Abstract
Epidemiological studies indicate that intake of statins decrease the risk of developing Alzheimer disease. Cellular and in vivo studies suggested that statins might decrease the generation of the amyloid β-peptide (Aβ) from the β-amyloid precursor protein. Here, we show that statins potently stimulate the degradation of extracellular Aβ by microglia. The statin-dependent clearance of extracellular Aβ is mainly exerted by insulin-degrading enzyme (IDE) that is secreted in a nonconventional pathway in association with exosomes. Stimulated IDE secretion and Aβ degradation were also observed in blood of mice upon peripheral treatment with lovastatin. Importantly, increased IDE secretion upon lovastatin treatment was dependent on protein isoprenylation and up-regulation of exosome secretion by fusion of multivesicular bodies with the plasma membrane. These data demonstrate a novel pathway for the nonconventional secretion of IDE via exosomes. The modulation of this pathway could provide a new strategy to enhance the extracellular clearance of Aβ.
Keywords: Alzheimer Disease, Amyloid, Cholesterol, Drug Action, Exocytosis, Insulin-degrading Enzyme, MVB Exosome Pathways, Statin, Amyloid Degradation, Microglia
Introduction
Alzheimer disease (AD)3 is associated with extracellular deposits of the amyloid β-peptide (Aβ) and intraneuronal aggregates of hyperphosphorylated Tau protein in the brain (1). Evidence suggests that the pathogenesis of AD involves deleterious neurotoxic effects of aggregated Aβ peptides (2), which are derived by sequential proteolytic processing of the β-amyloid precursor protein (APP) by β- and γ-secretases (3). APP can also be cleaved in a nonamyloidogenic pathway that involves initial cleavage by α-secretase within the Aβ domain that precludes the later generation of Aβ peptides (4). Brain Aβ levels are not only determined by the rate of production but also by different clearance mechanisms, including receptor-mediated endocytosis/phagocytosis and subsequent degradation in the endosomal/lysosomal compartment, transcytosis via the blood-brain barrier, as well as proteolytic degradation of extracellular Aβ by cell surface-localized and secreted proteases (5–10).
Several studies indicated a dysregulation of lipid metabolism as an important aspect of AD-associated neurodegeneration. In particular, increased cholesterol levels seem to correlate with increased AD risk. Retrospective studies revealed the beneficial effects of statins (11). However, molecular mechanisms by which statins could offer protection against AD remain to be characterized in detail (12, 13). Most studies with cultured cells and animal models indicate that statin-mediated inhibition of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) could decrease the generation of Aβ by promoting nonamyloidogenic processing of APP. Other studies also showed that extraction of cholesterol from cellular membranes by cyclodextrins differentially affects Aβ generation. Although strong reduction of cholesterol decreased Aβ generation, a moderate extraction rather promoted the secretion of Aβ. These effects were attributed to alterations in the distribution of APP and secretases within membrane microdomains (14–18). In addition to inhibition of cholesterol biosynthesis, statins also impair the generation of isoprenoids, including farnesyl pyrophosphate (FPP) and geranylgeranyl pyrophosphate that play important roles in the post-translational modifications of Rho and Rab proteins thereby regulating their localization and biological function. Isoprenoids have also been shown to affect Aβ generation, by modulating β-secretase as well as γ-secretase cleavage of APP (16, 17, 19).
Notably, apoE, a major genetic factor for sporadic AD, is one of the important lipoproteins involved in cholesterol shuttling between neurons and astrocytes and is involved in remodeling and reorganization of neuronal networks after injury (20). Extracellular Aβ in complex with lipoproteins such as apoE can be internalized via receptor-mediated endocytosis involving the low density lipoprotein (LDL) receptor or LDL receptor-related protein or via receptor-independent pinocytosis, followed by proteolytic degradation (21, 22). Here, we address the potential role of statins in Aβ clearance and demonstrate that statins promote the degradation of extracellular Aβ by stimulating the secretion of IDE in association with exosomes.
EXPERIMENTAL PROCEDURES
Antibodies, cDNA, and Chemicals
Primary antibodies were obtained from the indicated suppliers as follows: anti-IDE (Abcam); anti-Aβ antibodies 4G8 (Signet); 82E1 (IBL); anti-HMGCR (Upstate); anti-β-tubulin (Hybridoma Bank, University of Iowa); anti-flotillin-1 (BD Transduction Laboratories); anti-alix (Santa Cruz Biotechnology); anti-β-actin (Sigma); anti-calnexin (Santa Cruz Biotechnology); and anti-BiP (BD Transduction Laboratories). Polyclonal antibody 2964 against Aβ was described before (25). Itraconazole, lovastatin, farnesyltransferase inhibitor (FTI, B581), geranylgeranyltransferase inhibitor (BML-G225), FPP, and geranylgeranyl pyrophosphate were obtained from Axxora Inc., and zaragozic acid was from Sigma.
Cell Culture, Drug Treatments and Transfection
The respective cell lines were cultured in Dulbecco's modified Eagle's medium with Glutamax (Invitrogen) supplemented with 10% fetal calf serum and 1% PenStrep (Invitrogen). Incubation with chemical compounds was started at a confluency of 70%; control cells were treated with the respective carrier. Cell transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Analysis of Cellular Sterols
Lipids were extracted with chloroform/methanol (2:1, v/v) from cultured cells and dried to constant weight in a SpeedVac (Servant Instruments, Inc., Farmingdale, NY). 5α-Cholestane (Serva Electrophoresis Inc., Heidelberg), epicoprostanol (Sigma), and racemic 24(RS)-[23,23,24,25-2H4]OHchol (Medical Isotopes Inc., Pelham, NH) were added as internal standards. After saponification, extraction, and derivatization, sterols were determined as trimethylsilyl ethers by using gas-liquid chromatography-flame ionization detection to analyze cholesterol and GC-MS to measure concentrations of lanosterol and desmosterol, as described before (23).
Protein Analysis and Cell Fractionation
Proteins in cell supernatants and lysates were analyzed by Western immunoblotting. Aliquots of the cell culture media were either loaded directly or concentrated by precipitation with trichloroacetic acid (TCA). Cell lysates were prepared by lysing the cells in RIPA buffer (150 mm NaCl, 10 mm Tris, pH 8.0, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 5 mm EDTA) followed by centrifugation at 16,000 × g for 10 min. For the purification of cellular membranes, cells were incubated in hypotonic buffer (10 mm Tris/HCl, pH 7.5, 10 mm NaCl, 0.1 mm EGTA, 25 mm β-glycerophosphate, 1 mm DTT, 1× proteinase inhibitor) for 15 min on ice. After homogenization, nuclei and debris were removed by centrifugation at 200 × g for 10 min. Supernatant was then centrifuged at 16,000 × g to obtain a membrane pellet. The pellet was resuspended in STEN Lysis buffer (250 mm Tris/HCl, pH 7.6, 750 mm NaCl, 10 mm EDTA, 1% Igepal, 1% Triton X-100) and then subjected to SDS-PAGE.
