Abstract
Tumor necrosis factor-α (TNF) enhances osteoclast formation and activity leading to bone loss in various pathological conditions, but its precise role in osteoclastogenesis remains controversial. Although several groups showed that TNF can promote osteoclastogenesis independently of the receptor activator of NF-κB (RANK) ligand (RANKL), others demonstrated that TNF-mediated osteoclastogenesis needs permissive levels of RANKL. Here, we independently reveal that although TNF cannot stimulate osteoclastogenesis on bone slices, it can induce the formation of functional osteoclasts on bone slices in the presence of permissive levels of RANKL or from bone marrow macrophages (BMMs) pretreated by RANKL. TNF can still promote the formation of functional osteoclasts 2 days after transient RANKL pretreatment. These data have confirmed that TNF-mediated osteoclastogenesis requires priming of BMMs by RANKL. Moreover, we investigated the molecular mechanism underlying the dependence of TNF-mediated osteoclastogenesis on RANKL. RANK, the receptor for RANKL, contains an IVVY535–538 motif that has been shown to play a vital role in osteoclastogenesis by committing BMMs to the osteoclast lineage. We show that TNF-induced osteoclastogenesis depends on RANKL to commit BMMs to the osteoclast lineage and RANKL regulates the lineage commitment through the IVVY motif. Mechanistically, the IVVY motif controls the lineage commitment by reprogramming osteoclast genes into an inducible state in which they can be activated by TNF. Our findings not only provide important mechanistic insights into the action of RANKL in TNF-mediated osteoclastogenesis but also establish that the IVVY motif may serve as an attractive therapeutic target for bone loss in various bone disorders.
Keywords: Bone, Gene Expression, Osteoclast Acidification, TRAF, Tumor Necrosis Factor (TNF), IVVY Motif, RANKL, Osteoporosis
Introduction
TNF, a proinflammatory cytokine produced by many cells including fibroblasts, endothelial cells, and macrophages, plays a critical role in immune and inflammatory responses (1). Consequently, aberration in TNF expression and function has been associated with many autoimmune disorders and inflammatory complications (2). Moreover, this potent proinflammatory cytokine has also been shown to be a pro-osteoclastogenic factor and thus promotes bone loss in postmenopausal osteoporosis (3) and inflammatory conditions such as rheumatoid arthritis (4) and periodontitis (5) by stimulating osteoclast formation and function.
Osteoclasts, the sole bone-resorbing cells in the body, are multinucleated giant cells derived from mononuclear cells of the monocyte-macrophage lineage upon stimulation by the macrophage/monocyte colony-stimulating factor (M-CSF)2 and RANKL (6). RANKL (also known as OPGL, ODF, and TRANCE), identified as a member of the TNF superfamily (7, 8), stimulates osteoclast formation and function by activating its receptor RANK, a member of the TNF receptor (TNFR) superfamily (8). RANKL also has a soluble decoy receptor, osteoprotegerin (OPG), which inhibits RANKL functions by competing with RANK for binding RANKL (9). Members of the TNFR superfamily primarily employ TNF receptor-associated factors (TRAFs) to transmit downstream signaling (10). Indeed, RANK contains three functional TRAF-binding sites in its cytoplasmic tail (PFQEP369–373, PVQEET559–564, and PVEQG604–609) (11, 12) that activate six major signaling pathways (NF-κB, JNK, ERK, p38, NFATc1, and Akt) to regulate osteoclast formation, function, and/or survival (6, 13). Moreover, a TRAF-independent RANK cytoplasmic motif (IVVY535–538) has recently been identified and shown to play a vital role in osteoclastogenesis by committing BMMs to the osteoclast lineage in vitro (14) and stimulating the osteoclast formation and function in vivo (15). Consistently, truncating mutations causing loss of a region containing the IVVY motif has been reported to cause osteopetrosis in humans (16).
TNF was shown to regulate osteoclastogenesis indirectly by enhancing the expression of RANKL in stromal and osteoblastic cells (17, 18). However, the discovery of RANK as a member of the TNFR superfamily (8) and the subsequent observations that RANK also utilizes TRAFs to activate intracellular signaling pathways (19–21) have led to a hypothesis that TNF can directly target osteoclast precursors to stimulate osteoclastogenesis in vitro independently of the RANKL/RANK system (22, 23). Indeed, several studies showed that TNF can stimulate osteoclastogenesis in vitro independently of the RANKL/RANK system (22, 23). Nonetheless, TNF fails to induce hypercalcemia in vivo when administrated to RANK−/− mice, and rare appearance of TRAF-positive cells was seen only near the site of administration (24). Furthermore, OPG prevents inflammation-induced bone loss in animal model of arthritis (25). These in vivo studies have raised concerns about the reported ability of TNF to mediate osteoclastogenesis in vitro independently of RANKL. Consistent with the in vivo studies, other groups demonstrated that TNF cannot induce osteoclastogenesis in vitro unless assisted by permissive dosages of RANKL (26, 27). However, the molecular mechanism by which TNF-mediated osteoclastogenesis requires the permissive levels of RANKL has not yet been fully investigated. In this study, we have sought to address further the regulatory role of TNF in osteoclastogenesis and investigated the molecular basis of the requirement of TNF-mediated osteoclastogenesis for permissive levels of RANKL.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Chemicals were purchased from Sigma unless indicated otherwise. All synthetic oligonucleotides were from Sigma-Genosys (Woodlands, TX). Recombinant GST-RANKL was purified as described previously (28). Mouse M-CSF was prepared from a M-CSF-producing cell line, CMG14-12, which was constructed and kindly provided by Dr. Sunao Takeshita (29). Recombinant mouse TNF (410-TRNC-050) was obtained from R&D Systems. Anti-human Fas-activating antibody (Fas-AB) was purchased from Millipore. Anti-human Fas antibody conjugated with phycoerythrin (sc-21730PE) for flow cytometry was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 488 phalloidin (A12379) and Hoechst 33258 (H1398) were from Invitrogen.
In Vitro Osteoclastogenesis Assays in Tissue Culture Dish
BMMs were isolated from long bones of 4–6-week-old C3H mice from Harlan Industries (Indianapolis, IN) and were maintained in α-minimal essential medium (α-MEM) containing 10% heat-inactivated FBS in the presence of M-CSF (220 ng/ml) as described previously (30). Mice were maintained, and the experiments involving mice were performed in accordance with the regulations of the University of Alabama at Birmingham (UAB) institutional animal care and use committee. To generate osteoclasts from BMMs, 5 × 104 cells/well were plated in 24-well tissue culture plates and cultured in the presence of 44 ng/ml M-CSF and different concentrations of purified GST-RANKL as indicated in individual experiments. The osteoclastogenesis cultures were stained for tartrate resistant acid phosphatase (TRAP) activity with a Leukocyte Acid Phosphatase kit (387-A) from Sigma. All assays were performed in triplicate and repeated at least two times. A representative view from each condition is shown.
