Abstract
Cellular responses to drug treatment show tremendous variations. Elucidating mechanisms underlying these variations is critical for predicting therapeutic responses and developing personalized therapeutics. Using a small molecule screening approach, we discovered how a disease causing allele leads to opposing cell fates upon pharmacological perturbation. Diverse microtubule-depolymerizing agents protected mutant huntingtin-expressing cells from cell death, while being toxic to cells lacking mutant huntingtin or those expressing wild-type huntingtin. Additional neuronal cell lines and primary neurons from Huntington disease mice also showed altered survival upon microtubule depolymerization. Transcription profiling revealed that microtubule depolymerization induced the autocrine growth factor connective tissue growth factor and activated ERK survival signaling. The genotype-selective rescue was dependent upon increased RhoA protein levels in mutant huntingtin-expressing cells, because inhibition of RhoA, its downstream effector, Rho-associated kinase (ROCK), or a microtubule-associated RhoA activator, guanine nucleotide exchange factor-H1 (GEF-H1), all attenuated the rescue. Conversely, RhoA overexpression in cells lacking mutant huntingtin conferred resistance to microtubule-depolymerizer toxicity. This study elucidates a novel pathway linking microtubule stability to cell survival and provides insight into how genetic context can dramatically alter cellular responses to pharmacological interventions.
Keywords: Cell Death, Drug Action, Huntingtons Disease, Microtubules, Rho
Introduction
Different cells respond to identical environmental and physiological changes in diverse ways. Genetic alterations are often key determinants of cellular response to perturbations (1), with such changes modifying response to physiological stresses (2), vulnerability to infections (3), and responses to drugs (4). Understanding how genetic alterations determine differential cellular responses can help predict physiological and therapeutic response to drugs, and aid in the development of selective drugs that are only effective in diseased cells (5). Here, we sought to determine how a genetic alteration that causes a neurodegenerative disease modifies cell survival in response to perturbations.
Huntington disease (HD)2 is an autosomal dominant disease that is characterized by neuronal dysfunction and cell loss mainly in the striatum and cortex (6). HD is caused by a mutation in the huntingtin gene; the mutant allele expresses the mutant huntingtin (htt) protein with an expanded polyglutamine stretch (>36 glutamine repeats) in its amino-terminal region (7). Substantial differences have been observed between neurons of HD animal models or patients and those of normal individuals, including altered gene expression, cell signaling, and response to neuromodulators and stressors (8–10). To identify these alterations in cell survival mechanisms, we used the strategy of small molecule screening in a previously described HD model using immortalized rat striatal neurons (ST14A cells) (11). This model recapitulates several key features of HD. The cells are of striatal origin, the brain region most affected in HD (7), and the mutant transgene is expressed at comparable levels to endogenous wild-type (WT) protein, similar to physiological expression levels observed in HD mouse models and patients. These cells do not undergo spontaneous cell death in tissue culture, a phenotype shared by primary striatal neuronal cultures derived from transgenic HD mouse models (12, 13). Additional features relevant to HD have been demonstrated in this model, including altered caspase activation (11), JNK signaling (14), and adenosine A2 receptor activity (15). Finally, small molecules that are active in this model are efficacious in diverse HD models; some of these are in clinical trials (16).
Using a high-throughput screen we discovered that microtubule (MT)-depolymerizing agents prevented death in mutant htt-expressing cells, but enhanced death in cells lacking mutant htt or those expressing WT htt. Altered sensitivity to MT depolymerization was observed in two additional HD models. We identified a novel signaling pathway involving a microtubule-associated Rho activator, guanine nucleotide exchange factor-H1 (GEF-H1), downstream RhoA-ROCK signaling, that induced connective tissue growth factor (CTGF) and activated prosurvival ERK upon MT depolymerization in mutant htt cells. We thus elucidated a signaling pathway linking MT depolymerization to cell survival and demonstrated a mechanism whereby genetic context alters cell fate upon MT depolymerization.
EXPERIMENTAL PROCEDURES
High-throughput Screen
The high-throughput screening assay has been described previously (17). In brief, 1,500 cells were plated per well in 384-well plates (Costar) in medium containing 0.5% serum that we referred to as serum-deprived medium (SDM), incubated at 33 °C for 4 h, and compounds were added. All compounds were prepared in 384-well plates as 4 mg/ml of solutions in dimethyl sulfoxide (DMSO) except NINDS compounds, which were at 10 mm. “Daughter plates” were prepared from stock plates by a 1:50 dilution in serum-free DMEM (3 μl of compound to 147 μl of DMEM) in 384-well plates and compounds were tested at a final concentration of 4 μg/ml or 10 μm (NINDS compounds). Mutant htt cells were incubated at 39 °C for 3 days; calcein AM dye was added to the wells and fluorescence (excitation 485/emission 535) was measured 4 h later using a Victor3 plate reader (PerkinElmer Life Sciences). Hits were identified as compounds that increased fluorescence more than 50% above DMSO control-treated wells and were reconfirmed in concentration-response experiments.
Cell Culture and Generation of Puromycin-resistant Cell Populations
Rat striatal neuronal cell lines (parental ST14A, WT htt, or mutant htt) were cultured as described previously (17). The STHdhQ7 and STHdhQ111 cell lines were generated by replacing the endogenous mouse exon-1 of htt with a chimeric human-mouse exon 1 containing 7 (Q7) or 111 (Q111) polyglutamine repeats and grown as previously described (18). For generating the puromycin-resistant ST14A cell populations, a puromycin resistance plasmid vector encoding a non-targeting short-hairpin clone (MISSION SHC002, Sigma) was transduced using lentiviral infection into ST14A cells (5 × 104 cells/well of a 6-well plate) and after 2 days, cells were selected in puromycin (3 μg/ml) at the same concentration used to select mutant htt clones (11). This puromycin concentration was sufficient to kill 100% of untransfected ST14A cells by 2 days. After 4 days in puromycin, 10 puromycin-resistant pools of ST14A were selected, expanded, and cryopreserved. For viability studies, the 10 independent puromycin-resistant cell populations and the parental ST14A cells were plated in six-well plates in duplicate (105 cells/well) and treated with DMSO or Pdx (400 nm) in SDM at 39 °C and viability was determined after 2 days using the trypan blue dye exclusion assay.
Primary Striatal Neuronal Cultures (HD94)
Primary cortical neurons were cultured from P0 mice. In this model, mutant htt (exon-1 with 94 polyglutamine repeats) is inducibly expressed using the binary tetracycline-regulatable system (19). In this system, mutant htt expression can be conditionally eliminated by exposure to doxycycline. As control, primary neurons were derived from mice expressing only one of the two transgenes (tetO-exon1 httQ94), where mutant htt expression cannot be achieved and cultured as previously described (19). The genotype (control versus HD94) was assessed by X-Gal and confirmed by PCR genotyping as previously described (12, 19).