Isolation and Characterization of Exosomes
Exosomes of cultured cells were prepared as described earlier (24). Conditioned media collected from control and treated cells were subjected to sequential centrifugation steps (10 min at 3,000 × g, two times for 10 min at 4,000 × g, 30 min at 10,000 × g, and 1 h at 100,000 × g). Supernatants and resuspended pellets were analyzed by Western immunoblotting. Where indicated, the exosome (P100) fraction was further subjected to density centrifugation. A step gradient of sucrose (2.5, 2.25, 2.0, 1.75, 1.5, 1.25, 1.0, 0.75, 0.5, and 0.25 m) was loaded on top of the P100 fraction and ultracentrifuged at 200,000 × g for 3 h. 1 ml of each fraction was collected and precipitated with TCA.
Aβ Degradation Assays
BV-2 cells grown in a 24-well plate to 70% confluency were treated with 2.5 μm itraconazole, 5 μm lovastatin, 10 μm zaragozic acid, and 20 μm FTI, respectively, for 24 h. After changing the medium, cells were treated again with the respective compounds and incubated with 1 μm Aβ (EZ Biolabs). Media were collected at the indicated time points and separated on a 16% Tricine gel (Anamed). Cellular lysates were separated on a 4–12% BisTris gel (Invitrogen). To analyze Aβ degradation in cell-free conditioned media, BV-2 cells were incubated with respective drugs for 24 h. After removal of the media, cells were incubated for an additional 12 h in serum-free DMEM. The supernatant was then cleared by centrifugation and incubated with 1 μm Aβ for the indicated time periods in the absence or presence of 5 mm EDTA (Sigma), 10 μg/ml bacitracin A (Sigma), or 10 μm insulin (Sigma). Aβ levels were quantified by Western immunoblotting and ECL imaging. For degradation of Aβ with recombinant IDE, 80 ng/μl Aβ was incubated with 0.3 ng/μl IDE in Tris/NaCl buffer (50 mm Tris, 1 m NaCl, pH 7.5) for 1 h.
Aβ Immunoprecipitation
Polyclonal anti-Aβ antibody 2964 (25) and protein A-Sepharose beads were added to cell culture conditioned media and incubated at 4 °C for 12–16 h, followed by repetitive washing with PBS. After separation of immunoprecipitates by SDS-PAGE, Aβ was detected by Western immunoblotting.
Treatment of Mice with Lovastatin
12-Week-old female C57BL6 mice (Charles River Laboratories) were intraperitoneally injected with 1 mg of lovastatin or 200 μl of PBS. After 24 h, blood was collected and centrifuged at 800 × g to obtain the serum and cell pellet. The cell pellet was washed with PBS and lysed in STEN Lysis buffer (250 mm Tris/HCl, pH 7.6, 750 mm NaCl, 10 mm EDTA, 1% Igepal, 1% Triton X-100). IDE was detected in serum and cell lysates by Western immunoblotting. For Aβ degradation assay, 2 μl of serum was diluted to 200 μl in DMEM and incubated with 1 μm Aβ40 (EZ-Biolabs) for 8 h at 37 °C. Aβ levels were analyzed by Western immunoblotting.
siRNA-mediated Knockdown of IDE
N9 cells were transfected with 22.5 nm siRNA (target sequence GCCTGTTGTCAGAACTCAA) using HiPerFect transfection reagent (Qiagen) according to the supplier's instructions. Knockdown of IDE was analyzed after 48 h in cell lysates by Western immunoblotting. For Aβ degradation experiments, lovastatin treatment was started 24 h after siRNA transfection followed by 16 h of incubation in serum-free medium that was later used to study IDE-dependent Aβ degradation.
Electron Microscopy of Exosomes
The P100 fraction was fixed in 50 μl of 2% paraformaldehyde. 5 μl of the fixed exosomes were dropped on Formvar carbon-coated grids and left to dry at room temperature. The grid was washed in Sorensen's wash buffer (81% 0.1 m disodium hydrogen orthophosphate, 19% 0.1 m sodium dihydrogen phosphate) and fixed in 1% glutaraldehyde for 5 min. The grid was then washed with water before the exosomes were stained with saturated aqueous uranyl acetate. The samples were embedded in 0.4% uranyl acetate, 1.8% methylcellulose on ice. Excess liquid was removed, and the grid was dried at room temperature, before viewing in a transmission electron microscope (Zeiss-TEM900, Oberkochen, Germany).
Electron Microscopy of Cells
Cells were fixed in 3% glutaraldehyde for 3 h followed by washing with 0.1 m HEPES buffer. Cells were later progressively dehydrated in a graded series of ethanol (30–100%) and embedded in Epon. Ultrathin sections (50–60 nm) were cut using ultracut microtome and placed on copper grids for analysis. The grids stained contrasted with uranyl acetate and lead citrate, followed by examination with Zeiss TEM900 electron microscope.
MTT Test
MTT solution (5.5 mg/ml in phosphate-buffered saline) was added directly to the cells to give a final concentration of 0.5 mg/ml. Cells were incubated at 37 °C and 5% CO2 for 2 h. Subsequently, the medium was discarded, and 100 μl of DMSO was added to each well. After solubilization of the MTT precipitate, the absorbance at 540 nm was determined photometrically.
Neprilysin (NEP) Activity Assay
To test cell surface NEP activity, MEF, BV-2 cells were washed once with PBS and then incubated with 50 μm fluorogenic NEP substrate N-succinyl-Ala-Ala-Phe-7-amido-4-methylcoumarin (Sigma) in PBS with or without the specific NEP inhibitor thiorphan (10 μm; Sigma) for 30 min. The fluorescence at 460 nm (excitation 390 nm) was measured with a fluorescence spectrophotometer (Molecular Devices, Spectramax Gemini). To analyze NEP activity of secreted and cellular NEP, fluorogenic substrate was added to cell-free conditioned medium and cleared cell lysates, respectively, and measured as described above.
Mass Spectrometry Analysis
Snap-frozen samples from Aβ degradation assays were thawed and desalted using with C18 ZipTips (Millipore) and analyzed on a matrix-assisted laser desorption ionization-time of flight mass spectrometer (Bruker Daltonics) with α-cyano-4-hydroxycinnamic acid as the matrix. The instrument was operated in positive ion linear mode with mass range 1–5 kDa.
