetic background) is available in the public domain, enabling the loss-of-function analysis. Col-0 and Col-2 behaved similarly in terms of their pathogen sensitivity and the expression level of the endogenous AtYchF1 (supplemental Fig. S7). Subsequent experiments adopted Col-0 as the wild type control for comparison. A Northern blot analysis showed that the level of the AtYchF1 transcripts was barely detectable in the AtYchF1 knockdown mutant (Fig. 7A). When challenged with the bacterial pathogen P. syringae pv. tomato DC3000, the ectopic expression of OsGAP1 and the deficiency in AtYchF1 both caused increased resistance, whereas the ectopic expression of OsYchF1 led to the opposite result. For instance, pathogen titers (Fig. 7B) and disease symptoms (Fig. 7C) were alleviated in both the OsGAP1 transgenic A. thaliana and AtYchF1 knockdown mutant, when compared with the wild type Col-0. On the other hand, both parameters were aggravated in OsYchF1 transgenic A. thaliana. Consistent with these observations, under the challenge of P. syringae pv. tomato DC3000, the expression levels of the defense marker genes, PR1 and PR2 (markers for the salicyclic acid defense pathway against biotrophic pathogens) were also found to increase in the OsGAP1 transgenic line and the AtYchF1 knockdown mutant but decrease in the OsYchF1 transgenic lines (Fig. 7D). All of these results suggest a negative role of OsYchF1 in plant defense response, which is in turn negatively regulated by OsGAP1.
FIGURE 7.
Pathogen inoculation tests of the AtYchF1 knockdown mutant and transgenic A. thaliana expressing OsYchF1 and OsGAP1. A, Northern blot analysis showing the reduced level of AtYchF1 transcripts in the AtYchF1 knockdown mutant. B, pathogen titers (error bars, S.E. of five plants; one-way analysis of variance showed that all lines were significantly different from Col-0 at p < 0.05). C, disease symptoms (highlighted by red arrows). D, expression of defense marker genes (error bars, S.E. values of at least three samples) estimated by real time PCR. AtYchF1 knockdown mutant and transgenic A. thaliana expressing OsYchF1 and OsGAP1 were examined 5 days after inoculation of P. syringae pv. tomato DC3000 via syringe infiltration. The expression in Col-0 was set to 1 for reference. OsYchF1 overexpressors A and F are two independent transgenic lines. Two biological repeats were performed, and similar results were obtained.
DISCUSSION
The last systematic search for different families of P-loop GTPases was performed 8 years ago (2). The analysis included all major groups of P-loop GTPases and was based on structural and genome sequence information available at that time. YchF was identified as a subfamily with unknown function(s). The exponentially increasing genome data prompted us to perform a new phylogenetic analysis, focusing only on YchF proteins. The apparent absence of YchF homologues in archaea (2) was reconfirmed by examining the sequenced archaeal genomes. It could be attributable to an evolutionary loss in archaea or a horizontal gene transfer between bacteria and an ancestral eukaryote.
More detailed analyses of the domain eukarya showed that YchF proteins can be clustered into three lineages: plantae, animalia, and fungi (Fig. 1A). The apparent low branch bootstrap values indicate that YchF proteins are much conserved. OsYchF1 possesses all of the signatures of an YchF protein (Fig. 1B), and the phylogenetic analysis shows that it is a member in the lineage of monocot YchF proteins (Fig. 1A). All of these data support the notion that OsYchF1 is a descendent of an ancestral YchF protein that was present before the diversification of eukarya into different kingdoms.
It is proposed that all GTPases evolved from a single ancestral GTPase (2). Gene duplication and diversification might have led to new and specific functions (31). Another way to alter the function and regulation of GTPases is by the emergence of new regulatory proteins. Intriguingly, OsGAP1, a regulatory protein of OsYchF1 (11), seems to be more plant-specific, because no close homologues can be identified in other lineages. The appearance of OsGAP1 might have altered/regulated the functions of OsYchF1 so that it could cope with new tasks during the evolution of higher plants.
In most P-loop GTPases, the NKXD consensus sequence is a signature that provides specificity toward GTP. An alteration in this conserved sequence is a possible cause for the secondary loss of GTPase specificity or activity (2). Due to the absence of this consensus in YchF proteins, their specificity toward GTP was questioned. Experimental evidence showed that YchF proteins can also be ATPases (6, 10). This property was also confirmed in OsYchF1 (Fig. 2). The observation that OsGAP1 could activate the GTPase/ATPase activities of OsYchF1 makes OsGAP1 a putative candidate of a GTPase-activating protein. However, the -fold activation by OsGAP1 is relatively low compared with other GTPase-activating proteins (30). Because no other known GTPase-activating proteins that act on YchF proteins are available for comparison, whether OsGAP1 is a bona fide GTPase-activating protein remains unknown at this stage.