Cell Viability Assays
Trypan blue dye exclusion assay, calcein AM assay, and microscopic morphology used to assess cell viability assays have been described in detail previously (17). ATP-based cell viability assay was used to assess viability of STHdh cells. STHdhQ7 and STHdhQ111 cells were plated (15,000 cells per well) in collagen-coated 96-well plates (BD Biosciences). After 24 h, 2.5 μm of each compound was added in 100 μl of fresh medium (DMEM, 10% FBS, 1% penicillin/streptomycin, 500 μg/ml of G418). The cells were incubated with the compound for 24 h and then ATP levels were measured using a luminescence-based ATP detection assay (ATPlite 1step, PerkinElmer Life Sciences). For assaying cell death in HD94 primary neurons, a LIVE/DEAD inclusion/exclusion assay (Molecular Probes) was used in which dead cells incorporate the ethidium D1 homodimer in the nucleus (EthD1 positive cells). All experiments were performed 3 times, and a minimum of 100 neurons were counted per genotype per experiment. Images were analyzed using NIH IMAGE 5.0 and statistical analyses were performed using Statview 4.0.
Flow Cytometry
Cells (mutant htt, WT htt, and ST14A) were seeded at 5 × 105 cells/10-cm dishes in duplicate and grown overnight under permissive conditions. Cells were harvested by trypsinization at various times after serum deprivation and treatments as indicated, spun down once, washed in 1.5 ml of PBS, and resuspended in PBS (300 μl) after another spin. Cells were fixed in ice-cold ethanol (600 μl) and stored at −20 °C. For flow cytometry, cells were kept on ice, spun down, washed with PBS, and then resuspended in 300 μl of PBS containing RNase A (50 μg/ml) and propidium iodide (62.5 μg/ml) and analyzed by FACS (FACS excalibur, BD Biosciences). Data were analyzed by manually setting gates and calculating the percentage in each cell cycle phase.
Lentivirus Preparation and Infections
RhoA (WT and constitutive active RhoA14V) plasmids were kindly provided by Dr. Akiko Mammoto (Departments of Pathology and Surgery, Harvard Medical School, Boston) and lentiviruses were prepared as described (20). Lentiviral supernatant expressing each of the viruses was spin-transfected (2250 rpm, 1 h at 33 °C) using Polybrene (6 μg/ml) onto ST14A cells (5 × 104/well in 6-well plates). Control vector was a lentivirus expressing puromycin resistance gene along with a non-targeting short hairpin RNA (MISSION SHC002, Sigma). RhoA expression was confirmed by Western blotting and viability was determined using a trypan blue dye exclusion assay.
Chemical Libraries, Growth Factors, and Antibodies
The bioactive compound library (NINDS), containing 1,040 compounds, was obtained from Microsource Inc. Other compounds included 20,000 synthetic compounds from a combinatorial library (Comgenex International, Inc.), and 23,685 natural, semi-natural, and drug-like compounds of unknown biological activity from diverse sources (Timtec, Interbioscreen, and Chembridge). All chemicals were obtained from Sigma, unless otherwise indicated. Pdx was used at 400 nm in all experiments, unless otherwise indicated. CTGF (catalog number 120-19), ciliary neurotrophic factor (catalog number 450-50), and BDNF (catalog number 450-02), were obtained from Peprotec Inc. NGF (catalog number N0513) was from Sigma and EGF-1 (catalog number PMG006) was from Invitrogen. BOC-D-fmk (catalog number FK011) was obtained from MP Biomedicals. All phosphospecific antibodies and antibodies to the corresponding non-phosphorylated proteins were from Cell Signaling. The anti-htt antibody (MAB2166) was from Millipore, and the anti-CTGF (H-55, sc-25440, and L-20, sc-14939) and tubulin antibodies (sc-32293) were from Santa Cruz Biotechnology Inc. Alexa 488-conjugated goat anti-mouse secondary antibody was from Invitrogen (catalog number A11029). The cell-permeable Rho inhibitor C3 transferase (catalog number CT-4) was from Cytoskeleton Inc.
Western Blotting and Immunofluorescence
Western blotting was conducted as previously described (21). Image J was used for quantitation. For immunofluorescence, cells were fixed in acetone:methanol (1:1) and incubated with primary mouse anti-tubulin antibody (DM1α) followed by an Alexa 488-conjugated goat anti-mouse secondary antibody. Cells were viewed on a Diaphont 300 microscope (Nikon) and images were acquired using Digital Sight DS-2MBW camera (Nikon).
Microarray and Data Analysis
Mutant htt or ST14A cells were seeded at 0.5 × 106 cells/10-cm tissue culture dish and after overnight incubation at 33 °C were treated with Pdx (400 nm) or vehicle (DMSO, 0.1%) in SDM at 39 °C for 6 h. This early time point would allow sufficient time for MT depolymerization (∼30 min) by Pdx and early gene expression changes that rescue cell death to occur, but minimize late secondary gene expression changes. Total RNA was isolated using TRIzol (Invitrogen) after 6 h. The RNA was quantified (A260 nm), and the quality and integrity of RNA assayed by measuring the absorbance ratio (A260/A280) and gel electrophoresis. The cRNA prepared from the RNA was hybridized with the Rat 231A chip (Affymetrix) that contained 15,923 probe sets including all annotated rat genes and additional expressed sequence tags. Each experiment was performed in triplicate for N548 mutant cells and in duplicate for the ST14A cells, and the ratio of gene expression levels in DMSO relative to Pdx-treated samples was calculated after excluding low intensity transcripts (<50 arbitrary units) from the analysis. A threshold of p < 0.01 (Student's t test) was used as a cut-off to identify significantly altered transcripts. All transcripts that were altered more than 2-fold upon Pdx treatment (relative to DMSO) were considered for further study. The experiment was conducted at the Center for Microarray Technology, Whitehead Institute for Biomedical Research (Cambridge, MA).
siRNA and Transfections
siRNA oligonucleotides were obtained from Sigma, and their sequences were: RhoA, 5′-GUGAAUUAGGCUGUAACUAdTdT; GEF-H1 si#1, 5′-CAUUGCUGGACAUUUCAAUdTdT; si#2, 5′-CAGAUGUGCUGGUGUUUCUdTdT; Cdc42, 5′-GCCUAUUACUCCAGAGACUdTdT; Rac1, 5′-CCAAUACUCCCAUCAUCCUdTdT; and CTGF, 5′-CCUGUCAAGUUUGAGCUUUdTdT3. Control siRNA (Non-targeting siGenome RNA pool #1, catalog number D001206-13-05) was obtained from Dharmacon. Oligonucleotides were transfected with Lipofectamine 2000 (Invitrogen) using the protocol suggested by the manufacturer. For CTGF siRNA transfections, DharmaFECT 1 transfection reagent (ThermoScientific) was used to get adequate knockdown of CTGF because Lipofectamine transfections were inefficient. Briefly, 105 cells were plated per 60-mm dish. After overnight incubation, cells were transfected with 170 nm of each siRNA on two consecutive days. The medium was changed to SDM for the indicated times with or without addition of Pdx. Cells were then harvested for viability determination or protein lysates prepared for Western blotting.