Statistical Analyses of Western Immunoblots
ECL signals were quantified using an ECL imager (ChemiDoc XRS, Bio-Rad) and the Quantity One software (Bio-Rad). Statistical analysis of three independent experiments (n = 3) was carried out using Student's t test. The following p values were used: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
RESULTS
Inhibition of HMGCR Promotes Degradation of Aβ by Microglia
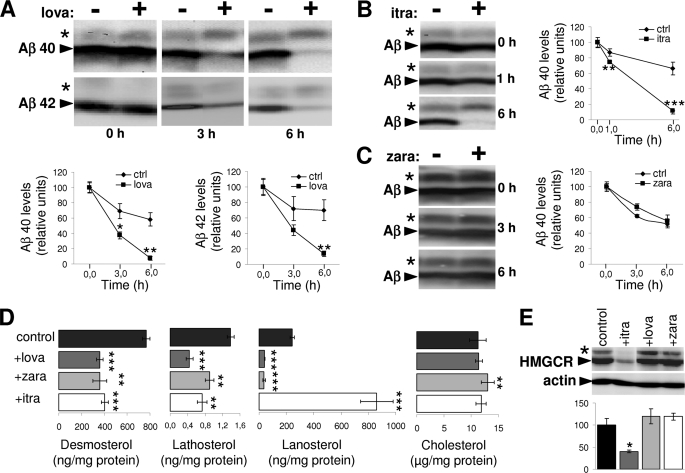
We first analyzed the effect of lovastatin on the degradation of extracellular Aβ by mouse microglial BV-2 cells. Preincubation of BV-2 cells with statin strongly increased the degradation of exogenous Aβ. This effect was observed for both major Aβ variants, Aβ40 and Aβ42 (Fig. 1A). Inhibition of HMGCR by statins impairs cholesterol as well as isoprenoid biosynthesis (supplemental Fig. 1). To assess the potential involvement of isoprenoids in the regulation of Aβ clearance, we also used zaragozic acid and itraconazole that target squalene synthase and CYP51, respectively (23, 26), and therefore would allow isoprenoid synthesis (supplemental Fig. 1). Interestingly, although itraconazole also promoted Aβ clearance similar to lovastatin, zaragozic acid had no significant effect on this process (Fig. 1, B and C). MTT assays revealed that neither lovastatin nor itraconazole and zaragozic acid affected the viability of BV-2 cells (supplemental Fig. 2).
FIGURE 1.
Lovastatin and itraconazole enhance Aβ clearance by microglia. A–C, BV-2 cells were treated with 5 μm lovastatin (lova) (A), 2.5 μm itraconazole (itra) (B), 10 μm zaragozic acid (zara) (C), or solvent alone (control) for 24 h. Aβ40 (A–C) or Aβ42 (A) was added to the cells for the indicated time periods. At the end of incubation, conditioned media were collected, and Aβ was detected by Western immunoblotting. Levels of Aβ at each time point were quantified by ECL imaging. Asterisks indicate Aβ dimers. D, determination of cellular sterol levels. BV-2 cells were treated with 5 μm lovastatin, 10 μm zaragozic acid, 2.5 μm itraconazole, or solvent alone (control) for 24 h. Cellular sterol levels were estimated after extraction as described under “Experimental Procedures.” E, expression analysis of HMGCR in cellular membranes from control and treated BV-2 cells. The asterisk indicates a minor (likely phosphorylated) variant of HMGCR. Actin was detected as loading control. Quantification of HMGCR expression was done with ECL imaging.
A comprehensive analysis of cellular sterols revealed that the inhibitors decreased the levels of cholesterol precursors desmosterol and lathosterol, indicating efficient inhibition of cholesterol biosynthesis by these drugs (Fig. 1D). Lanosterol levels were reduced by lovastatin and zaragozic acid, whereas itraconazole induced a strong accumulation of lanosterol, demonstrating efficient inhibition of CYP51. Cellular cholesterol concentrations were not strongly affected by either drug under the experimental conditions, which is consistent with feedback control mechanisms like decreased secretion or increased lipoprotein-mediated uptake of extracellular cholesterol to ensure cellular cholesterol homeostasis (27). Consistent with the role of lanosterol as a negative regulator of HMGCR (28), we found significantly decreased expression of HMGCR in BV-2 cells upon treatment with itraconazole (Fig. 1E). Taken together, these data strongly indicate that stimulation of Aβ clearance by lovastatin and itraconazole is not caused by altered concentrations of cellular cholesterol but rather might involve the inhibition of HMGCR-dependent isoprenoid biosynthesis.
Statin and Itraconazole Stimulate Secretion of IDE
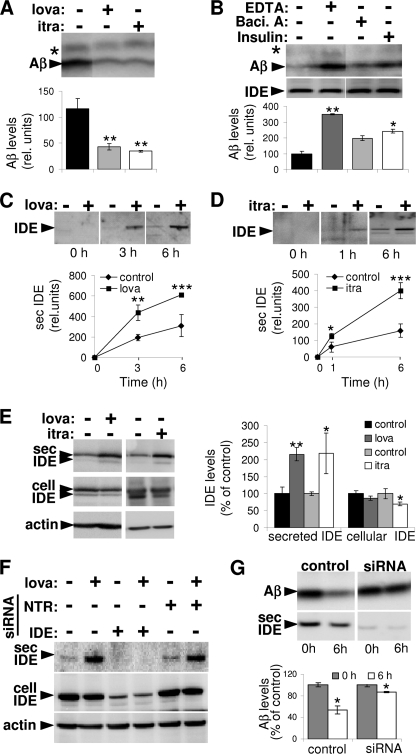
Notably, pretreatment of cells with lovastatin or itraconazole also increased the degradation of exogenous Aβ in cell-free conditioned media (Fig. 2A). These data indicate a prominent role of secreted enzyme activity in increased degradation of extracellular Aβ. This was further supported by experiments where cell-derived Aβ was mixed with cell-free conditioned media of statin-treated or control cells (supplemental Fig. 3). IDE and NEP have been shown to be released and are known to contribute to Aβ degradation in the extracellular region and at the cell surface (29–32). To characterize the protease responsible for increased Aβ degradation, we analyzed the clearance of Aβ in cell-free conditioned media of lovastatin-treated cells in the absence or presence of EDTA, bacitracin A, or insulin. The broad metalloprotease inhibitor EDTA strongly decreased the degradation of Aβ, indicating the involvement of a secreted metalloprotease in the degradation of extracellular Aβ. Additionally, the inhibitor of the IDE, bacitracin A, as well as the cognate IDE and NEP substrate insulin also decreased Aβ degradation (Fig. 2B). In contrast, inhibition of NEP by thiorphan did not affect Aβ degradation mediated by extracellular proteases (supplemental Fig. 4). Lack of detectable NEP activity in control as well as lovastatin-treated BV-2 cells either at the cell surface and within the conditioned media (supplemental Fig. 5A) or the cellular lysates (supplemental Fig. 5B) further ruled out significant contribution of NEP under the experimental conditions. On the other hand, abundant NEP activity was found to be present at the cell surface and within the cellular lysates in MEF cells, which could be efficiently blocked by thiorphan (supplemental Fig. 5, A–C). Because thiorphan was incubated for 6 h in conditioned media obtained from BV-2 cells to assess its impact on Aβ degradation (supplemental Fig. 4), we confirmed the ability of thiorphan to inhibit NEP under similar experimental conditions in MEF cells (supplemental Fig. 5C). In accordance with activity measurement, NEP was not detected in BV-2 cells by Western immunoblotting, whereas MEF cells revealed robust cellular expression of this protein (supplemental Fig. 5D). Similarly, we also failed to detect secreted NEP within the conditioned media of BV-2 as well as MEF cells. However, robust levels of IDE were present in conditioned medium in both cell types (supplemental Fig. 5D). Moreover, levels of secreted IDE appeared to be significantly increased upon lovastatin treatment, suggesting an involvement of IDE in lovastatin-stimulated Aβ degradation.