OsYchF1 has a TGS domain at the C terminus, a common feature found in Obg GTPase family and guanosine polyphosphate phosphohydrolases/synthetases (e.g. threonyl-tRNA synthetase) that may play a role in nucleotide binding and regulation (7). We demonstrated the binding of the 26 S RNA, but not the 18 S RNA or the 5.8 S RNA, to OsYchF1 (Fig. 3). However, we do not exclude the possibility that OsYchF1 can bind to other RNA species.
The TGS domain alone is sufficient for the binding of OsYchF1 to the 26 S RNA (Fig. 3C). OsGAP1 can compete for the binding site on the TGS domain to prevent OsYchF1 from binding to the 26 S RNA (Fig. 3D). This is the first report to identify an RNA target for YchF proteins and to explain how such a process can be regulated.
It is speculated that the ancestral GTPase for all of the members of GTPases had a generic regulatory role in translation (2). Some translation factors, such as EF-Tu and EF-G, are GTPases that can bind to tRNA (32) and 4.5 S RNA (33), respectively. Furthermore, some small GTPases may even participate in the ribosome assembly (34). The 26 S RNA binding capacity of OsYchF1 shown in this report is consistent with the hypothesis that YchF proteins may be uncharacterized translation factors (6, 10).
Using bimolecular fluorescence complementation techniques, we confirmed the interaction between OsYchF1 and OsGAP1 in vivo (supplemental Fig. S6). We then went on to study the possible regulation of the subcellular localization of OsYchF1 by OsGAP1. This may help to determine the function and activity of regulatory proteins (35). For example, some small GTPases, such as the Rho GTPases, cycle between the plasma membrane and the cytosol (36), and the cycling process is regulated by a guanine nucleotide dissociation inhibitor. Rice phospholipase D was found to be clustered at the sites adjacent to the infecting Xanthomonas cells while distributed evenly along the plasma membrane in normal conditions (37). Using immunogold electron microscopy, we observed a parallel cycling of OsYchF1 and OsGAP1 between the cytosol and the plasma membrane (Figs. 4 and 5). The wounding of plant cells sends a signal that is perceived by cells away from the damaged region (3 cm from the wounding site in this study) and leads to the localization of both OsYchF1 and OsGAP1 to the plasma membrane. Such regulations by wounding may be related to their functions in the plant defense response (see below).
Neither OsYchF1 nor OsGAP1 possesses transmembrane domains. OsGAP1 has a C2 domain (38, 39) that may interact with phospholipids of the plasma membrane. The C2 domains found in some GTPase-activating proteins may also play important roles in the GTPase activation (40). In vitro binding experiments using total phospholipids extracted from plant leaves (Fig. 6) suggested that OsYchF1 may associate with phospholipids of the plasma membrane via its interaction with OsGAP1.
Despite the fact that YchF proteins are ubiquitous in eukarya and bacteria, their functions remain elusive. One report suggested that a bacterial YchF homologue might be involved in iron utilization, and it functioned as a regulator of the Ton system (41). In the protista Trypanosoma cruzi, the inactivation of an YchF homologue could inhibit the growth of procyclic forms of the parasite (10). A recent report on the human YchF homologue suggested a role in the suppression of the antioxidant response via non-transcriptional mechanisms (42). The physiological roles of YchF proteins in plants are completely unknown.
Using the model plant A. thaliana, we show that OsYchF1 and its Arabidopsis homologue AtYchF1 may play a negative role in plant defense response (Fig. 7). When the YchF protein level was low (i.e. in the AtYchF1 knockdown mutant), a higher resistance toward the pathogen P. syringae pv. tomato DC3000 was observed, including reduced disease symptoms and pathogen titers as well as increased expressions of defense marker genes. Similar effects were obtained by overexpressing OsGAP1. However, when YchF proteins were in excess (i.e. in the OsYchF1 overexpressor), we had the opposite results.
Linking the function of OsYchF1 to the plant defense response may shed light on the physiological roles of this group of novel GTPases. The involvement by other families of GTPases in the plant defense response has been reported. In the Ras-like superfamily, overexpression of a Ras-related GTPase in tobacco enhanced the level of salicylic acid and induced PR gene expressions (43). Moreover, the OsRac1 from rice is involved in the resistance toward rice bacterial blight and fungal blast via its regulatory role in the production of reactive oxygen species that trigger the hypersensitive response (44, 45).