RESULTS
MT Depolymerizing Agents Rescue Cell Death in Mutant htt-expressing Cells
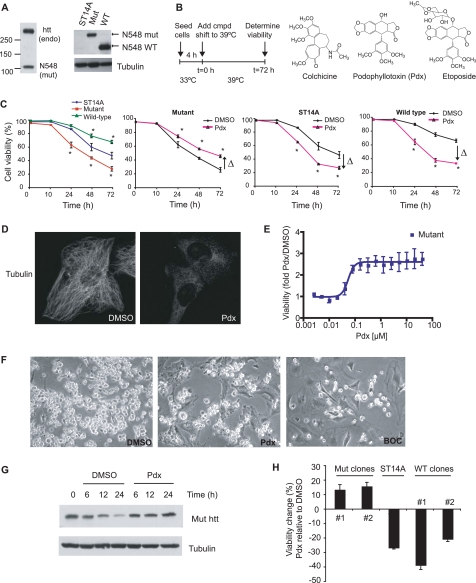
Mutant htt cell lines were obtained from parental ST14A cells by expressing an N-terminal 548-amino acid fragment of mutant human htt containing 128 glutamine repeats, at levels comparable with endogenous htt (Fig. 1A) (11). We used the parental line and a parental derived cell line expressing the WT N-terminal 548-amino acid htt fragment containing 15 glutamine repeats as controls. These cell lines proliferate comparably at the permissive temperature (33 °C) but differentiate upon shifting to the non-permissive temperature of 39 °C (22). Mutant htt cells do not undergo appreciable spontaneous cell death under permissive conditions (11), and neither do primary neurons from HD mice (13). However, serum deprivation, a stress known to sensitize cells to polyglutamine toxicity (11), induced death in mutant htt cells at an enhanced rate relative to parental ST14A cells (Fig. 1C). In contrast, consistent with an established cytoprotective effect of WT htt (23), the WT htt cell line was resistant to serum deprivation-induced cell death (Fig. 1C).
FIGURE 1.
MT depolymerizers rescue mutant htt cell death. A, full-length endogenous WT (endo) and mutant htt (N-terminal 548-amino acid) fragment were detected by Western blotting using the MAB2166 antibody in mutant htt cells (left panel). Mutant htt fragment has decreased electrophoretic mobility compared with the WT htt fragment due to more polyglutamine repeats (Gln128 in mutant versus Gln15 in WT htt, right panel). B, schematic of the design of high-throughput screen. 1,500 cells were plated per well in 384-well plates in serum-deprived medium. After 4 h at 33 °C the cells were shifted to 39 °C. The shift to 39 °C is time (t) 0 for determining cell viability. Structures of two MT-depolymerizing agents (colchicine and podophyllotoxin (Pdx)), and the topoisomerase inhibitor etoposide, a structural analog of Pdx (right panel) are shown. C, mutant, WT htt, and parental ST14A cells were serum deprived and cell viability was assayed by trypan blue dye exclusion assay (left panel). In parallel, these cell lines were serum deprived, treated with Pdx (400 nm), and cell viability determined at the indicated time points. The data are the average ± S.D. for an experiment performed in duplicate. Arrows indicate the direction of change (Δ) in viability of Pdx treated, relative to DMSO (0.1%) treated cells (*, p < 0.05 Student's t test). D, tubulin immunofluorescence in mutant htt cells treated with DMSO or Pdx (400 nm) for 6 h. E, cell viability of a dose dilution of Pdx-treated relative to DMSO-treated mutant htt cells was determined by calcein AM assay, a fluorescence based viability assay (see “Experimental Procedures”). The assay was performed 3 days after serum deprivation. The data are the mean ± S.D. of an experiment performed in triplicate. F, phase-contrast images of mutant htt cells treated with DMSO, Pdx (400 nm), or the pan-caspase inhibitor BOC-D-fmk (BOC, 50 μm) for 2 days. Dying cells detach and are rounded and brighter than live cells. We confirmed that detached, rounded cells were mostly dead (94%) compared with 17% cell death in attached cells using the trypan blue dye exclusion assay. G, cells were treated with DMSO or Pdx (400 nm) over 24 h and mutant htt protein levels were determined by Western blotting. H, cell viability change due to Pdx treatment was determined in parental cells and two N548 mutant and WT htt fragment expressing clones. Cells were incubated at 33 °C overnight, treated with Pdx (400 nm) in SDM, and viability was determined after 3 days at 39 °C. Data are the mean ± S.D. of an experiment in duplicate.
We screened 44,725 compounds, including known bioactive compounds, in a high-throughput viability assay (17) where cell death was induced by a change to 0.5% serum containing medium (SDM), and a shift to non-permissive temperature, 39 °C (Fig. 1B, left panel). We found that structurally diverse MT depolymerizers (24), such as colchicine, vincristine, and podophyllotoxin (Pdx) (see Fig. 1B for structures of colchicine and Pdx), suppressed cell death in mutant htt cells, whereas enhancing death in both parental ST14A cells and WT htt cells (Fig. 1C). In contrast, a pan-caspase inhibitor, BOC-D-fmk, rescued both parental and mutant htt cell lines (17), indicating that MT depolymerization targets a survival mechanism unique to mutant htt cells. Furthermore, etoposide, a structural analog of Pdx (Fig. 1B) that is a topoisomerase inhibitor and does not depolymerize MT (25), and cytochalasin D, an actin depolymerizer (26), were ineffective at rescue (supplemental Fig. S1A and data not shown). Pdx depolymerized MT between 0.5 and 1 h (supplemental Fig. S2 and Fig. 1D) at concentrations that rescued cell death (Fig. 1E). The EC50 for rescue was comparable with the reported EC50 for MT depolymerization by these compounds (∼25 nm for Pdx) (24). We confirmed rescue using three independent cell viability assays (Fig. 1, C, E, and F). We also excluded decreased mutant htt expression upon Pdx treatment as an explanation for the rescue (Fig. 1G). We noted that in cells undergoing cell death, mutant htt levels decreased relative to control cells not induced to undergo cell death; Pdx prevented this decrease in mutant htt levels (Fig. 1G). This relative stabilization of mutant htt correlated with rescue of cell death upon Pdx treatment, and was likely due to suppression of mutant htt degradation by caspases because htt is an established substrate of caspase (27, 28) and these cells undergo caspase-dependent cell death (17). Furthermore, caspase inhibition using BOC-D-fmk also prevented the decrease in mutant htt (data not shown). These results indicated that the rescue was linked to MT depolymerization.
MT Depolymerization-induced Rescue Is Linked to Mutant htt Expression
MT depolymerization rescued cell death in multiple clonal cell lines expressing N548 mutant htt, whereas enhancing cell death in parental and WT htt expressing cell lines (Fig. 1H). Pdx was also protective in a full-length mutant htt cell line, but toxic in a full-length WT htt expressing cell line (29) (supplemental Fig. S3). Puromycin selection used to generate mutant htt cell lines from parental ST14A cells did not alter cell survival upon MT depolymerization; 10 puromycin-resistant cell populations generated from ST14A cells (see “Experimental Procedures”) showed comparable cell death to parental cells upon serum deprivation, and Pdx did not rescue these cells (supplemental Fig. S4). These results indicated that mutant htt expression was involved in the rescue upon MT depolymerization.