FIGURE 2.
Increased IDE secretion by lovastatin and itraconazole promotes extracellular degradation of Aβ. A, conditioned media were collected from BV-2 cells after 24 h of incubation in the presence or absence of 5 μm lovastatin (lova) or 2.5 μm itraconazole (itra), and Aβ40 (1 μm) was added to the cell-free conditioned media for 6 h. At the end of incubation, Aβ levels were analyzed by Western immunoblotting and ECL imaging. B, Aβ clearance was analyzed in the conditioned media obtained from lovastatin-treated BV-2 cells in the presence or absence of EDTA (10 μm), bacitracin A (10 μm, Baci. A), or recombinant insulin (10 μm) by ECL imaging. Western immunoblotting showed equal amounts of IDE in all samples. C and D, BV-2 cells were treated with 5 μm lovastatin (C) or 2.5 μm itraconazole (D) for 24 h. IDE in the conditioned media was analyzed at the indicated time points by Western immunoblotting and by ECL imaging. E, BV-2 cells were treated with 5 μm lovastatin (lova) or 2.5 μm itraconazole (itra) for 24 h. Cellular and secreted IDE levels were analyzed by Western immunoblotting and ECL imaging. Cellular actin was detected as loading control. F, N9 cells were transfected with IDE targeting or nontargeting (NTR) control siRNA as indicated. 24 h after transfection, cells were treated with 5 μm lovastatin or DMSO (control) for 24 h. Expression of cellular and secreted IDE was analyzed with Western immunoblotting. Cellular actin was detected as a loading control. G, control and IDE-suppressed (using siRNA) N9 cells were treated with 5 μm lovastatin for 24 h. Conditioned media obtained from these cells were then incubated with 1 μm Aβ40 for 6 h. Aβ levels in the beginning and at the end of incubation period were analyzed by Western immunoblotting and ECL imaging.
To prove a statin-dependent release of IDE, BV-2 cells were treated with lovastatin for 3, 6, 12, and 24 h, and secreted IDE in conditioned medium was analyzed. Significant stimulation of IDE release was observed in statin-treated cells after 24 h (supplemental Fig. 6). In the next experiments, cells were pretreated with lovastatin (Fig. 2C) or itraconazole (Fig. 2D) for 24 h, and secretion of IDE was monitored after additional incubation with the respective drugs. The secretion of IDE was still significantly increased after prolonged cell incubation (Fig. 2, C and D), indicating a sustained effect of statins and itraconazole. In addition, the analysis of cellular IDE revealed that lovastatin and itraconazole treatment slightly decreased levels of cell-associated IDE, further supporting an increased secretion of IDE rather than overall increase in cellular IDE expression (Fig. 2E). We also found increased IDE secretion in BV-2 cells upon treatment with another statin (simvastatin; supplemental Fig. 7). To further establish the contribution of IDE to statin-mediated Aβ degradation, we analyzed the lovastatin-stimulated degradation of Aβ after siRNA-mediated knockdown of IDE expression. Because IDE expression could not be efficiently suppressed in BV-2 (not shown), we tested another mouse microglia cell line N9. First, we confirmed the effect of lovastatin to promote IDE release in these cells (Fig. 2F). siRNA-mediated knockdown of IDE strongly reduced the expression of cellular as well as secreted IDE in control and lovastatin-treated cells, whereas nontarget siRNA transfection did not affect the IDE expression and release (Fig. 2F). After knockdown, IDE levels were strongly decreased in cell lysates (control, 100 ± 6.35%; IDE siRNA, 23.49 ± 6.08%) and in conditioned media (control, 100 ± 6.97%; IDE siRNA, 27.40 ± 5.51%). Importantly, lovastatin-stimulated degradation of Aβ in the conditioned media was strongly decreased upon suppression of IDE expression (Fig. 2G). The residual Aβ-degrading activity correlated with the residual levels of IDE upon siRNA knockdown, again indicating that IDE plays the major role in statin-stimulated Aβ degradation.
We also compared the Aβ proteolysis pattern obtained after its degradation by recombinant IDE (supplemental Fig. 9) and by conditioned media from lovastatin-treated BV-2 cells (supplemental Fig. 10). Very similar and strongly overlapping MALDI-TOF spectra contained the peaks that correspond to individual Aβ fragments generated after its degradation predominantly by IDE (supplemental Table 1) (33) in both cases. This further supports the crucial role of IDE in lovastatin-mediated Aβ degradation; however, data do not rule out the involvement of other proteases. Additionally, we also checked the direct effect of lovastatin on Aβ degradation in cell-free conditioned media. Lovastatin did not affect the Aβ degradation significantly when present only during degradation, indicating its specificity toward IDE release (supplemental Fig. 11).
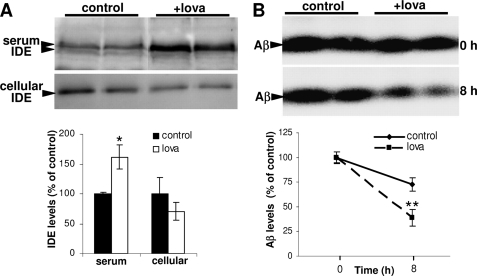
To test the in vivo relevance of these findings, lovastatin was administered to mice, and IDE was detected after 24 h in blood serum and cell pellet. Secreted IDE in the serum was significantly increased as compared with controls, whereas IDE levels in the cell pellets obtained after serum removal were reduced (Fig. 3A). Importantly, the degradation of Aβ was significantly enhanced in the serum of statin-treated mice (Fig. 3B). Thus, inhibition of HMGCR using statins or itraconazole promotes clearance of extracellular Aβ in both cell culture models and the circulation of mice by stimulating the release of the Aβ-degrading enzyme IDE.
FIGURE 3.
Lovastatin stimulates release of IDE in the circulation of mice. A, wild type mice were injected intraperitoneally with 1 mg of lovastatin (+lova, n = 6) dissolved in PBS (5 mg/ml) or 200 μl of PBS (control, n = 6), and sacrificed after 24 h. IDE levels in the sera and cell pellets were analyzed by Western immunoblotting and ECL imaging. B, aliquots of the serum from control and lovastatin-treated mice were incubated with 1 μm Aβ40 for indicated time periods at 37 °C. Aβ was detected by Western immunoblotting and ECL imaging.