We postulate that under normal conditions, both OsGAP1 and OsYchF1 are mainly localized in the cytosol of rice cells. The level of OsGAP1 is low (11), and therefore most OsYchF1 molecules are in their active form, binding to the 26 S RNA. Without pathogen challenges, OsYchF1 may act as a negative regulator to prevent unnecessary provoking of the detrimental defense response. When a triggering signal is perceived (e.g. wounding), the increased level of OsGAP1 (11) will alter the function of OsYchF1 by 1) activating its GTPase/ATPase activities, so that OsYchF1 will be converted into an inactive form; 2) blocking the binding of OsYchF1 to the 26 S RNA; and 3) removing OsYchF1 from the cytosol by assisting its relocalization to the plasma membrane. More experimental data are needed to confirm this hypothetical model.
In short, we provide the first report on the possible functions of plant YchF proteins, which are descendents of an ancient P-Loop GTPase/ATPase. We also demonstrate that a higher plant-specific regulatory protein can moderate the functions of YchF proteins in plants. These results bring a new angle to the studying and understanding of the evolution and regulation of this ubiquitous and ancient group of P-loop GTPases, which will in turn provide an important piece of the jigsaw puzzle that is the complex cellular regulatory network.
Acknowledgments
We thank J.-Y. Chu, J. He, and P. Harper for proofreading the manuscript and Y. Shen, W.-K. Kwok, and S. W. Tong for technical assistance. C. Lo, L. Jiang, and J. He kindly provided Pseudomonas syringae pv. tomato DC3000, rice suspension culture, and bimolecular fluorescence complementation vectors, respectively.
This work was supported by Hong Kong Research Grants Council General Research Fund Grant 467608 (to H.-M. L.), Hong Kong University Grants Committee Area of Excellence Plant and Agricultural Biotechnology Project AoE-B-07/09, and the Shanghai-Hong Kong-Anson Research Foundation Grant (to H.-M. L. and S. S.-M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
- Mant-GTP
- 2′,3′-O-(N-methyl-anthraniloyl) guanosine 5′-triphosphate, trisodium salt
- DIG
- digoxigenin.
REFERENCES
- 1.Koonin E. V., Wolf Y. I., Aravind L. (2000) Adv. Protein Chem. 54, 245–275 [DOI] [PubMed] [Google Scholar]
- 2.Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002) J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 3.Bourne H. R., Sanders D. A., McCormick F. (1990) Nature 348, 125–132 [DOI] [PubMed] [Google Scholar]
- 4.Jones A. M., Assmann S. M. (2004) EMBO Rep. 5, 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittenhuber G. (2001) J. Mol. Microbiol. Biotechnol. 3, 21–35 [PubMed] [Google Scholar]
- 6.Koller-Eichhorn R., Marquardt T., Gail R., Wittinghofer A., Kostrewa D., Kutay U., Kambach C. (2007) J. Biol. Chem. 282, 19928–19937 [DOI] [PubMed] [Google Scholar]
- 7.Wolf Y. I., Aravind L., Grishin N. V., Koonin E. V. (1999) Genome Res. 9, 689–710 [PubMed] [Google Scholar]
- 8.Caldon C. E., Yoong P., March P. E. (2001) Mol. Microbiol. 41, 289–297 [DOI] [PubMed] [Google Scholar]
- 9.Teplyakov A., Obmolova G., Chu S. Y., Toedt J., Eisenstein E., Howard A. J., Gilliland G. L. (2003) J. Bacteriol. 185, 4031–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gradia D. F., Rau K., Umaki A. C., de Souza F. S., Probst C. M., Correa A., Holetz F. B., Avila A. R., Krieger M. A., Goldenberg S., Fragoso S. P. (2009) Int. J. Parasitol. 39, 49–58 [DOI] [PubMed] [Google Scholar]
- 11.Cheung M. Y., Zeng N. Y., Tong S. W., Li W. Y., Xue Y., Zhao K. J., Wang C., Zhang Q., Fu Y., Sun Z., Sun S. S., Lam H. M. (2008) New. Phytol. 179, 530–545 [DOI] [PubMed] [Google Scholar]
- 12.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 13.Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. (2008) Planta. 227, 1343–1349 [DOI] [PubMed] [Google Scholar]
- 14.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 16.Rohn T. T., Nelson L. K., Davis A. R., Quinn M. T. (1999) Free Radic. Biol. Med. 26, 1321–1331 [DOI] [PubMed] [Google Scholar]