Mutant htt Cells Are Selectivity Resistant to MT Depolymerizer Toxicity
The selective effect of MT depolymerization in mutant htt cells was not limited to serum deprivation conditions. Mutant htt cells were resistant to the toxicity of MT depolymerizers compared with parental cells under permissive growth conditions at 33 °C and in 10% serum containing medium (supplemental Fig. S5A). In contrast, the MT-stabilizing agent Taxol (30), slightly enhanced toxicity in mutant compared with parental cells. The limited toxicity of Taxol was not due to inadequate concentrations; the concentrations used were severalfold greater than the reported EC50 for MT stabilization (31) and Taxol-induced MT stabilization in mutant htt cells (supplemental Fig. S1C). Both cell lines were equally sensitive to cytotoxic agents, such as Etoposide (a DNA-damaging agent), actinomycin D (a transcriptional inhibitor), and staurosporine (a nonspecific kinase inhibitor) (supplemental Fig. S5A) indicating that mutant htt cells are not generally resistant to cytotoxic agents. Finally, 3-nitropropionic acid, a mitochondrial complex II inhibitor used to chemically induce an HD-like phenotype (7), was more toxic in mutant htt relative to parental cells, suggesting the relevance of this model to disease mechanisms (supplemental Fig. S5A). These results indicated that mutant htt cells were selectively resistant to MT depolymerization.
Mutant htt Attenuates Toxicity of MT Depolymerizers in Diverse HD Models
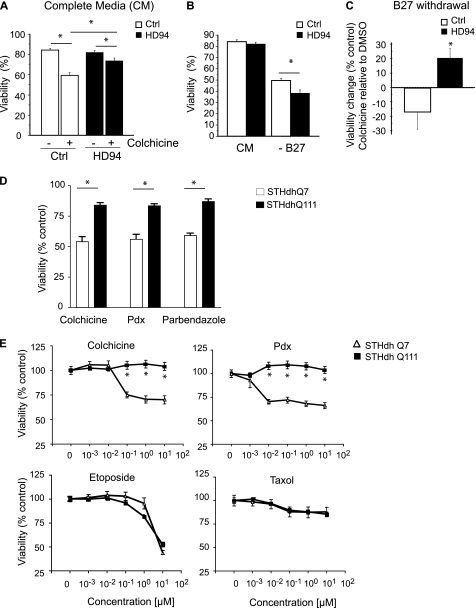
To determine the relevance of these findings, we tested the survival response upon MT depolymerization in two additional HD models. In the first model, mutant huntingtin (exon-1 with 94 polyglutamine repeats) was conditionally expressed in neurons using a tetracycline-regulatable system in a mouse HD model (see “Experimental Procedures”) (19). Control primary neurons were derived from mice lacking mutant htt expression (19). Under baseline conditions, neurons from HD94 and control animals demonstrated comparable rates of cell death (Fig. 2A), similar to the ST14A model. However, HD94 neurons were markedly resistant to MT depolymerizer toxicity compared with control neurons (p = 0.0006) (Fig. 2A). We also tested whether MT depolymerization was neuroprotective in this model. Because primary neurons are routinely cultured in serum-free medium with growth supplement, we achieved trophic factor deprivation by withdrawing the B27 supplement from the medium. B27 withdrawal was more toxic in HD94 neurons than control neurons (p = 0.0074) (Fig. 2B). Importantly, this toxicity was suppressed by MT depolymerization in HD94 neurons, but exacerbated in control neurons (p < 0.05) (Fig. 2C), similar to the result in ST14A cell culture model.
FIGURE 2.
Diverse HD models demonstrate altered sensitivity to MT depolymerization. A, primary striatal neurons from HD mice (HD94) are more resistant to colchicine-induced toxicity than control neurons. After 16 days in vitro (DIV16) neurons were administered colchicine (10 μm) or vehicle (DMSO) and cell death was assessed 96 h later (see “Experimental Procedures”). Colchicine was significantly less toxic to HD94 neurons (analysis of variance, p = 0.0006) (*, p < 0.05). B, B27 (medium supplement) withdrawal is more toxic in HD94 neurons than control neurons: medium from DIV14 neurons was exchanged with complete medium or medium lacking B27, and assessed for cell death 48 h later. Fisher post hoc analysis revealed that under complete medium (CM), cell death was comparable in control (Ctrl) and HD94 neurons (p = 0.3276). B27 withdrawal decreased viability in both genotypes (p < 0.0001); however, it was significantly more toxic in HD94 neurons than control (*, p = 0.0074). C, medium from DIV14 neurons was exchanged with medium lacking B27 in the presence or absence of colchicine (0.1 μm) and cell viability was assessed 48 h later. Viability is shown relative to DMSO-treated cells. Analysis of variance revealed B27 withdrawal induced toxicity was significantly (*, p < 0.05) suppressed by colchicine treatment in HD94, but not in control neurons. D, immortalized striatal neurons from HD knock-in mice (WT, STHdhQ7, and mutant, STHdhQ111) were treated with DMSO or various MT inhibitors, each at 2.5 μm, and cell viability was determined after 24 h using a luminescence-based ATP assay (see “Experimental Procedures”). Cell viability was normalized to DMSO-treated STHdhQ7 and STHdhQ111 cells and the data are the mean ± S.E., of an experiment performed in triplicate. E, STHdhQ7 and STHdhQ111 were treated with a dilution series of colchicine, Pdx, etoposide, or Taxol and viability was determined as in D. Data are the mean ± S.E. of an experiment performed in triplicate (*, p < 0.05 Student's t test).
In parallel, we identified MT depolymerizing agents in an unbiased small molecule screen performed in striatal cell lines derived from a knock-in HD mouse model. In this model endogenous murine htt was replaced either with htt containing normal (Gln7) or pathogenic (Gln111) glutamine repeats to generate STHdhQ7 and STHdhQ111 cell lines (18). STHdhQ111 cells were selectively resistant to diverse MT depolymerizing agents at doses that correlated with concentrations known to cause MT depolymerization (Fig. 2, D and E). However, STHdhQ111 cells were not generally resistant to cytotoxic agents such as Taxol and etoposide (Fig. 2E). These data demonstrate an altered response to MT depolymerization in three distinct HD models.