Immunofluorescence Analysis of in Vitro Osteoclastogenesis on Bone Slices
BMMs (5 × 104 cells/well) were seeded on bovine cortical bone slices plated in 24-well tissue culture plates and then cultured under conditions indicated in individual experiments for 4 days to stimulate osteoclastogenesis. The bone slices were then fixed with 3.7% formaldehyde solution in PBS for 10 min at room temperature. They were then treated with 0.1% Triton X-100 in PBS for 8 min and stained with Alexa Fluor 488 phalloidin and Hoechst 33258 for 15 min for actin ring and nucleic staining, respectively. The bone slices were analyzed and imaged using a Leica DMIRBE inverted UV SP1 confocal microscope system with Leica confocal software at the Imaging Facility of UAB. The assays were performed in triplicate, and a representative view from each assay is shown.
In Vitro Bone Resorption Assays
BMMs (5 × 104 cells/well) were seeded on bovine cortical bone slices plated in 24-well culture plates and were cultured under conditions indicated in individual experiments to promote osteoclast formation and bone resorption. The bone slices were then harvested, and the cells were subsequently removed with 0.25 m ammonium hydroxide and mechanical agitation. Bone slices were subjected to scanning electron microscopy for analysis using a Philips 515 scanning electron microscope in the Material Engineering Department at UAB. The assays were performed in triplicate, and a representative view from each assay is shown. The data were quantified by measuring the percentage of the resorbed areas in three random resorption sites. The percentage of the resorbed area was determined using ImageJ analysis software obtained from the National Institutes of Health.
Preparation of Retrovirus and Infection of BMMs
293GPG cells were cultured in DMEM with 10% heat-inactivated FBS supplemented with tetracycline, puromycin, G418, and penicillin/streptomycin as described previously (31). The chimeric receptor constructs, pMX-puro-hFas-RANK (Ch1) and pMX-puro-hFas-PM3 (Ch2), were generated in previous studies (32). pMX-puro-GFP (GFP) was prepared by inserting a GFP cDNA into pMX-puro vector. The retroviral vectors were transiently transfected into 293GPG cells using Lipofectamine Plus reagent (Invitrogen). Virus supernatant was collected at 48, 72, and 96 h after transfection. BMMs were then infected with virus for 24 h in the presence of M-CSF (220 ng/ml) and 8 μg/ml Polybrene. Cells were further cultured in the presence of M-CSF (220 ng/ml) and 2 μg/ml puromycin for selection and expansion of transduced cells. Selected cells were subsequently used for various studies. For assays involving the activation of the chimeric receptor, human Fas-AB was added at the concentrations indicated in individual assays.
Flow Cytometric Analysis
Retrovirally infected BMMs (1 × 106) were suspended in 200 ml of α-MEM containing 10% heat-inactivated FBS supplemented with M-CSF (44 ng/ml). Cells were then blocked with 1 μg of 2.4G2 antibody (33) for 30 min at 4 °C. Under dim light, 10 μl of human Fas-AB conjugated with phycoerythrin was added to the cell suspension, and cells were incubated for an additional 30 min at 4 °C before centrifuging at 2,000 rpm for 5 min. Cells were then washed three times in 1 ml of α-MEM. Cells were suspended in 1 ml of α-MEM for cytometric analysis. Flow cytometric analysis was performed using a BD LSR II flow cytometer at the Center for AIDS Research at UAB.
Semiquantitative Reverse Transcription (RT)-PCR
Total RNA was isolated from BMMs using TRIzol reagent (Invitrogen). 1 μg of total RNA was reversed-transcribed to cDNA with oligo(dT) using the ThermoScriptTM RT-PCR system (Invitrogen). The RT was carried out in a 20-μl volume at 55 °C for 55 min followed by enzyme inactivation and RNA H digestion. After completion, 40 μl of H2O was added to the reaction to bring the volume to 60 μl. 5 μl was used for PCR amplification of the MMP9, cathepsin K (Ctsk), TRAP, and GAPDH cDNAs and 8 μl for carbonic anhydrase II (Car2) cDNA using the following condition: preheating at 95 °C for 2 min, (denaturing 95 °C for 30 s, annealing 58 °C for 30 s, and extension 72 °C for 30 s) × 20 cycles for MMP9, Ctsk, TRAP, and GAPDH transcripts or × 25 cycle for Car2, followed by final extension 72 °C for 5 min. PCR was performed with Go-Taq DNA polymerase from Promega (Madison, WI) in a 50-μl reaction volume. PCR primers are 5′-CTTCTTCTCTGGACGTCAAATG-3′ and 5′-CATTTTGGAAACTCACACGCC-3′ for MMP9, 5′-AGAGAACTGGCACAAGGACTT-3′ and 5′-CCTCCTTTCAGCACTGCATTGT-3′ for Car2, 5′-GATGCTTACCCATATGTGGGC-3′ and 5′-CATATCCTTTGTTTCCCCAGC-3′ for Ctsk, 5′-GCCAAGATGGATTCATGGGTGG-3′ and 5′-CAGAGACATGATGAAGTCAGCG-3′ for TRAP, 5′-ACATCATCCCTGCATCCACTG-3′ and 5′-TCATTGAGAGCAATGCCAGC-3′ for GAPDH. 20 μl of PCR mixture was loaded on 2% agarose gel for electrophoretic analysis. All semiquantitative RT-PCR assays were independently repeated twice.
RESULTS
TNF Cannot Induce Osteoclastogenesis Partially Due to Its Inability to Activate the Expression of Osteoclast Genes
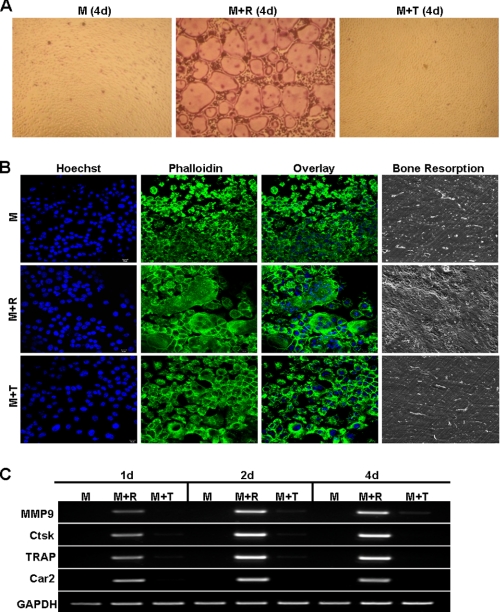
Given the controversy on the role of TNF in osteoclastogenesis, we first independently examined whether TNF can mediate osteoclastogenesis in the presence of M-CSF. Although BMMs treated with M-CSF and RANKL formed numerous osteoclasts in tissue culture dish, those treated with M-CSF and TNF did not form any osteoclast (Fig. 1A), replicating a finding from the previous study that TNF cannot replace RANKL in mediating osteoclastogenesis in tissue culture dish (26). To address this issue further, we switched to a more physiologically relevant assay to determine whether we can reproduce this finding on bone slices (Fig. 1B). As a negative control, BMMs cultured on bone slices with M-CSF alone remained mononuclear, did not form actin ring, and were unable to resorb bone (top row, Fig. 1B). As a positive control, M-CSF and RANKL promoted osteoclastogenesis on bone slices, which was substantiated by the presence of multinucleation and actin ring, two important features of mature osteoclasts, and numerous resorption pits (center row, Fig. 1B). However, although M-CSF and TNF treatment induced the formation of actin ring, the cells still remained mononuclear (bottom row, Fig. 1B), revealing that TNF cannot promote osteoclastogenesis on bone slices. The inability of TNF to induce osteoclastogenesis on bone slices is further substantiated by lack of resorption pits on bone slices (far right panel, Fig. 1B). Taken together, these results confirm that TNF cannot replace RANKL in promoting osteoclastogenesis.