Protein Farnesylation Regulates IDE Secretion and Aβ Degradation
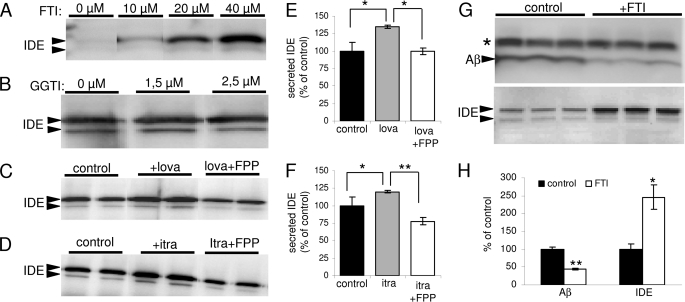
The above data indicate an involvement of the isoprenoid pathways in the degradation of extracellular Aβ by secreted IDE. Isoprenoids, FPP, and geranylgeranyl pyrophosphate are used for the post-translational modification of intracellular proteins, including members of the G protein family, thereby regulating their membrane localization and activity (34). To further investigate the role of protein isoprenylation in IDE release and Aβ degradation, we used selective inhibitors of enzymes that are involved in transfer of isoprenoids to target proteins. Notably, treatment with FTI efficiently stimulated the secretion of IDE, whereas the geranylgeranyltransferase inhibitor had little if any effect (Fig. 4, A and B). Furthermore, FPP attenuated the effect of statin (Fig. 4, C and E) and itraconazole (Fig. 4,D--F) on IDE secretion. This result suggested that indeed the impaired biosynthesis of FPP is responsible for increased IDE release in statin-treated cells. Next, we also analyzed the effect of farnesyltransferase inhibition on Aβ degradation. FTI treatment promoted the degradation of Aβ with the concomitant increase of IDE secretion in the conditioned media of microglia (Fig. 4, G and H). Cell viability was found not to be affected by FTI treatment at the concentrations used (supplemental Fig. 2).
FIGURE 4.
Regulation of IDE secretion by protein farnesylation. A and B, BV-2 cells were treated with FTI (A) or geranylgeranyltransferase inhibitor (GGTI; B) for 24 h with the indicated concentrations, and secreted IDE in conditioned media was detected by Western immunoblotting. C–F, BV-2 cells were treated with 5 μm lovastatin (C and E) and 2.5 μm itraconazole (D and F) either alone or together with 20 μg/ml FPP for 24 h as indicated. Secreted IDE was analyzed as above. G, BV-2 cells were treated with 20 μm FTI for 24 h, and degradation of synthetic Aβ40 was analyzed in conditioned media (upper panel). Secreted IDE in conditioned media was also detected by Western immunoblotting (lower panel). H, Aβ levels at the end of incubation and secreted IDE expression were quantified with ECL imaging.
Statins Stimulate the Release of IDE in Association with Exosomes
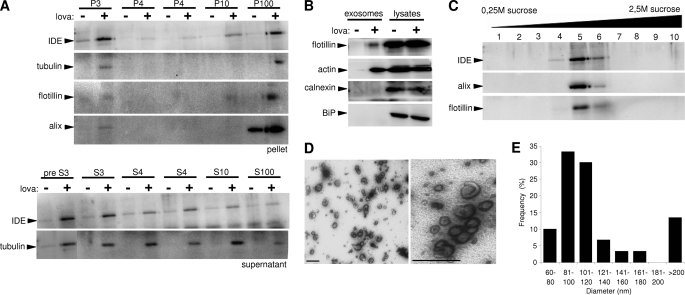
The mechanism of IDE secretion is enigmatic. IDE lacks a signal sequence that targets proteins to the classical secretory pathway (8, 35). Therefore, IDE together with fibroblast growth factors, interleukins, and galectins belongs to a group of proteins that follow nonconventional pathways of protein secretion (36). Different mechanisms, including direct translocation across plasma membrane as a molten globule (37), secretion in association with membrane-derived vesicles (38), as well as caspase-1-dependent inflammasome-mediated secretions (39), have been proposed. We first tested whether secreted IDE is soluble or associated with membrane vesicles. Differential centrifugation of conditioned media revealed the presence of IDE in the final pellet (P100). Notably, lovastatin treatment strongly increased the amount of secreted IDE in the P100 fraction (Fig. 5A). A similar sedimentation behavior was observed for secreted forms of tubulin, flotillin-1, and alix, which are known to be associated with exosomes (24). In contrast, the endoplasmic reticulum resident proteins calnexin and BiP were not found in this fraction (Fig. 5B). To further characterize the exosome association of IDE, isolated exosomes were separated by sucrose gradient density centrifugation as described previously (24). IDE co-fractioned with flotillin-1 and alix, two proteins known to be associated with exosomes (Fig. 5C). Moreover, electron microscopy revealed high enrichment of membrane vesicles with diameters of 80–120 nm, which is typical for exosomes (Fig. 5E). Visualization of characteristic saucer-shaped vesicles confirmed the presence of exosomes in this fraction (Fig. 5D).
FIGURE 5.
Secreted IDE is associated with exosomes. A, conditioned cell culture media obtained from control and lovastatin (5 μm)-treated BV-2 cells was subjected to subsequent centrifugation steps. The indicated proteins were detected in the respective pellets (P, upper panel) and supernatants (S, lower panel). B, characterization of proteins in exosome preparations from conditioned cell culture media of control and lovastatin-treated (5 μm, 24 h) BV-2 cells. Total cell lysates served as controls. C, co-fractionation of exosomal proteins and IDE. The P100 fraction of conditioned media from lovastatin-treated BV-2 cells (see A) was separated in a sucrose gradient by ultracentrifugation. Resulting fractions were analyzed for IDE, alix, and flotillin-1. D, electron micrographs of P100 fractions. Purified exosomes from conditioned media of statin-treated BV-2 cells were resuspended in 2% paraformaldehyde and negatively stained with uranyl acetate (scale bars, 200 nm). E, quantitative analysis of vesicle sizes in the P100 fraction. The distribution of mean diameter of vesicles was analyzed by scoring 10 randomly selected electron micrographs.