MT Transport, G2/M Arrest, and htt Aggregation Are Not Involved in Rescue
Because htt is highly associated with MT (32), we tested several previously described mechanisms linking MT to htt function. Mutant htt is thought to contribute to toxicity by decreasing MT-based transport (33, 34). It is clear that MT depolymerization could not possibly improve MT-based transport defects reported in HD models, because MT depolymerizers disrupt all MT-based transport. Inhibition of MT-based motor protein Eg5 (35), using Monastrol, was ineffective at rescue (supplemental Fig. S1A). Thus augmenting axonal transport was not relevant to the rescue upon MT depolymerization. Furthermore, simply altering MT stability did not rescue cell death. Taxol, a MT stabilizing agent (24), enhanced cell death in mutant htt cells at concentrations that stabilized MT, as determined by tubulin immunofluorescence (supplemental Fig. S1, A–C). Furthermore, Taxol could not prevent the rescue caused by Pdx. In cells co-treated with Pdx and Taxol, Pdx was dominant and induced depolymerization (supplemental Fig. S1, B and C).
Because Taxol is similar to MT depolymerizing agents in inducing cell cycle arrest (31), the above result also suggested that the selective rescue of mutant cells was independent of growth differences between mutant and parental or WT htt cell lines. Consistent with a previous report (11), we found similar proliferation rates of these cell lines based on cell cycle analysis and cell growth under permissive conditions (supplemental Fig. S6, A and C). We confirmed a similar degree of G2/M phase cell cycle arrest upon MT depolymerizer treatment in these cell lines, indicating the differential rescue is not due to altered cell cycle arrest of mutant htt cells (supplemental Fig. S6, A and B). We also considered that mutant htt toxicity may occur in a cell cycle phase distinct from G2/M and therefore arresting cells in G2/M prevents toxicity. To test this hypothesis, we pretreated mutant htt cells with Taxol or Pdx for 24 h before serum deprivation. This treatment caused a G2/M arrest in mutant htt cells, the percentage of cells in G2/M increased from 13% in cycling cells to 36 and 44% in Taxol and Pdx-pretreated cells, respectively (supplemental Fig. S7A). However, serum deprivation of cells pre-arrested in G2/M did not enhance the protective effect of Pdx, and Taxol was still toxic relative to cycling cells (supplemental Fig. S7B), indicating that cell cycle arrest in G2/M per se was not involved in enhancing survival in mutant cells. Furthermore, pre-arresting cells at G1/S by hydroxyurea treatment (36) for 24 h before serum deprivation did not prevent rescue by MT depolymerization (supplemental Fig. S7D), despite the fact that hydroxyurea-treated cells did not proliferate or progress to G2/M (supplemental Fig. S7, E and F). These results indicate that the rescue was independent of arrest in G2/M. Finally, it has been suggested that mutant htt aggregation can be affected by MT stability (37, 38). Mutant htt showed diffuse cytoplasmic localization with occasional cells (<1%) showing visible aggregates (supplemental Fig. S8) (21). This is consistent with studies showing low incidence of aggregates in cells expressing large (>N-terminal 528 amino acids) mutant htt fragments (39, 40). The percentage of cells with aggregates was unaffected by MT depolymerization (data not shown), indicating that aggregation was unlikely to play a role in toxicity or rescue upon MT depolymerization in this model.
MT Depolymerization Alters Gene Expression in Mutant htt Cells
Altered transcription is implicated in HD pathophysiology (6), and MT dynamics can regulate gene expression (41). We tested if gene expression changes upon MT depolymerization affected cell survival. DNA microarray analysis (see “Experimental Procedures”) revealed that 0.5% of transcripts were significantly (p < 0.01) altered upon MT depolymerization in mutant htt cells. Genes whose expression changed more than 2-fold upon Pdx treatment are listed in supplemental Table S1. We chose the ST14A cell lines as controls for mutant htt rather than WT htt cells because ST14A and mutant htt cells had more similar cell death kinetics in contrast to WT htt cells, which were protected from cell death upon serum deprivation (Fig. 1C). We were concerned that the large difference in cell death between mutant and WT htt cells could make it difficult to distinguish gene expression changes due to Pdx from those due to differential cell death. Comparative profiles of Pdx-treated mutant and ST14A cells revealed few transcripts that were similarly altered in both cell lines, indicating differential transcriptional response to MT depolymerization in these cell lines (supplemental Table S1).
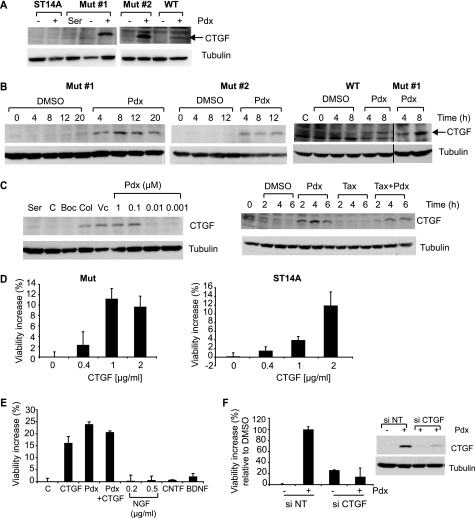
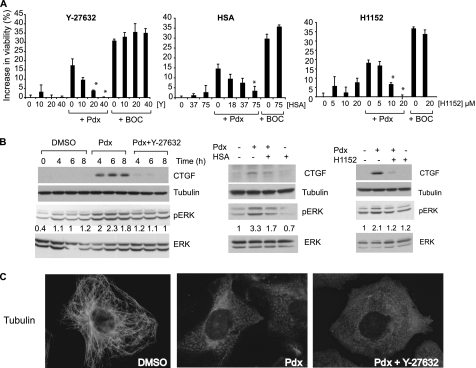
CTGF was one of the most highly induced transcripts (>4-fold increase) upon MT depolymerization in mutant cells, but was not significantly increased in ST14A cells (p > 0.01) (supplemental Table S1). CTGF protein induction was confirmed in mutant htt cells treated with Pdx (400 nm), but was not detectable in parental cells (Fig. 3A). CTGF is involved in cell migration, extracellular matrix formation, and cell survival (42). Because CTGF can enhance cell survival in several models (43), we focused on this factor. CTGF induction upon Pdx treatment correlated with rescue, as demonstrated by the following observations. First, CTGF was induced in two mutant htt-expressing cell lines that were rescued by Pdx (400 nm) treatment, but was not substantially induced in ST14A. Although in WT htt cells, CTGF was induced upon Pdx treatment, the levels were lower compared with mutant htt cells (Fig. 3, A and B). Second, several structurally diverse MT depolymerizing agents induced CTGF, whereas the MT stabilizer, Taxol (1 μm) did not (Fig. 3C); Taxol also could not prevent the induction of CTGF by Pdx that was consistent with the dominance of Pdx over Taxol in destabilizing MT (supplemental Fig. S1C). Third, the pan-caspase inhibitor, BOC-D-fmk (50 μm), rescued cell death (17) (Fig. 1F), but without inducing CTGF, showing that CTGF induction was not simply a consequence of cell survival (Fig. 3C). Fourth, the concentration-response for rescue by Pdx paralleled that for CTGF induction; lower concentrations (10 nm or lower) that did not rescue cell death were also ineffective in inducing CTGF (Figs. 1E and 3C). Finally, CTGF induction occurred by 4 h (Fig. 3B), and preceded cell death, which begins ∼10 h after serum deprivation (Fig. 1C).