FIGURE 1.
TNF can neither induce osteoclastogenesis nor activate osteoclast genes. A, BMMs were treated with M-CSF (M, 44 ng/ml), M-CSF (44 ng/ml) and RANKL (100 ng/ml) (M+R), or M-CSF (44 ng/ml) and TNF (10 ng/ml) (M+T) in a tissue culture dish for 4 days (d). The cultures were then stained for TRAP activity. B, BMMs on bone slices were cultured with M-CSF (44 ng/ml), M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and TNF (10 ng/ml) for 4 days, and bone slices were then stained with Hoechst 33258 (Hoechst) or Alexa Fluor 488 phalloidin (Phalloidin). A separate set of cultures was continued for 4 additional days to perform bone resorption assays. C, BMMs were treated with M-CSF (44 ng/ml), M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and TNF (10 ng/ml) in a tissue culture dish for 1, 2, or 4 days. Gene expression was determined by semiquantitative RT-PCR.
Previous gene expression profiling studies have established that RANKL stimulates osteoclast differentiation by altering the expression of numerous genes (34–36). In particular, RANKL dramatically up-regulates the expression of the genes encoding MMP9, Car2, Ctsk, and TRAP (34, 35), which play important roles in osteoclast formation and/or function and are thus widely used as markers for osteoclasts (37, 38). To investigate the molecular basis of the differential roles of RANKL and TNF in osteoclastogenesis, we examined the effect of TNF on the expression of the four osteoclast genes (Fig. 1C). Whereas one-day RANKL treatment significantly increased the expression of these genes, TNF treatment as long as 4 days failed to do so. This finding indicates that the inability of TNF to activate the expression of genes required for osteoclast differentiation accounts for its failure to promote osteoclastogenesis.
TNF Is Capable of Activating the Expression of Osteoclast Genes in the Presence of Permissive Levels of RANKL
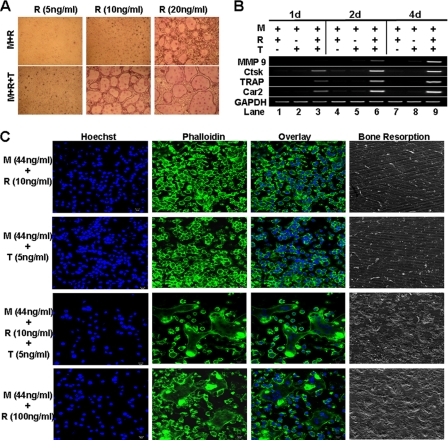
Lam et al. demonstrated that TNF can induce osteoclastogenesis in tissue culture dish in the presence of permissive levels of RANKL (26), suggesting that permissive doses of RANKL may play a critical role in rendering osteoclast genes responsive to TNF. To address this possibility, we first treated BMMs with M-CSF plus different concentrations of RANKL in the absence or presence of TNF for 4 days to identify subosteoclastogenic doses of RANKL that can promote osteoclastogenesis in concert with TNF (Fig. 2A). We further demonstrated that the longer treatment (up to 8 days) of BMMs with the subosteoclastogenic dose of RANKL (10 ng/ml) alone failed to promote osteoclast formation (supplemental Fig. 1). These data replicate a finding from the previous study that low levels of RANKL, although unable to promote osteoclastogenesis, can fully induce osteoclastogenesis in tissue culture dish when attended by TNF (26). To determine whether TNF can activate the expression of the four genes in the presence of permissive levels of RANKL, we treated BMMs with RANKL (10 ng/ml) and/or TNF (5 ng/ml) in the presence of M-CSF for 1, 2, or 4 days (Fig. 2B). 10 ng/ml RANKL barely activated the expression of the genes in the presence of M-CSF (44 ng/ml) (lanes 1, 4, and 7). Similarly, TNF (5 ng/ml) was unable to activate the osteoclast genes in the presence of M-CSF (44 ng/ml) (lanes 2, 5, and 8). However, TNF (5 ng/ml) and RANKL (10 ng/ml) together significantly up-regulated the expression of Ctsk, TRAP, and Car2 genes at day 1 (lane 3) and the MMP9 gene at day 2 (lane 6). Thus, the data indicate that permissive doses of RANKL play a crucial role in TNF-mediated osteoclastogenesis by committing BMMs into osteoclast lineage, which in part involves rendering osteoclast genes responsive to TNF.
FIGURE 2.
TNF can induce osteoclastogenesis and activate osteoclast genes in the presence of permissive levels of RANKL. A, BMMs were cultured with 44 ng/ml M-CSF (M) plus different doses of RANKL (R) with or without TNF (T, 5 ng/ml) for 4 days (d) in a tissue culture dish. The cultures were then stained for TRAP activity. B, BMMs were treated with M-CSF (44 ng/ml) and RANKL (10 ng/ml), M-CSF (44 ng/ml) and TNF (5 ng/ml), or M-CSF (44 ng/ml) + RANKL (10 ng/ml) + TNF (5 ng/ml) for 1, 2, or 4 days. Gene expression was determined by semiquantitative RT-PCR. C, BMMs on bone slices were treated with M-CSF (44 ng/ml) and RANKL (10 ng/ml), M-CSF (44 ng/ml) and TNF (5 ng/ml), M-CSF (44 ng/ml) + RANKL (10 ng/ml) + TNF (5 ng/ml), or M-CSF (44 ng/ml) and RANKL (100 ng/ml) for 4 days, and bone slices were stained with Hoechst 33258 (Hoechst) or Alexa Fluor 488 phalloidin (Phalloidin). A separate set of cultures was continued for 4 additional days to perform bone resorption assays.
Although we have recapitulated the previous observation that TNF can promote osteoclastogenesis in tissue culture dish in the presence of permissive levels of RANKL (Fig. 2A), it is still unclear whether TNF can do so on the bone surface, which represents physiological substratum for osteoclasts. More importantly, it is also unknown whether osteoclasts derived from stimulation with TNF and permissive levels of RANKL are functional. We extended our study to address these issues. We found that neither 10 ng/ml RANKL nor 5 ng/ml TNF alone was able to mediate osteoclastogenesis on bone slices in the presence of M-CSF, as evidenced by lack of multinucleation and resorption pits (top two rows, Fig. 2C). In contrast, 5 ng/ml TNF plus 10 ng/ml RANKL promoted osteoclastogenesis, which was substantiated by the presence of multinucleation, actin ring, and resorption pits (the third row from top, Fig. 2C). As positive control, 100 ng/ml RANKL stimulated the formation of functional osteoclasts on bone slices (bottom row, Fig. 2C). These data demonstrate that TNF can promote osteoclastogenesis on the bone surface in the presence of permissive levels of RANKL, and more importantly, osteoclasts formed under this condition are functional.