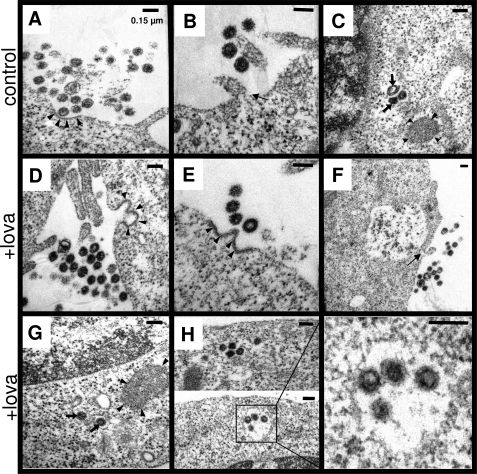
Next, we also analyzed exosome release by electron microscopy. We observed secreted exosomes in the extracellular space of control (Fig. 6, A and B) and statin-treated (Fig. 6, D–F) cells in close proximity to the plasma membrane. Importantly, the numbers of such cells engaged in exosome release were markedly increased upon statin treatment, supporting an increase in the total number of exosomes released in statin-treated cells. Exosomes can derive from intraluminal vesicles (ILVs) after fusion of multivesicular bodies (MVBs) with the plasma membrane (40). MVBs are morphologically distinctive late endosomes with a mean diameter of 200–500 nm and possess electron lucent matrix, whereas ILVs have diameters of 60–80 nm (41). Both MVBs as well as ILVs have a double membrane layer. The assessment of intracellular lysosomal organelles revealed the presence of heterolysosomes with diameter ranging from 400 to 2,000 nm and smaller double membrane vesicles with a diameter of 40–150 nm in control (Fig. 6 C) and statin-treated (Fig. 6G) cells. Notably, MVBs were selectively observed in statin-treated cells, which might be due to either increased generation or decreased fusion of MVBs to lysosomes (Fig. 6H). Similar changes in the MVB-exosome pathway were also observed in HeLa cells upon statin treatment. Together, these findings establish an involvement of statins in the up-regulation of the MVB-exosome pathway, which results in increased degradation of extracellular Aβ by secreted IDE.
FIGURE 6.
Ultrastructural analysis of exosome release and MVBs. A, B, and D–F, exosome release in control (A and B) and statin-treated (D–F) BV-2 cells was analyzed by electron microscopy. Exosomes from a recent MVB fusion with plasma membrane is seen in control and statin-treated cells. Probable MVB fusion area at plasma membrane can be distinguished by double membrane layer and electron dense region (indicated by arrowheads). Individual cells also show exosome release without complete MVB fusion with plasma membrane, with small opening (indicated by arrow) of MVB into the extracellular space. C and G, presence of heterolysosomes (indicated by arrowheads) and smaller vesicles likely representing autophagosomes or lysosomes (indicated by arrows) is observed in control and statin-treated BV-2 cells. H, intracellular MVBs are observed with electron microscopy selectively in statin-treated BV-2 cells (indicated by box). Presence of ILVs is indicated by arrowheads. Scale bar, 150 nm.
DISCUSSION
Epidemiological studies suggest that elevated levels of cholesterol increase the risk for AD. In line with this, certain studies indicated the beneficial effects of statins against AD (11, 42). On the contrary, some studies did not reveal any beneficial effects of statins (43). Such variable conclusions might stem from the fact that most of these studies were not conducted in the form of randomized clinical trials and usage of different statins with the differential ability to cross blood-brain barriers. Thus, the ability of statins to protect against AD is not fully elucidated, and further prospective randomized clinical trials might be necessary to establish or rule out the beneficial role of statins in protection against AD (44).
In addition to reduction in blood cholesterol levels, statins have also been shown to affect several important physiological and systemic processes such as endothelial functioning, inflammation, atherosclerosis, oxidative stress, blood coagulation, cellular proliferation, and differentiation in a favorable manner (45, 46). Many of these pleiotropic effects of statin are attributed to inhibition of isoprenoid biosynthesis and subsequent impairment of protein isoprenylation. Although the beneficial effects of statins in AD are still debatable, there are plenty of data to support the alteration of APP processing, amyloid plaque deposition, and neurofibrillary tangle burden by statin treatment in cell culture and animal models. Initial studies demonstrated that lowering of membrane cholesterol by statins results in decreased generation of Aβ by altering the subcellular transport, localization, and dimerization of either APP, secretases, or both (47–50). Recent studies also support a predominant role of protein isoprenylation in the statin-mediated effects on APP processing and plaque pathology (16, 17, 19). However, there are also data indicating the lack of any positive effects of statins on Aβ levels, amyloid pathology, and Tau metabolism. These studies suggest that the beneficial effects of statins might be due to its favorable pleiotropic effects on the immune and vascular system as well as reduced oxidative stress (51, 52).
The present data provide the first evidence for a novel mechanism by which statins could affect the metabolism of Aβ. Statins strongly enhanced the degradation of extracellular Aβ by stimulation of IDE release from microglial cells. Moreover, statin treatment of mice enhanced the levels of IDE in the blood serum, while IDE in the cell pellet was decreased, also indicating a selective stimulation of IDE secretion in peripheral cells. It will therefore be interesting to further dissect the relative contribution of increased Aβ degradation and decreased Aβ production to amyloid pathology in vivo upon statin treatment.
The role of microglia in AD pathology remains controversial (53, 54). Microglia depletion did not significantly affect amyloid deposition into plaques (55). However, levels of soluble Aβ were found to be increased in the brains of microglia-depleted mice, indicating a role of this cell type in the metabolism of extracellular Aβ. Furthermore, AD-associated neuron loss was also shown to be dependent on microglia (56). Because we showed that statins promoted the secretion of IDE in other cell types (supplemental Fig. 8), additional brain cells beside microglia or peripheral cells could contribute to the degradation of soluble extracellular Aβ in vivo. NEP, another Aβ-degrading enzyme, is an integral membrane protein, and only very little amounts are found in extracellular fluids (57). However, our results indicate that NEP activity was not increased by statins. Rather our siRNA data convincingly prove major contributions of IDE to increased Aβ degradation by statins.
IDE was shown to degrade extracellular monomeric Aβ in vitro and in vivo (8, 31), but the mechanism of its secretion was unknown. IDE lacks a classical signal for secretion and does not follow the conventional secretory pathway (35). Our data demonstrate that inhibition of HMGCR strongly increased the secretion of IDE. Interestingly, we found secreted IDE in association with exosomes, which is consistent with a recent report also demonstrating IDE release with exosomes from neuroblastoma cells (58). Although the topology of IDE in the secreted exosomes remains unclear, insulin-degrading activity has been shown in purified exosomes (58). Importantly, we also demonstrate that the degradation of cell-derived Aβ is increased upon stimulation of exosome-associated IDE release by statin treatment. Thus, IDE could access cell-derived Aβ. However, further studies are needed to analyze the sorting of IDE that is generated in the cytosol to the surface of exosomes. Our data also do not rule out the possibility that Aβ itself ruptures the exosome membrane to release luminal IDE. Indeed, Aβ has been shown to interact with membrane lipids and interfere with membrane integrity by forming pores (59, 60).
Exosomes could originate from endosome-derived MVBs. An association of IDE with endosomes and detergent-resistant membrane microdomains, which are enriched in endosomal compartments, has been reported recently (61). MVBs are intrinsic components of the endosomal system and formed by the inward budding of endosomal membranes (38, 40). The invagination of endosomal membranes is mediated by endosomal sorting complexes required for transport (ESCRT) that could deform endosomal limiting membranes (62). Ceramide, which is generated by sphingomyelinase-dependent degradation of sphingomyelin, could also trigger such invagination and exosome formations (24). It will therefore be interesting to analyze whether statins modulate ESCRT complexes and/or sphingomyelinase. Because exosomes also could contain APP together with β-site APP-cleaving enzyme and γ-secretase components, there might be generation of Aβ within these vesicles (63, 64). Together with our findings, these data indicate that exosomes could contribute to generation as well as degradation of Aβ.