- 17.Spangler C., Spangler C. M., Spoerner M., Schäferling M. (2009) Anal. Bioanal. Chem. 394, 989–996 [DOI] [PubMed] [Google Scholar]
- 18.Ma L., Xie B., Hong Z., Verma D. P., Zhang Z. (2008) Plant Physiol. 148, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venables J. P., Ruggiu M., Cooke H. J. (2001) Nucleic Acids Res. 29, 2479–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitchcock C., Nichols B. W. (1971) Plant Lipid Biochemistry: The Biochemistry of Fatty Acids and Acyl Lipids with Particular Reference to Higher Plants and Algae, pp. 279–305, Academic Press, Inc., New York [Google Scholar]
- 21.Stevenson J. M., Perera I. Y., Boss W. F. (1998) J. Biol. Chem. 273, 22761–22767 [DOI] [PubMed] [Google Scholar]
- 22.Jensen R. B., Lykke-Andersen K., Frandsen G. I., Nielsen H. B., Haseloff J., Jespersen H. M., Mundy J., Skriver K. (2000) Plant Mol. Biol. 44, 799–814 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q., Shi A. N., Yang W. C., Wang C. L. (1996) Acta Agric. Sin. 22, 135–141 [Google Scholar]
- 24.Lam S. K., Siu C. L., Hillmer S., Jang S., An G., Robinson D. G., Jiang L. W. (2007) Plant Cell 19, 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou L., Cheung M. Y., Zhang Q., Lei C. L., Zhang S. H., Sun S. S., Lam H. M. (2009) Plant Cell Environ. 32, 1804–1820 [DOI] [PubMed] [Google Scholar]
- 26.Brears T., Liu C., Knight T. J., Coruzzi G. M. (1993) Plant Physiol. 103, 1285–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bechtold N., Pelletier G. (1998) Methods Mol. Biol. 82, 259–266 [DOI] [PubMed] [Google Scholar]
- 28.Katagiri F., Thilmony R., He S. Y. (2002) in The Arabidopsis Book (Last R., Chang C., Jander G., Kliebenstein D., McClung R., Millar H. eds) pp. 28–35, American Society of Plant Biologists, Rockville, MD [Google Scholar]
- 29.Scamardella J. M. (1999) Int. Microbiol. 2, 207–216 [PubMed] [Google Scholar]
- 30.Scheffzek K., Ahmadian M. R. (2005) Cell Mol. Life Sci. 62, 3014–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ober D. (2005) Trends Plant Sci. 10, 444–449 [DOI] [PubMed] [Google Scholar]
- 32.Louie A., Ribeiro N. S., Reid B. R., Jurnak F. (1984) J. Biol. Chem. 259, 5010–5016 [PubMed] [Google Scholar]
- 33.Suzuma S., Hayashi K., Nakamura K., Yamane K. (1999) FEMS Microbiol. Lett. 180, 271–277 [DOI] [PubMed] [Google Scholar]
- 34.Wicker-Planquart C., Foucher A. E., Louwagie M., Britton R. A., Jault J. M. (2008) J. Bacteriol. 190, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byun-McKay S. A., Geeta R. (2007) Trends Ecol. Evol. 22, 338–344 [DOI] [PubMed] [Google Scholar]
- 36.Schmidt A., Hall A. (2002) Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 37.Young S. A., Wang X., Leach J. E. (1996) Plant Cell 8, 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopka J., Pical C., Hetherington A. M., Müller-Röber B. (1998) Plant Mol. Biol. 36, 627–637 [DOI] [PubMed] [Google Scholar]
- 39.Wang T., Pentyala S., Elliott J. T., Dowal L., Gupta E., Rebecchi M. J., Scarlata S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7843–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pena V., Hothorn M., Eberth A., Kaschau N., Parret A., Gremer L., Bonneau F., Ahmadian M. R., Scheffzek K. (2008) EMBO Rep. 9, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danese I., Haine V., Delrue R. M., Tibor A., Lestrate P., Stevaux O., Mertens P., Paquet J. Y., Godfroid J., De Bolle X., Letesson J. J. (2004) Infect. Immun. 72, 5783–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Rubio V., Lieberman M. W., Shi Z. Z. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15356–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Q., Dröge-Laser W., Dixon R. A., Lamb C. (1996) Curr. Opin. Genet. Dev. 6, 624–630 [DOI] [PubMed] [Google Scholar]
- 44.Ono E., Wong H. L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suharsono U., Fujisawa Y., Kawasaki T., Iwasaki Y., Satoh H., Shimamoto K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]