TNF Can Promote Osteoclastogenesis from BMMs Transiently Exposed to RANKL
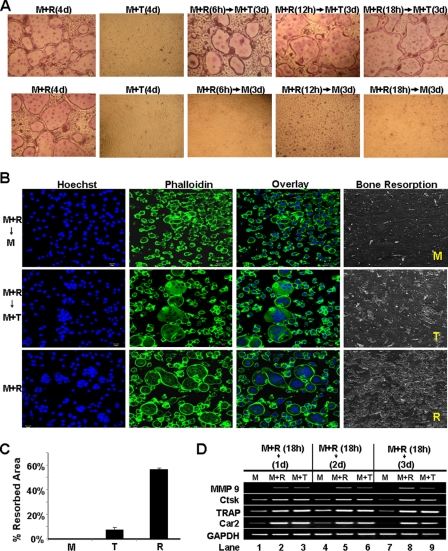
Next, we investigated whether TNF is able to stimulate osteoclastogenesis from BMMs previously treated with RANKL in tissue culture dish (Fig. 3A) and bone slices (Fig. 3B). RANKL pretreatment as short as 6 h rendered BMMs capable of forming osteoclasts in the tissue culture dish in response to subsequent TNF stimulation (top panel, Fig. 3A). However, without subsequent TNF stimulation, BMMs pretreated with RANKL for as long as 18 h failed to form osteoclasts (bottom panel, Fig. 3A). The finding replicates the conclusion from a previous study that TNF can promote osteoclastogenesis from BMMs pretreated with RANKL (26). Conversely, we also examined whether TNF can prime BMMs to form osteoclasts in response to subsequent RANKL stimulation. BMMs were treated with 5 ng/ml TNF for 18 h, and then the cultures were continued with 100 ng/ml RANKL for 3 days (supplemental Fig. 2). The data indicate that, unlike RANKL, TNF cannot prime BMMs because BMMs pretreated with TNF for 18 h formed smaller osteoclasts in response to subsequent 3-day RANKL treatment (supplemental Fig. 2), compared with the assay involving 18-h RANKL pretreatment followed by 3-day TNF stimulation (Fig. 3A). Furthermore, whereas 18h RANKL pretreatment without subsequent TNF stimulation induced no osteoclastogenesis (top row, Fig. 3B), cultures continued with M-CSF plus TNF for 3 days formed osteoclasts on the bone slices (center row, Fig. 3B). Nonetheless, osteoclasts derived from RANKL pretreatment and subsequent TNF stimulation gave rise to fewer resorption pits compared with control osteoclasts resulting from the continuous RANKL treatment (Fig. 3, B and C), indicating that although the TNF-induced osteoclasts are functional, they do not resorb bone as efficiently as those formed by RANKL. Taken together, these findings reveal that TNF can promote osteoclastogenesis from BMMs transiently exposed to 100 ng/ml RANKL, but osteoclasts formed under this condition have lower capacity to resorb bone.
FIGURE 3.
TNF can promote osteoclastogenesis and activate osteoclast genes in BMMs pretreated by RANKL. A, BMMs were primed with M-CSF (M, 44 ng/ml) and RANKL (R, 100 ng/ml) for 6, 12, or 18 h and then continued with M-CSF (44 ng/ml) alone or M-CSF (44 ng/ml) plus TNF (T, 5 ng/ml) for 3 days (d). BMMs treated with M-CSF (44 ng/ml) plus RANKL (100 ng/ml) or M-CSF (44 ng/ml) plus TNF (5 ng/ml) served as positive and negative controls, respectively. All cultures were then stained for TRAP activity. B, BMMs on bone slices were primed with M-CSF (44 ng/ml) and RANKL (100 ng/ml) for 18 h and then treated with M-CSF (44 ng/ml) alone, M-CSF (44 ng/ml) and TNF (5 ng/ml), or M-CSF (44 ng/ml) and RANKL (100 ng/ml) for 4 days, and bone slices were then stained with Hoechst 33258 (Hoechst) or Alexa Fluor 488 phalloidin (Phalloidin). A separate set of cultures was continued for 4 additional days to perform bone resorption assays. C, quantification of the bone resorption assays is shown. Bars show averages ± S.D. D, BMMs were primed with M-CSF (44 ng/ml) and RANKL (100 ng/ml) for 18 h and then cultured with M-CSF (44 ng/ml) alone, M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and TNF (5 ng/ml) for 1, 2, or 3 days. Gene expression was assessed by semiquantitative RT-PCR.
We then examined the effect of RANKL pretreatment and subsequent TNF stimulation on the expression of the four osteoclast genes (Fig. 3D). The 18-h RANKL treatment rendered all four genes persistently responsive to TNF (lane 1 versus 3, 4 versus 6, and 7 versus 9, Fig. 3D). Moreover, MMP9 transcripts were not detectable 1 day after the pretreatment (lane 1, Fig. 3D), and the expression levels of the Ctsk, TRAP, and Car2 genes decreased with time (lanes 1, 4, and 7, Fig. 3D), indicating that 18-h RANKL pretreatment only transiently turns on these genes. However, once these genes are turned on by RANKL pretreatment, their expression can be further up-regulated and maintained by TNF to promote osteoclast formation. These findings support the notion that RANKL primes BMMs into osteoclast lineage through rendering osteoclast genes responsive to TNF.
RANKL-mediated Osteoclast Lineage Commitment Is Durable and Involves Long Term Reprogramming of Osteoclast Genes into an Inducible State
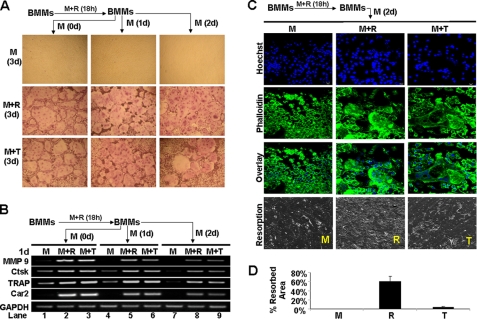
TNF can promote osteoclastogenesis from BMMs previously exposed to RANKL (Fig. 3), which suggests that RANKL-mediated lineage commitment is durable. We performed additional assays to address this possibility further (Fig. 4). The results reveal that TNF can still promote osteoclastogenesis 2 days after 18-h RANKL pretreatment (bottom right panel, Fig. 4A), indicating that the RANKL-mediated lineage commitment is not transient. Moreover, our gene expression assays show that the expression of the genes decreases to almost undetectable levels 3 days after the RANKL pretreatment (lanes 1, 4, and 7, Fig. 4B). However, TNF can strongly restore the expression of the four genes (lanes 3, 6, and 9, Fig. 4B), indicating that the RANKL-mediated inducible state of the four osteoclast genes is long term.
FIGURE 4.
RANKL mediated-BMM priming is durable. A, BMMs were treated with M-CSF (M, 44 ng/ml) and RANKL (R, 100 ng/ml) for 18 h and then cultured with M-CSF alone for 0, 1, or 2 days (d) before treating with M-CSF (44 ng/ml) alone, M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and TNF (T, 5 ng/ml) for 3 days. The cultures were stained for TRAP activity. B, BMMs were treated with M-CSF (44 ng/ml) and RANKL (100 ng/ml) for 18 h and then cultured with M-CSF alone for 0, 1 or 2 days before treating with M-CSF (44 ng/ml) alone, M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and TNF (5 ng/ml) for 1 day. Gene expression was determined by semiquantitative RT-PCR. C, BMMs on bone slices were treated with M-CSF (44 ng/ml) and RANKL (100 ng/ml) for 18 h and then cultured with M-CSF alone for 2 days before treating with M-CSF (44 ng/ml) alone, M-CSF (44 ng/ml) and RANKL (100 ng/ml), or M-CSF (44 ng/ml) and TNF (5 ng/ml) for 3 days, and bone slices were then stained with Hoechst 33258 (Hoechst) or Alexa Fluor 488 phalloidin (Phalloidin). A separate set of cultures was continued for 4 additional days to perform bone resorption assays. D, quantification of the bone resorption assays is shown. Bars show averages ± S.D.