The stimulation of IDE secretion in association with exosomes by statins could be due to increased sorting of IDE to MVBs or more general up-regulation of the MVB-exosome pathway. The enhanced secretion of characteristic exosomal proteins together with increased exosome release events observed by electron microscopy in statin-treated cells strongly support a general up-regulation of the exosome secretion upon inhibition of protein isoprenylation. Although MVBs are rare cell organelles, we demonstrate increased number of MVBs in BV-2 cells upon statin treatment, also suggesting an up-regulation of MVB biogenesis. The persistent increase in IDE secretion up to 36 h after statin treatment also supports up-regulation of MVB generation as well as subsequent fusion with plasma membrane. However, the possibility of decreased fusion of MVBs with lysosomes leading to increased stability of MVBs in the presence of statin cannot be ruled out.
Isoprenylation is of particular importance in the regulation of small GTPases, including Rho and Rab proteins that are known to regulate protein and vesicle trafficking (65, 66). Indeed, Rab11, Rab27a, and Rab27b are also involved in the regulation of docking and fusion of MVBs and exosome secretion (67), and a recent study demonstrated the involvement of Rab11 in the sorting of IDE to exosomes (58). Thus, changes in the isoprenylation of Rab and Rho proteins might underlie the increased secretion of exosome-associated IDE upon cell treatment with statin or itraconazole.
Together, the present data indicate that protein prenylation and the MVB-exosome pathway could be targeted to stimulate degradation of extracellular Aβ. We further demonstrate a fundamental role of protein prenylation in nonconventional protein secretion via exosomes. Importantly, these findings might have strong relevance to therapeutic application of statins as they reveal a general molecular mechanism that might contribute to the pleiotropic effects of statins.
Acknowledgments
We thank Dr. Sascha Weggen for advice with the transfection of N9 cells. The E7 antibody against β-tubulin developed by Dr. Klymkowsky was obtained from the Developmental Studies Hybridoma Bank developed under auspices of the NICHD, National Institutes of Health, and maintained by the Department of Biology, University of Iowa, Iowa City, IA 52242.
This work was supported in part by the Deutsche Forschungsgemeinschaft, Grants SFB645 and KFO177, the Federal Ministry for Education and Research (BMBF), Grant 01GI0708, and the BONFOR program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–11 and Table 1.
- AD
- Alzheimer disease
- APP
- β-amyloid precursor protein
- Aβ
- amyloid-β-peptide
- IDE
- insulin-degrading enzyme
- MVB
- multivesicular body
- HMGCR
- hydroxy-3-methylglutaryl-coenzyme-A reductase
- FPP
- farnesyl pyrophosphate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- FTI
- farnesyltransferase inhibitor
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NEP
- neprilysin
- ILV
- intraluminal vesicle
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1.Masters C. L., Beyreuther K. (2006) Brain 129, 2823–2839 [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J. (2008) Behav. Brain Res. 192, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haass C., De Strooper B. (1999) Science 286, 916–919 [DOI] [PubMed] [Google Scholar]
- 4.Kojro E., Fahrenholz F. (2005) Subcell. Biochem. 38, 105–127 [DOI] [PubMed] [Google Scholar]
- 5.Iwata N., Tsubuki S., Takaki Y., Shirotani K., Lu B., Gerard N. P., Gerard C., Hama E., Lee H. J., Saido T. C. (2001) Science 292, 1550–1552 [DOI] [PubMed] [Google Scholar]
- 6.Iwata N., Higuchi M., Saido T. C. (2005) Pharmacol. Ther. 108, 129–148 [DOI] [PubMed] [Google Scholar]
- 7.Bateman R. J., Munsell L. Y., Morris J. C., Swarm R., Yarasheski K. E., Holtzman D. M. (2006) Nat. Med. 12, 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selkoe D. J. (2001) Neuron 32, 177–180 [DOI] [PubMed] [Google Scholar]
- 9.Bell R. D., Sagare A. P., Friedman A. E., Bedi G. S., Holtzman D. M., Deane R., Zlokovic B. V. (2007) J. Cereb. Blood Flow Metab. 27, 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deane R., Sagare A., Hamm K., Parisi M., Lane S., Finn M. B., Holtzman D. M., Zlokovic B. V. (2008) J. Clin. Invest. 118, 4002–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haag M. D., Hofman A., Koudstaal P. J., Stricker B. H., Breteler M. M. (2009) J. Neurol. Neurosurg. Psychiatry 80, 13–17 [DOI] [PubMed] [Google Scholar]
- 12.Wolozin B., Manger J., Bryant R., Cordy J., Green R. C., McKee A. (2006) Acta Neurol. Scand. Suppl. 185, 63–70 [DOI] [PubMed] [Google Scholar]
- 13.Whitfield J. F. (2007) Expert Opin. Ther. Targets 11, 1257–1260 [DOI] [PubMed] [Google Scholar]
- 14.Taylor D. R., Hooper N. M. (2007) Semin. Cell Dev. Biol. 18, 638–648 [DOI] [PubMed] [Google Scholar]
- 15.Ledesma M. D., Dotti C. G. (2006) FEBS Lett. 580, 5525–5532 [DOI] [PubMed] [Google Scholar]
- 16.Cole S. L., Vassar R. (2006) Neurobiol. Dis. 22, 209–222 [DOI] [PubMed] [Google Scholar]
- 17.Ostrowski S. M., Wilkinson B. L., Golde T. E., Landreth G. (2007) J. Biol. Chem. 282, 26832–26844 [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y., Suram A., Venugopal C., Prakasam A., Lin S., Su Y., Li B., Paul S. M., Sambamurti K. (2008) FASEB J. 22, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons R. B., Price G. C., Farrant J. K., Subramaniam D., Adeagbo-Sheikh J., Austen B. M. (2006) Biochem. J. 399, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bales K. R., Dodart J. C., DeMattos R. B., Holtzman D. M., Paul S. M. (2002) Mol. Interv. 2, 363–375 [DOI] [PubMed] [Google Scholar]
- 21.Jiang Q., Lee C. Y., Mandrekar S., Wilkinson B., Cramer P., Zelcer N., Mann K., Lamb B., Willson T. M., Collins J. L., Richardson J. C., Smith J. D., Comery T. A., Riddell D., Holtzman D. M., Tontonoz P., Landreth G. E. (2008) Neuron 58, 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brendza R. P., Bales K. R., Paul S. M., Holtzman D. M. (2002) Mol. Psychiatry 7, 132–135 [DOI] [PubMed] [Google Scholar]