FIGURE 3.
MT depolymerization-induced CTGF up-regulation rescues cell death. A, CTGF protein levels were monitored by Western blotting in parental ST14A cells, two mutant htt-expressing (Mut) cell lines, and a WT htt-expressing cell line with or without Pdx (400 nm) treatment for 6 h in SDM. Mutant cells in 10% serum containing medium (Ser) served as a control for no cell death. Tubulin was used as a loading control. B, CTGF protein levels were determined at the indicated times after Pdx (400 nm) treatment in two N548 mutant htt expressing clones (Mut#1 and Mut#2) and in the comparable N548 WT htt cell line. WT htt cells in 10% serum served as controls (C). C, mutant htt cells were in serum-containing medium (Ser) or were serum-deprived and treated with vehicle DMSO (C), caspase inhibitor BOC-D-fmk (Boc, 50 μm), colchicine (Col, 1 μm), vincristine (Vc, 1 μm), and a dose series of Pdx. CTGF levels were determined by Western blotting after 6 h (left panel). Mutant htt cells were treated with Pdx (400 nm) or Taxol (1 μm) alone, or in combination, and CTGF levels were monitored by Western blotting (right panel). D, mutant htt or parental cells were treated with recombinant CTGF and cell viability was assessed after 2 days in SDM. Data are mean ± S.D. of an experiment performed in duplicate and representative of two independent experiments. E, mutant htt cells were treated with CTGF (1 μg/ml), Pdx (400 nm), CTGF (1 μg/ml) + Pdx (400 nm), nerve growth factor (NGF, 0.5 μg/ml), ciliary neurotrophic growth factor (CNTF, 0.2 μg/ml), or BDNF (0.2 μg/ml) and cell viability was determined after 2 days in SDM. F, mutant htt cells were transfected with non-targeting (NT) or CTGF siRNA for 2 days, and medium was changed to SDM with DMSO or Pdx (400 nm). Cell viability was determined after an additional 2 days and expressed on a scale relative to DMSO set as 0% and Pdx as 100%. Data are mean ± S.D. of an experiment performed in duplicate and representative of two independent experiments. In parallel, mutant htt cells transfected with indicated siRNAs were treated with Pdx (400 nm) or DMSO for 6 h and CTGF levels were determined by Western blotting. Tubulin was a loading control.
Next, we tested if exogenous CTGF could enhance survival. In these experiments, we assumed that adding CTGF to the outside of cells had similar effects as endogenously produced CTGF. Exogenous recombinant CTGF enhanced survival in a concentration-dependent manner in both mutant htt and ST14A cells indicating that mutant htt cells were not selectively responsive to CTGF (Fig. 3D). CTGF treatment did not increase cell survival in WT htt cells (data not shown); this could be due to the fact that these cells show little cell death upon serum deprivation and thus a proportionately smaller degree of protection would be more difficult to detect (Fig. 1C). The rescue by different batches of recombinant CTGF varied, ranging from 10 to 65% of the rescue obtained with Pdx (data not shown). In contrast, other growth factors, such as nerve growth factor (NGF), ciliary neurotrophic factor, and brain-derived growth factor (BDNF), at concentrations known to activate their respective receptors (44, 45), did not substantially rescue cell death (Fig. 3E). Although BDNF is neuroprotective in certain HD models, we observed only a small increase (∼5%) in viability (Fig. 3E and data not shown) and this may be related to the absence in vitro of diverse mechanisms that are implicated in the in vivo protective effect of BDNF (46). Finally, we tested if CTGF induction was required for rescue. Abrogation of CTGF induction using siRNA prevented the rescue upon MT depolymerization (Fig. 3F). These data indicate that CTGF induction correlates with, and contributes to rescue upon MT depolymerization.
MT Depolymerization Activates Pro-survival ERK Signaling in Mutant htt Cells
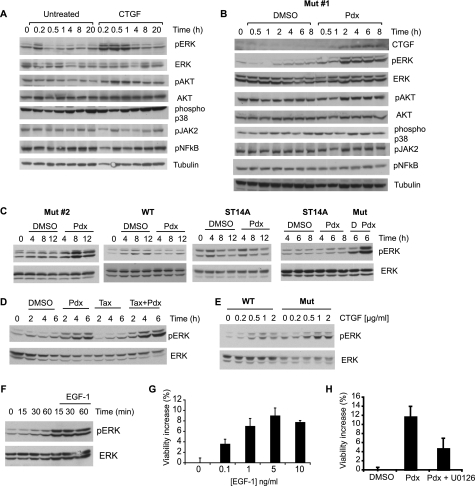
CTGF is a secreted protein that affects cellular signaling via diverse extracellular receptors, including the insulin-like growth factor-1 and EGF-1 receptors (42). We tested the effect of exogenous CTGF, assuming similarity of effects as endogenous CTGF, on several signaling pathways in mutant htt cells. We used antibodies that detect activating phosphorylations of ERK, AKT, p38, NFκB, and Jak2 and detected specific activation of pro-survival ERK, and a reproducible, but lesser amount of AKT; the other pathways probed were not activated (Fig. 4A). We hypothesized that if CTGF mediated the protective effects of MT depolymerization, ERK activation should follow MT depolymerization. A time course experiment revealed MT depolymerization by 1 h (supplemental Fig. S2) and CTGF induction, increased ERK phosphorylation, and to a lesser extent AKT phosphorylation, by 2 h after Pdx treatment (Fig. 4B). ERK was activated upon MT depolymerization in another mutant htt cell line (Mut#2) but not in parental ST14A or WT htt cells (Fig. 4C). In contrast, MT stabilization by Taxol treatment did not activate ERK (Fig. 4D). We also found that recombinant CTGF activated ERK to a lesser extent in WT htt compared with mutant htt cells (Fig. 4E). This suggested that a certain threshold of activated ERK was required for survival, and that lower CTGF induction in WT htt cells (Fig. 3B), together with decreased ERK pathway activation by CTGF may contribute to the lack of survival upon Pdx treatment in WT htt cells. Finally, we confirmed that ERK pathway activation was indeed protective in mutant htt cells, as previously reported (21). EGF-1, a physiological ERK pathway activator, increased ERK phosphorylation and enhanced mutant htt cell survival. Conversely, pharmacological ERK inhibition suppressed the rescue induced by Pdx (Fig. 4, F–H). Together, these results indicated that differential ERK activation upon MT depolymerization selectively enhanced mutant htt cell survival.
FIGURE 4.