Next, we determined whether TNF induces the formation of functional osteoclasts on bone slices 2 days after the 18-h RANKL pretreatment (Fig. 4C). TNF can promote osteoclastogenesis on bone slices 2 days after RANKL pretreatment, as indicated by the presence of multinucleation (right panels, Fig. 4C). More importantly, these osteoclasts are capable of resorbing bone, but their bone-resorbing capacity is much lower than those derived from RANKL stimulation (bottom row, Fig. 4, C and D). Thus, we conclude that RANKL-mediated osteoclast lineage commitment is durable and involves long term reprogramming of osteoclast genes into an inducible state in which they can be activated by TNF.
RANK IVVY535–538 Motif Plays a Critical Role in TNF-mediated Osteoclastogenesis by Rendering Osteoclast Genes Responsive to TNF
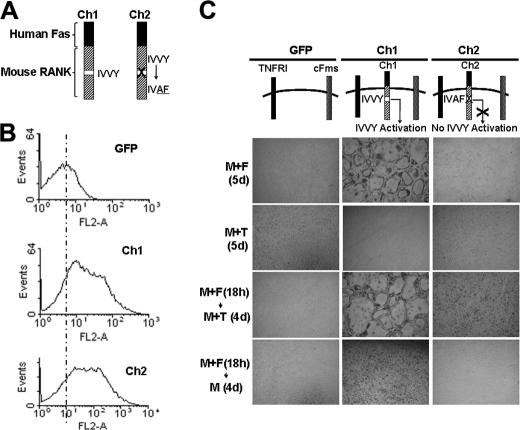
We have previously shown that the RANK IVVY535–538 plays an essential role in priming BMMs to the osteoclast lineage in RANKL-mediated osteoclastogenesis (14). Thus, we investigated whether this IVVY motif also plays a role in priming BMMs in TNF-induced osteoclastogenesis. To this end, we utilized two chimeric receptors (Ch1 and Ch2) that were previously developed and validated (14, 32). Specifically, the chimeras were generated by linking the external domain of human Fas to the transmembrane and cytoplasmic domains of normal mouse RANK (Ch1) or RANK-bearing inactivating mutations in the IVVY motif (Ch2) (Fig. 5A) (32). These chimeras can be activated by a specific Fas-AB to activate RANK exclusively without affecting endogenous mouse Fas (32). Flow cytometric analysis showed that Ch1 and Ch2 were expressed at comparable levels on the surface of infected cells (Fig. 5B). Although Ch1-expressing cells formed osteoclasts in response to 5-day M-CSF and Fas-AB treatment, those expressing Ch2 generated no osteoclasts (top row, Fig. 5C), confirming our previous observation that the IVVY motif plays an essential role in osteoclastogenesis (14). When Ch1- or Ch2-expressing cells were treated with M-CSF and TNF for 5 days, they both failed to form osteoclasts (second row from top, Fig. 5C). However, in the chimera-expressing cells that were treated with M-CSF plus Fas-AB for 18 h prior to the TNF stimulation, the Ch1-expressing cells, but not Ch2-expressors, formed osteoclasts (third row from top, Fig. 5C). As a control, the chimera-expressing cells pretreated with M-CSF plus Fas-AB for 18 h failed to induce osteoclastogenesis in response to 4-day stimulation with M-CSF alone (bottom row, Fig. 5C). These results indicate that the IVVY motif is specifically responsible for priming BMMs.
FIGURE 5.
RANK IVVY535–538 motif plays a critical role in TNF-mediated osteoclastogenesis. A, schematic diagram of Ch1 and Ch2. B, flow cytometric analysis of BMMs infected with virus encoding GFP, Ch1, or Ch2. C, BMMs expressing GFP, Ch1, or Ch2 were treated with M-CSF (M, 44 ng/ml) and Fas-AB (F, 100 ng/ml) for 18 h. The cultures were then continued with M-CSF (44 ng/ml) alone or M-CSF (44 ng/ml) and TNF (T, 5 ng/ml) for 4 days (d). Infected BMMs treated with M-CSF (44 ng/ml) plus Fas-AB (100 ng/ml) or M-CSF (44 ng/ml) plus TNF (T, 5 ng/ml) for 5 days served as positive and negative controls, respectively. The cultures were then stained for TRAP activity.
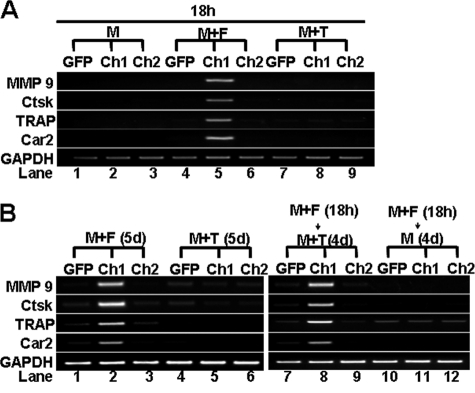
Next, we examined the role of the RANK IVVY motif in activating the expression of the MMP9, Ctsk, TRAP, and Car2 genes using the chimeric receptor approach. The expression of the four genes was activated in the Ch1-expresors but not in Ch2-expressors following 18-h Fas-AB priming (lanes 5 and 6, Fig. 6A), demonstrating that the IVVY motif plays a crucial role in the activation of the genes. These results also indicate that the IVVY mediates BMM priming partly by activating the osteoclast genes. We then determined whether the IVVY motif is involved in rendering these genes responsive to TNF (Fig. 6B). We found that the expression of the four genes was activated in BMMs expressing Ch1 but not Ch2, indicating that the RANK IVVY motif is specifically response for reprogramming genes into the inducible state.
FIGURE 6.
RANK IVVY535–538 motif renders osteoclast genes responsive to TNF. A, BMMs expressing GFP, Ch1, or Ch2 were treated with M-CSF (M, 44 ng/ml) alone, M-CSF (44 ng/ml) and Fas-AB (F, 100 ng/ml), or M-CSF (44 ng/ml) and TNF (T, 5 ng/ml) for 18 h. B, BMMs expressing GFP, Ch1, or Ch2 were treated with M-CSF (44 ng/ml) and Fas-AB (100 ng/ml) for 18 h, and the cultures were then continued with M-CSF (44 ng/ml) alone or M-CSF (44 ng/ml) and TNF (5 ng/ml) for 4 days (d). Infected BMMs treated with M-CSF (44 ng/ml) plus Fas-AB (100 ng/ml) or M-CSF (44 ng/ml) plus TNF (5 ng/ml) for 5 days served as positive and negative controls, respectively. Gene expression in A and B was determined by semi-quantitative RT-PCR.