- 23.Tamboli I. Y., Prager K., Thal D. R., Thelen K. M., Dewachter I., Pietrzik C. U., St George-Hyslop P., Sisodia S. S., De Strooper B., Heneka M. T., Filippov M. A., Müller U., van Leuven F., Lütjohann D., Walter J. (2008) J. Neurosci. 28, 12097–12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 25.Tamboli I. Y., Prager K., Barth E., Heneka M., Sandhoff K., Walter J. (2005) J. Biol. Chem. 280, 28110–28117 [DOI] [PubMed] [Google Scholar]
- 26.Bergstrom J. D., Kurtz M. M., Rew D. J., Amend A. M., Karkas J. D., Bostedor R. G., Bansal V. S., Dufresne C., VanMiddlesworth F. L., Hensens O. D., et al. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown M. S., Goldstein J. L. (1997) Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 28.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 29.Saito T., Iwata N., Tsubuki S., Takaki Y., Takano J., Huang S. M., Suemoto T., Higuchi M., Saido T. C. (2005) Nat. Med. 11, 434–439 [DOI] [PubMed] [Google Scholar]
- 30.Iwata N., Mizukami H., Shirotani K., Takaki Y., Muramatsu S., Lu B., Gerard N. P., Gerard C., Ozawa K., Saido T. C. (2004) J. Neurosci. 24, 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu W. Q., Walsh D. M., Ye Z., Vekrellis K., Zhang J., Podlisny M. B., Rosner M. R., Safavi A., Hersh L. B., Selkoe D. J. (1998) J. Biol. Chem. 273, 32730–32738 [DOI] [PubMed] [Google Scholar]
- 32.Hama E., Shirotani K., Masumoto H., Sekine-Aizawa Y., Aizawa H., Saido T. C. (2001) J. Biochem. 130, 721–726 [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee A., Song E., Kihiko-Ehmann M., Goodman J. P., Jr., Pyrek J. S., Estus S., Hersh L. B. (2000) J. Neurosci. 20, 8745–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McTaggart S. J. (2006) Cell. Mol. Life Sci. 63, 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J., Li L., Leissring M. A. (2009) Mol. Neurodegener. 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nickel W., Seedorf M. (2008) Annu. Rev. Cell. Dev. Biol. 24, 287–308 [DOI] [PubMed] [Google Scholar]
- 37.Cleves A. E., Kelly R. B. (1996) Curr. Biol. 6, 276–278 [DOI] [PubMed] [Google Scholar]
- 38.Stoorvogel W., Kleijmeer M. J., Geuze H. J., Raposo G. (2002) Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 39.Keller M., Rüegg A., Werner S., Beer H. D. (2008) Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 40.Février B., Raposo G. (2004) Curr. Opin. Cell Biol. 16, 415–421 [DOI] [PubMed] [Google Scholar]
- 41.Fader C. M., Colombo M. I. (2009) Cell Death Differ. 16, 70–78 [DOI] [PubMed] [Google Scholar]
- 42.Wolozin B., Kellman W., Ruosseau P., Celesia G. G., Siegel G. (2000) Arch. Neurol. 57, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 43.McGuinness B., Passmore P. (2010) J. Alzheimers. Dis. 20, 925–933 [DOI] [PubMed] [Google Scholar]
- 44.Kandiah N., Feldman H. H. (2009) J. Neurol. Sci. 283, 230–234 [DOI] [PubMed] [Google Scholar]
- 45.Miida T., Hirayama S., Nakamura Y. (2004) J. Atheroscler. Thromb. 11, 253–264 [DOI] [PubMed] [Google Scholar]
- 46.Takemoto M., Liao J. K. (2001) Arterioscler. Thromb. Vasc. Biol. 21, 1712–1719 [DOI] [PubMed] [Google Scholar]
- 47.Boimel M., Grigoriadis N., Lourbopoulos A., Touloumi O., Rosenmann D., Abramsky O., Rosenmann H. (2009) J. Neuropathol. Exp. Neurol. 68, 314–325 [DOI] [PubMed] [Google Scholar]
- 48.Simons K., Ehehalt R. (2002) J. Clin. Invest. 110, 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abad-Rodriguez J., Ledesma M. D., Craessaerts K., Perga S., Medina M., Delacourte A., Dingwall C., De Strooper B., Dotti C. G. (2004) J. Cell Biol. 167, 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parsons R. B., Farrant J. K., Price G. C., Subramaniam D., Austen B. M. (2007) Biochem. Soc. Trans. 35, 577–582 [DOI] [PubMed] [Google Scholar]
- 51.Höglund K., Blennow K. (2007) CNS Drugs 21, 449–462 [DOI] [PubMed] [Google Scholar]
- 52.van der Most P. J., Dolga A. M., Nijholt I. M., Luiten P. G., Eisel U. L. (2009) Prog. Neurobiol. 88, 64–75 [DOI] [PubMed] [Google Scholar]
- 53.Farfara D., Lifshitz V., Frenkel D. (2008) J. Cell. Mol. Med. 12, 762–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boche D., Nicoll J. A. (2008) Brain Pathol. 18, 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grathwohl S. A., Kälin R. E., Bolmont T., Prokop S., Winkelmann G., Kaeser S. A., Odenthal J., Radde R., Eldh T., Gandy S., Aguzzi A., Staufenbiel M., Mathews P. M., Wolburg H., Heppner F. L., Jucker M. (2009) Nat. Neurosci. 12, 1361–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuhrmann M., Bittner T., Jung C. K., Burgold S., Page R. M., Mitteregger G., Haass C., LaFerla F. M., Kretzschmar H., Herms J. (2010) Nat. Neurosci. 13, 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwata N., Higuchi M., Saido T. C. (2005) Pharmacol. Ther. 108, 129–148 [DOI] [PubMed] [Google Scholar]
- 58.Bulloj A., Leal M. C., Xu H., Castaño E. M., Morelli L. (2010) J. Alzheimers Dis. 19, 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eckert G. P., Wood W. G., Müller W. E. (2005) Subcell. Biochem. 38, 319–337 [DOI] [PubMed] [Google Scholar]
- 60.Jang H., Zheng J., Lal R., Nussinov R. (2008) Trends Biochem. Sci. 33, 91–100 [DOI] [PubMed] [Google Scholar]
- 61.Bulloj A., Leal M. C., Surace E. I., Zhang X., Xu H., Ledesma M. D., Castaño E. M., Morelli L. (2008) Mol. Neurodegener. 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lakkaraju A., Rodriguez-Boulan E. (2008) Trends Cell. Biol. 18, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharples R. A., Vella L. J., Nisbet R. M., Naylor R., Perez K., Barnham K. J., Masters C. L., Hill A. F. (2008) FASEB J. 22, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 64.Rajendran L., Honsho M., Zahn T. R., Keller P., Geiger K. D., Verkade P., Simons K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11172–11177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stenmark H. (2009) Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 66.Fukuda M. (2008) Cell. Mol. Life Sci. 65, 2801–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fader C. M., Colombo M. I. (2006) Autophagy 2, 122–125 [DOI] [PubMed] [Google Scholar]