ERK survival signaling is activated by CTGF and MT depolymerization in mutant htt cells. A, mutant htt cells were treated with CTGF (1 μg/ml), or untreated in SDM, and the activity of diverse signaling pathways was monitored by Western blotting using phosphospecific antibodies. B, mutant htt cells were treated with DMSO or Pdx (400 nm), and CTGF levels and activity of several signaling pathways were monitored by Western blotting using phosphospecific antibodies. C, Pdx selectively activates ERK in mutant htt but not in ST14A or WT htt cells. Mutant htt clone 2 (Mut#2), WT htt and ST14A cells were treated with DMSO (D) or Pdx (400 nm) under non-permissive conditions and ERK activity was determined using a phospho-ERK specific antibody. D, mutant htt cells were treated with Pdx (400 nm) or Taxol (1 μm) alone, or in combination. ERK activity was monitored using Western blotting. E, WT htt cells show attenuated ERK activation upon CTGF treatment. Mutant htt and WT htt cells were treated with a dose dilution of CTGF and ERK activity was monitored after a 1-h treatment. F, mutant htt cells were untreated or treated with EGF-1 (5 ng/ml) and ERK activity was monitored using a phospho-ERK specific antibody. G, dose response for increase in mutant htt cell viability upon EGF-1 treatment. Viability was determined using trypan blue dye exclusion assay after under 2 days in SDM. H, mutant htt cells were treated with Pdx (400 nm) alone or with U0126 (0.5 μg/ml), an inhibitor of ERK activation, and cell viability was determined after 2 days in SDM as in G. Data are mean ± S.D. of an experiment performed in duplicate.
Rho Kinase Inhibitors Suppress CTGF Induction, ERK Activation, and Survival
Because CTGF induction contributed to survival, we sought to identify pathways involved in CTGF induction to gain further insight into the survival mechanism. Rho-associated kinase (ROCK) is implicated in cytoskeleton-based cellular signaling, migration, apoptosis (47), and in CTGF induction upon mechanical stress (48). We treated mutant htt cells with three specific ROCK inhibitors Y-27632, H1152, and hydroxyfasudil (49, 50), either alone, or in combination with Pdx or the pan-caspase inhibitor BOC-D-fmk. These inhibitors alone had little effect on cell viability. However, they specifically abrogated the rescue of cell death by Pdx, but not by BOC-D-fmk (Fig. 5A). They also prevented CTGF induction and ERK activation caused by Pdx (Fig. 5B), but without preventing MT depolymerization (data shown for Y-27632, Fig. 5C). The fact that similar results were observed using these structurally distinct ROCK inhibitors (Fig. 5, A and B) suggests that this suppression was unlikely an off-target effect of these inhibitors. Additionally, the resistance to microtubule-depolymerizing agents observed in STHdhQ111 cells was partially abrogated upon treatment with Y-27632 (supplemental Fig. S5B), suggesting that ROCK or a closely related kinase was responsible for resistance to MT depolymerization in this model as well.
FIGURE 5.
Rho kinase (ROCK) inhibitors suppress rescue upon MT depolymerization. A, mutant htt cells were treated with a dilution series of three ROCK inhibitors Y-27632 (Y), hydroxyfasudil (HSA), or H1152 alone or in combination with Pdx (400 nm) or BOC-D-fmk (BOC, 50 μm) and cell viability was determined by a trypan blue dye exclusion assay. The increase in cell viability relative to DMSO-treated cells was determined after 2 days in SDM. Data are the mean ± S.D. of an experiment performed in duplicate. (*, p < 0.05, Student's t test). B, mutant htt cells were treated with DMSO, Pdx (400 nm), or Pdx (400 nm) in combination with individual ROCK inhibitors (Y-27632, 40 μm; hydroxyfasudil (HSA), 75 μm; H1152, 20 μm) and levels of CTGF, phosphorylated and total ERK were determined by Western blotting at the indicated time points for Y-27632, or 6 h after treatment, for hydroxyfasudil and H1152 treatments. Tubulin was a loading control. The experiments are representative of at least two independent experiments for each treatment. pERK and ERK were quantitated using Image J (NIH) and the level of pERK was normalized to ERK and the 6-h time in SDM was set as 1 in each treatment. pERK levels relative to the 6-h time point are provided below the blots. C, mutant htt cells were treated with DMSO, Pdx (400 nm), or Pdx (400 nm) + Y-27632 (40 μm) and the tubulin network was visualized by immunofluorescence 6 h after treatment.
Increased RhoA Protein in Mutant htt Cells Mediates Selective Survival
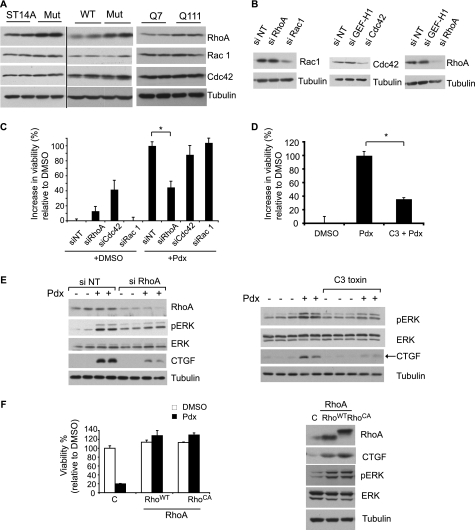
To further assess if a ROCK pathway-dependent mechanism was involved in the rescue upon MT depolymerization, we tested if RhoA, the upstream activator of ROCK (51) was required. RhoA is a member of a family of small GTPase proteins that includes Rac1 and Cdc42. These proteins act as molecular switches that transduce signals by cycling between inactive (GDP-bound) and active (GTP-bound) forms, where the GTP-bound active RhoA activates effectors such as ROCK (51). RhoA protein levels were higher in mutant htt relative to parental or WT htt cells, whereas the other Rho family proteins had similar levels (Fig. 6A). RhoA levels were also increased in the STHdhQ111 cell line compared with STHdhQ7 cells (Fig. 6A).
FIGURE 6.
RhoA signaling is required for rescue induced by MT depolymerization. A, the levels of Rho GTPases in parental ST14A, WT, and mutant htt cells (left and middle panels) and in STHdhQ7 and STHdhQ111 cells were determined by Western blotting (right panel). Tubulin was the loading control. B, mutant htt cells were transfected either with siRNA oligonucleotides directed against the indicated Rho GTPases or a non-targeting (NT) siRNA pool, and the levels of the respective proteins were assessed by Western blotting. C, mutant htt cells were transfected with the indicated siRNAs for 2 days, and the medium was changed to SDM with DMSO or Pdx (400 nm). Cell viability was determined after an additional 2 days and expressed on a scale relative to DMSO set as 0% and Pdx as 100% (*, p < 0.05, Student's t test). D, mutant htt cells were treated with Pdx (400 nm) alone or in combination with C3 Rho inhibitor (2 μg/ml) and cell viability was determined as in C (*, p < 0.05, Student's t test). E, mutant htt cells were either transfected with siRNA or treated with C3 transferase. The cells were then treated with Pdx (400 nm) or DMSO in duplicate for 4 h and the levels of the indicated proteins determined by Western blotting. F, ST14A cells were lentivirally transduced with expression vectors for control (puromycin resistance gene), WT (RhoAWT), or a constitutive active RhoA (RhoACA), and incubated for 2 days at 33 °C. Cell viability was determined after an additional 2 days of serum deprivation at 39 °C with or without Pdx (400 nm) treatment (left panel). RhoA, CTGF, pERK, and ERK levels were determined by Western blotting. RhoACA has lower electrophoretic mobility compared with RhoAWT. Tubulin was a loading control (right panel).