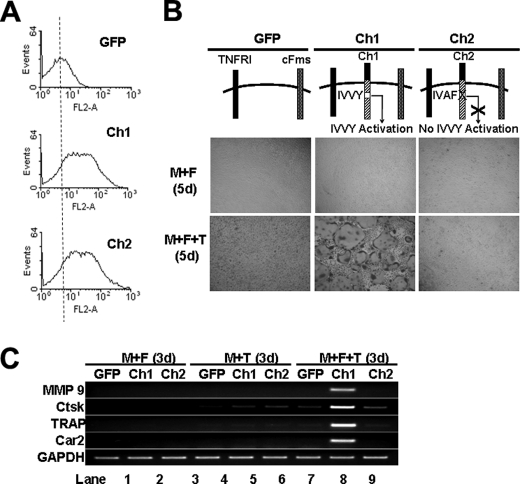
We further investigated the role of the IVVY motif in TNF-induced osteoclastogenesis in the co-treatment setting described in Fig. 2. BMMs expressing comparable levels of Ch1 or Ch2 (Fig. 7A) formed no osteoclasts when treated with Fas-AB (10 ng/ml) and M-CSF for 5 days (top row, Fig. 7B). However, Ch1-expressing cells, but not those expressing Ch2, formed osteoclasts when stimulated with Fas-AB (10 ng/ml) plus TNF (5 ng/ml) in the presence of M-CSF for 5 days (bottom row, Fig. 7B). These data further demonstrate that the IVVY motif plays a critical role in TNF-mediated osteoclastogenesis. Consistently, the expression of the four osteoclast genes was activated in Ch1-expressing cells, but not in those expressing Ch2, when stimulated with Fas-AB (10 ng/ml) plus TNF (5 ng/ml) in the presence of M-CSF for 3 days (lanes 8 and 9, Fig. 7C). These results further demonstrate that the IVVY motif plays a critical role in TNF-mediated osteoclastogenesis by reprogramming osteoclast genes into an inducible state in which they can be activated by TNF.
FIGURE 7.
TNF-induced osteoclastogenesis attended by permissive Fas-AB dosage requires RANK IVVY535–538-mediated signaling. A, flow cytometric analysis of BMMs infected with virus encoding GFP, Ch1, or Ch2 is shown. B, BMMs expressing GFP, Ch1, or Ch2 were treated with M-CSF (M, 44 ng/ml) and Fas-AB (F, 10 ng/ml) with or without TNF (T, 5 ng/ml) for 5 days (d). The cultures were then stained for TRAP activity. C, BMMs expressing GFP, Ch1, or Ch2 were treated with M-CSF (44 ng/ml) and Fas-AB (10 ng/ml), M-CSF (44 ng/ml) and TNF (5 ng/ml), or M-CSF (44 ng/ml) + Fas-AB (10 ng/ml) + TNF (5 ng/ml) for 3 days. Gene expression was determined by semiquantitative RT-PCR.
DISCUSSION
It was initially shown almost a quarter of century ago that TNF stimulates bone resorption in organ cultures (fetal rat bones) in vitro (39). This important observation prompted a question of whether estrogen exerts its bone-sparing effect by modulating the production of this cytokine (40). Indeed, subsequent studies demonstrated that human peripheral blood monocytes from ovariectomized premenopausal women secrete increased amounts of TNF (41). Moreover, it was also shown that estrogen inhibits the expression of TNF in osteoblasts and stromal cells (42). These findings support the notion that estrogen exerts its protective effect on bone by suppressing the expression of TNF. The pathological role for TNF in postmenopausal osteoporosis was further supported by animal model studies demonstrating that knock-out mice deficient in either TNF or TNFR1 are resistant to ovariectomy-induced bone loss (43). As a potent proinflammatory cytokine implicated in the pathogenesis of rheumatoid arthritis (4) and periodontitis (5), TNF has later been demonstrated to play a role in the bone loss associated with these inflammatory conditions (4, 5). Therefore, an effective therapy of the TNF-induced bone loss certainly requires a precise understanding of the mechanism by which TNF promotes bone loss.
Early studies indicated that TNF induces bone loss indirectly by stimulating osteoblasts to produce numerous factors including IL-6, M-CSF, and granulocyte-macrophage colony stimulating factor, which in turn promote osteoclast formation and function (40, 44). The discovery of the RANKL/RANK/OPG system has led to a revelation that RANKL is also among the factors produced by osteoblasts in response to TNF stimulation (17, 18), which further supports the indirect role of this cytokine in osteoclastogenesis. Importantly, more recent investigations have found that TNF also directly targets osteoclast precursors and mature osteoclasts to modulate osteoclast formation, function, and survival (22, 23, 45). However, how exactly TNF directly regulates osteoclastogenesis remains controversial. Whereas some groups showed that TNF can promote osteoclastogenesis in vitro independently of the RANKL/RANK system (22, 23), others demonstrated that TNF cannot induce osteoclastogenesis in vitro unless assisted by permissive levels of RANKL (26, 27).
Our current studies have further confirmed the previous finding that although TNF cannot stimulate osteoclastogenesis, it can induce the formation of functional osteoclasts in the presence of permissive levels of RANKL. More importantly, we have also shown that TNF can promote the formation of functional osteoclasts 2 days after transient RANKL pretreatment. These findings have important implications for not only better understanding the pathological roles of TNF and RANKL in various bone disorders but also devising better therapeutic strategies. For instance, although estrogen deficiency leads to an elevation in the expression of TNF and RANKL in bone marrow cells (41, 43, 46), the magnitude and temporal pattern of the changes in the expression of these two cytokines may differ among patients. Given that RANKL is a potent osteoclastogenic factor that alone suffices to stimulate osteoclastogenesis in the presence of M-CSF, a persistent and high elevation in RANKL expression, with or without a significant increase in TNF levels, can lead to the development of osteoporosis. Based on the current findings, it is reasonable to assume that either a low but persistent elevation or a transient but significant increase in RANKL levels, in concert with a dramatic increase in TNF levels, can promote the development of osteoporosis in certain patients. Consequently, although blockage of RANKL should be an effective therapeutic approach in the former circumstance, inhibition of either RANKL or TNF can be used to prevent and treat bone loss in the latter case.
Our second goal was to investigate the molecular basis of the RANKL-mediated priming for TNF-mediated osteoclastogenesis. It was shown previously that RANKL and TNF, through the transient activation of the JNK and NF-κB pathways, exhibit a synergistic effect which may underlie the priming of BMMs by RANKL (26). Our new data indicate that RANKL-mediated priming is durable because TNF can still promote osteoclastogenesis 2 days after the 18-h RANKL priming (Fig. 4, A–C). Thus, we suspected that other mechanisms may also be involved in priming BMMs. Although RANKL also activate several other signaling pathways such as Akt, ERK, and p38 (6, 13), these pathways are unlikely to mediate the RANKL-mediated BMM priming because they are also transiently activated by RANKL. Previous studies have established that RANKL promotes osteoclastogenesis by regulating the expression of a large set of genes (34–36). Hence, we decided to investigate the molecular basis of the RANKL-mediated priming at the gene expression level by examining the role of RANKL in the expression of four osteoclast genes (MMP9, Ctsk, TRAP, and Car2). We found that although TNF cannot activate these genes, the presence of permissive levels of RANKL in the cultures or 18-h RANKL pretreatment of BMMs renders these genes responsive to TNF treatment. Most importantly, we have also revealed that the RANK IVVY535–538 motif is responsible for the RANKL-mediated priming and reprogramming the selected osteoclast genes into an inducible state. Although we only chose four highly recognized osteoclast genes for the study, it is reasonable to conclude that RANKL primes BMMs in part by reprogramming some of osteoclast genes into an inducible state in which they can be activated by TNF. It is noted that although TNF can promote the formation of multinucleated TRAP-positive cells 2 days after the 18-h RANKL priming, these cells are less efficient in resorbing bone (Figs. 3B and 4C). Thus, it is likely that RANKL can reprogram only a subset of osteoclast genes into an inducible state which is sustainable.
As discussed above, an effective blockage of RANKL can be used to prevent and treat bone loss in various bone disorders. Because the RANKL/RANK/OPG axis not only plays a pivotal role in osteoclast formation and function (19) but is also involved in other biological processes such as dendritic cell survival and activation (47), T cell activation (48), and B cell differentiation (48), use of agents that function to inhibit RANKL-RANK interaction may cause deleterious effects on immune responses (49). Previously, we demonstrated that the IVVY motif plays an essential role in RANKL-mediated osteoclastogenesis by committing BMMs into the osteoclast lineage (14). In the current study, we have shown that the IVVY motif is also crucially involved in TNF-mediated osteoclastogenesis. Moreover, it has been shown that IVVY is not involved in the activation of the known RANK signaling pathways (14, 15). Thus, we believe that the RANK IVVY motif and its downstream signaling pathway(s) may have the potential to serve as better therapeutic targets for bone loss associated with various pathological conditions.
In summary, our key findings are as follows. (i) although TNF cannot stimulate osteoclastogenesis on bone slices, it can induce the formation of functional osteoclasts on bone slices in the presence of permissive levels of RANKL. (ii) TNF can also promote the formation of functional osteoclasts 2 days after transient RANKL pretreatment. (iii) TNF cannot stimulate osteoclastogenesis in part because of its inability to activate the expression of osteoclast genes. (iv) RANKL plays a crucial role in TNF-induced osteoclastogenesis by reprogramming genes into an inducible state in which they can be activated by TNF. (v) the RANK IVVY535–538 motif mediates the reprogramming of osteoclast genes into an inducible state. These observations provide important new insights into the molecular mechanism by which RANKL regulates TNF-mediated osteoclastogenesis. Our work has further addressed the controversy regarding the regulatory role of TNF in osteoclastogenesis, which helps better understand the pathological role of TNF in bone loss in various bone disorders. Significantly, the IVVY motif, due to its critical role in TNF-mediated osteoclastogenesis, has the potential to serve as a new therapeutic target for bone disorders. Interestingly, a recent study has shown that that the IVVY motif engages Vav3 indirectly via an unknown adaptor protein to regulate osteoclastogenesis (15). Thus, one potential mechanism by which RANKL primes BMMs may involve the reorganization of cytoskeleton by the IVVY motif-mediated signaling, leading to the constitutive activation of the RANK signaling required for osteoclastogenesis. Alternatively, because the IVVY motif is involved in reprogramming osteoclast genes into an inducible state, the IVVY motif-mediated signaling may prime BMMs by inducing epigenetic changes in the promoters of osteoclast genes, rendering them inducible by TNF. Hence, future studies aimed at identifying the unknown adaptor protein directly interacting with the IVVY motif may facilitate the unraveling of the molecular mechanisms by which RANKL primes BMMs and/or reprograms osteoclast genes into an inducible state.
This work was supported by National Institutes of Health Grant AR47830 through the NIAMS (to X. F.). This work was also supported by a National Institutes of Health/NIAMS graduate research supplement to AR47830 (to J. J.), a Within Our Reach innovative basic research grant from Research and Education Foundation of American College of Rheumatology (to X. F.), and National Institutes of Health/NIAMS University of Alabama at Birmingham Core Center for Basic Skeletal Research (CCBSR) Grant 5P30 AR046031.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- M-CSF
- macrophage/monocyte colony-stimulating factor
- BMM
- bone marrow macrophage
- Car2
- carbonic anydrase II
- Ch1
- chimeric receptor with normal RANK cytoplasmic domain
- Ch2
- chimeric receptor with mutated RANK IVVY535–538 motif (VY is replaced by AF)
- Ctsk
- cathepsin K
- Fas-AB
- human Fas-activating antibody
- hFas
- human Fas external domain
- MMP9
- matrix metalloproteinase 9
- OPG
- osteoprotegerin
- RANK
- receptor activator of nuclear factor-κB
- RANKL
- receptor activator of nuclear factor-κB ligand
- TNFR
- TNF receptor
- TRAF
- TNFR-associated factor
- TRAP
- tartrate resistant acid phosphatase
- UAB
- University of Alabama at Birmingham.
REFERENCES
- 1.Beyaert R., Fiers W. (1998) in Cytokines (Mire-Slus A., Thorpe R. eds) pp. 335–360, Academic Press, Orlando, FL [Google Scholar]
- 2.Vassalli P. (1992) Annu. Rev. Immunol. 10, 411–452 [DOI] [PubMed] [Google Scholar]
- 3.Jilka R. L. (1998) Bone 23, 75–81 [DOI] [PubMed] [Google Scholar]
- 4.Goldring S. R. (2003) Rheumatology 42, (Suppl. 2) ii11–ii16 [DOI] [PubMed] [Google Scholar]
- 5.Graves D. T., Cochran D. (2003) J. Periodontol. 74, 391–401 [DOI] [PubMed] [Google Scholar]
- 6.Boyle W. J., Simonet W. S., Lacey D. L. (2003) Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 7.Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 8.Anderson D. M., Maraskovsky E., Billingsley W. L., Dougall W. C., Tometsko M. E., Roux E. R., Teepe M. C., DuBose R. F., Cosman D., Galibert L. (1997) Nature 390, 175–179 [DOI] [PubMed] [Google Scholar]
- 9.Simonet W. S., Lacey D. L., Dunstan C. R., Kelley M., Chang M. S., Lüthy R., Nguyen H. Q., Wooden S., Bennett L., Boone T., Shimamoto G., DeRose M., Elliott R., Colombero A., Tan H. L., Trail G., Sullivan J., Davy E., Bucay N., Renshaw-Gegg L., Hughes T. M., Hill D., Pattison W., Campbell P., Sander S., Van G., Tarpley J., Derby P., Lee R., Boyle W. J., Boyle W. J. (1997) Cell 89, 309–319 [DOI] [PubMed] [Google Scholar]
- 10.Locksley R. M., Killeen N., Lenardo M. J. (2001) Cell 104, 487–501 [DOI] [PubMed] [Google Scholar]
- 11.Armstrong A. P., Tometsko M. E., Glaccum M., Sutherland C. L., Cosman D., Dougall W. C. (2002) J. Biol. Chem. 277, 44347–44356 [DOI] [PubMed] [Google Scholar]
- 12.Liu W., Xu D., Yang H., Xu H., Shi Z., Cao X., Takeshita S., Liu J., Teale M., Feng X. (2004) J. Biol. Chem. 279, 54759–54769 [DOI] [PubMed] [Google Scholar]
- 13.Feng X. (2005) Gene 350, 1–13 [DOI] [PubMed] [Google Scholar]
- 14.Xu D., Wang S., Liu W., Liu J., Feng X. (2006) J. Biol. Chem. 281, 4678–4690 [DOI] [PubMed] [Google Scholar]
- 15.Kim H., Choi H. K., Shin J. H., Kim K. H., Huh J. Y., Lee S. A., Ko C. Y., Kim H. S., Shin H. I., Lee H. J., Jeong D., Kim N., Choi Y., Lee S. Y. (2009) J. Clin. Invest. 119, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrini M. M., Sobacchi C., Cassani B., Abinun M., Kilic S. S., Pangrazio A., Moratto D., Mazzolari E., Clayton-Smith J., Orchard P., Coxon F. P., Helfrich M. H., Crockett J. C., Mellis D., Vellodi A., Tezcan I., Notarangelo L. D., Rogers M. J., Vezzoni P., Villa A., Frattini A. (2008) Am. J. Hum. Genet. 83, 64–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn J. M., Horwood N. J., Elliott J., Gillespie M. T., Martin T. J. (2000) J. Bone Miner. Res. 15, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 18.Kitaura H., Sands M. S., Aya K., Zhou P., Hirayama T., Uthgenannt B., Wei S., Takeshita S., Novack D. V., Silva M. J., Abu-Amer Y., Ross F. P., Teitelbaum S. L. (2004) J. Immunol. 173, 4838–4846 [DOI] [PubMed] [Google Scholar]
- 19.Hsu H., Lacey D. L., Dunstan C. R., Solovyev I., Colombero A., Timms E., Tan H. L., Elliott G., Kelley M. J., Sarosi I., Wang L., Xia X. Z., Elliott R., Chiu L., Black T., Scully S., Capparelli C., Morony S., Shimamoto G., Bass M. B., Boyle W. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darnay B. G., Ni J., Moore P. A., Aggarwal B. B. (1999) J. Biol. Chem. 274, 7724–7731 [DOI] [PubMed] [Google Scholar]
- 21.Galibert L., Tometsko M. E., Anderson D. M., Cosman D., Dougall W. C. (1998) J. Biol. Chem. 273, 34120–34127 [DOI] [PubMed] [Google Scholar]
- 22.Azuma Y., Kaji K., Katogi R., Takeshita S., Kudo A. (2000) J. Biol. Chem. 275, 4858–4864 [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi K., Takahashi N., Jimi E., Udagawa N., Takami M., Kotake S., Nakagawa N., Kinosaki M., Yamaguchi K., Shima N., Yasuda H., Morinaga T., Higashio K., Martin T. J., Suda T. (2000) J. Exp. Med. 191, 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Sarosi I., Yan X. Q., Morony S., Capparelli C., Tan H. L., McCabe S., Elliott R., Scully S., Van G., Kaufman S., Juan S. C., Sun Y., Tarpley J., Martin L., Christensen K., McCabe J., Kostenuik P., Hsu H., Fletcher F., Dunstan C. R., Lacey D. L., Boyle W. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong Y. Y., Feige U., Sarosi I., Bolon B., Tafuri A., Morony S., Capparelli C., Li J., Elliott R., McCabe S., Wong T., Campagnuolo G., Moran E., Bogoch E. R., Van G., Nguyen L. T., Ohashi P. S., Lacey D. L., Fish E., Boyle W. J., Penninger J. M. (1999) Nature 402, 304–309 [DOI] [PubMed] [Google Scholar]
- 26.Lam J., Takeshita S., Barker J. E., Kanagawa O., Ross F. P., Teitelbaum S. L. (2000) J. Clin. Invest. 106, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P., Schwarz E. M., O'Keefe R. J., Ma L., Boyce B. F., Xing L. (2004) J. Bone Miner. Res. 19, 207–213 [DOI] [PubMed] [Google Scholar]
- 28.McHugh K. P., Hodivala-Dilke K., Zheng M. H., Namba N., Lam J., Novack D., Feng X., Ross F. P., Hynes R. O., Teitelbaum S. L. (2000) J. Clin. Invest. 105, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeshita S., Kaji K., Kudo A. (2000) J. Bone Miner. Res. 15, 1477–1488 [DOI] [PubMed] [Google Scholar]
- 30.Feng X., Novack D. V., Faccio R., Ory D. S., Aya K., Boyer M. I., McHugh K. P., Ross F. P., Teitelbaum S. L. (2001) J. Clin. Invest. 107, 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ory D. S., Neugeboren B. A., Mulligan R. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu D., Shi Z., McDonald J., Pan G., Cao X., Yu X., Feng X. (2004) Biochem. J. 383, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unkeless J. C. (1979) J. Exp. Med. 150, 580–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cappellen D., Luong-Nguyen N. H., Bongiovanni S., Grenet O., Wanke C., Susa M. (2002) J. Biol. Chem. 277, 21971–21982 [DOI] [PubMed] [Google Scholar]
- 35.Rho J., Altmann C. R., Socci N. D., Merkov L., Kim N., So H., Lee O., Takami M., Brivanlou A. H., Choi Y. (2002) DNA Cell Biol. 21, 541–549 [DOI] [PubMed] [Google Scholar]
- 36.Ishida N., Hayashi K., Hoshijima M., Ogawa T., Koga S., Miyatake Y., Kumegawa M., Kimura T., Takeya T. (2002) J. Biol. Chem. 277, 41147–41156 [DOI] [PubMed] [Google Scholar]
- 37.Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 38.Teitelbaum S. L., Ross F. P. (2003) Nat. Rev. Genet. 4, 638–649 [DOI] [PubMed] [Google Scholar]
- 39.Bertolini D. R., Nedwin G. E., Bringman T. S., Smith D. D., Mundy G. R. (1986) Nature 319, 516–518 [DOI] [PubMed] [Google Scholar]
- 40.Horowitz M. C. (1993) Science 260, 626–627 [DOI] [PubMed] [Google Scholar]
- 41.Pacifici R., Brown C., Puscheck E., Friedrich E., Slatopolsky E., Maggio D., McCracken R., Avioli L. V. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5134–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komm B. S., Terpening C. M., Benz D. J., Graeme K. A., Gallegos A., Korc M., Greene G. L., O'Malley B. W., Haussler M. R. (1988) Science 241, 81–84 [DOI] [PubMed] [Google Scholar]
- 43.Roggia C., Gao Y., Cenci S., Weitzmann M. N., Toraldo G., Isaia G., Pacifici R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson B. M., Mundy G. R., Chambers T. J. (1987) J. Immunol. 138, 775–779 [PubMed] [Google Scholar]
- 45.Abu-Amer Y., Erdmann J., Alexopoulou L., Kollias G., Ross F. P., Teitelbaum S. L. (2000) J. Biol. Chem. 275, 27307–27310 [DOI] [PubMed] [Google Scholar]
- 46.Eghbali-Fatourechi G., Khosla S., Sanyal A., Boyle W. J., Lacey D. L., Riggs B. L. (2003) J. Clin. Invest. 111, 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong B. R., Josien R., Lee S. Y., Sauter B., Li H. L., Steinman R. M., Choi Y. (1997) J. Exp. Med. 186, 2075–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong Y. Y., Yoshida H., Sarosi I., Tan H. L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A. J., Van G., Itie A., Khoo W., Wakeham A., Dunstan C. R., Lacey D. L., Mak T. W., Boyle W. J., Penninger J. M. (1999) Nature 397, 315–323 [DOI] [PubMed] [Google Scholar]
- 49.Fouque-Aubert A., Chapurlat R. (2008) Joint Bone Spine 75, 5–10 [DOI] [PubMed] [Google Scholar]