We tested if RhoA was relevant to rescue upon MT depolymerization. RNAi-mediated knockdown of RhoA, but not of Cdc42 or Rac1, attenuated the rescue by Pdx (Fig. 6, B and C). We noted that Cdc42 knockdown enhanced survival in DMSO-treated cells, but did not further enhance survival induced by Pdx. The reasons for increased viability by Cdc42 knockdown but a lack of further increase with Pdx are unclear. Because Cdc42 and RhoA are often functionally antagonistic (52), Cdc42 knockdown may enhance RhoA function, the pathway activated by Pdx and thus not confer additional protection. Alternatively, Pdx may interfere with the survival pathway activated by Cdc42 knockdown. Next, we found that C3 transferase, a toxin that selectively inhibits RhoA (53) attenuated rescue (Fig. 6D). Both treatments to inhibit RhoA attenuated downstream CTGF induction and ERK activation (Fig. 6E). Conversely, increasing or activating RhoA in ST14A cells by expressing WT or constitutively active RhoA (RhoA14V) conferred resistance to the toxic effects of MT depolymerizing agents, making their response similar to mutant cells (Fig. 6F). RhoA overexpression also induced CTGF and activated ERK (Fig. 6F). These results suggest that MT depolymerization-induced survival is mediated at least in part via RhoA-ROCK, and that elevated RhoA levels in mutant htt cells relative to parental cells contribute to the selective rescue upon MT depolymerization.
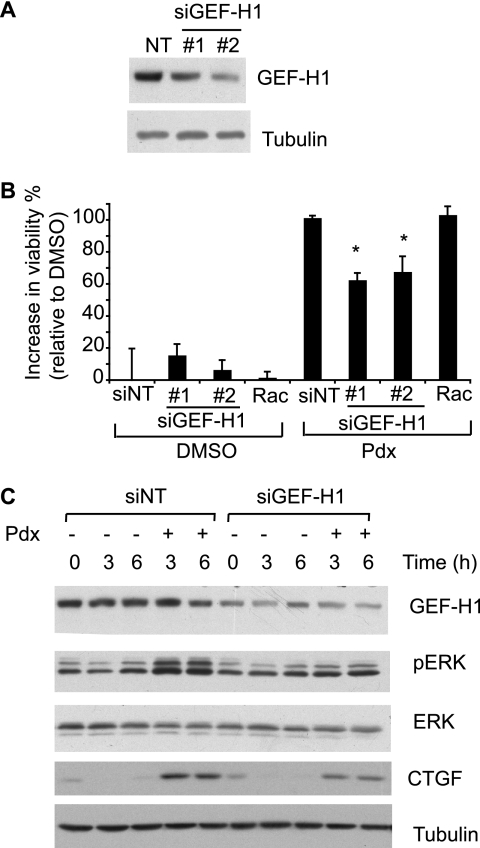
MT-associated Rho Activator GEF-H1 Is Required for Rescue
Finally, we sought to identify the link between RhoA and MTs. GEF-H1 is a MT-associated RhoA activator that is released and activated upon MT depolymerization (54). GEF-H1 activates RhoA by enhancing the rate of exchange of bound GDP for GTP. RNAi-mediated GEF-H1 knockdown (over 2 days) using two different siRNA oligonucleotides suppressed rescue upon Pdx treatment (Fig. 7, A and B). Knockdown by siRNA treatment for longer duration (3 days) caused more complete abrogation of rescue (data not shown). Furthermore, GEF-H1 knockdown attenuated ERK activation and CTGF induction (Fig. 7C). Together, these data indicate that the GEF-H1-RhoA-ROCK signaling pathway links MT stability to cell survival by inducing CTGF and activating ERK (Fig. 8).
FIGURE 7.
GEF-H1 knockdown attenuates MT depolymerization-induced survival. A, mutant htt cells were transfected with two distinct siRNAs directed against GEF-H1 (#1 and #2) or equal amounts of non-targeting siRNA pool (NT) and the knockdown assessed by Western blotting. B, mutant htt cells were transfected with the indicated siRNAs for 2 days and the medium was changed to SDM with DMSO or Pdx (400 nm). Cell viability was determined after an additional 2 days and expressed on a scale relative to DMSO (0%) and Pdx (100%). Both non-targeting (NT) and Rac-1 siRNAs served as negative controls. Data are mean ± S.D. of an experiment performed in duplicate and representative of two independent experiments (*, p < 0.05, Student's t test). C, mutant htt cells were transfected with GEF-H1 (siRNA #1) or NT siRNA. Transfected cells were treated with Pdx (400 nm) or DMSO for the indicated times and levels of the indicated proteins were determined by Western blotting. The results are representative of two independent experiments.
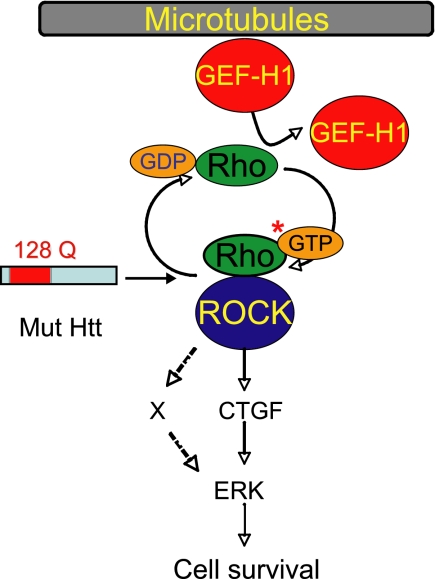
FIGURE 8.
Model of the cell survival pathway activated in mutant htt cells upon MT depolymerization. MT depolymerization releases GEF-H1 that activates RhoA by inducing exchange of GTP for GDP on RhoA. GTP-bound RhoA activates downstream ROCK, which up-regulates CTGF and activates ERK survival signaling. It is possible that mediators (X), in addition to CTGF, link GEF-H1-Rho-ROCK to ERK.
DISCUSSION
Understanding how genetic context alters cellular response to perturbations is important, especially for predicting response to drugs and developing selective therapeutics (1). Using a chemical screening approach, we discovered how genetic context leads to opposing cell fates. Mutant htt-expressing cells that were induced to undergo cell death by serum deprivation were protected by MT depolymerization; in contrast, the same treatment was cytotoxic in cells lacking mutant htt or those overexpressing an identical WT htt fragment (Fig. 1C and supplemental Fig. S3). We observed a similarly altered cell survival response to MT depolymerizing agents in two additional HD models (Fig. 2, A–E). The genotype-selective rescue was at least partially dependent upon increased RhoA protein levels in mutant cells relative to ST14A cells (Fig. 6A). This increase in RhoA selectively activated pro-survival ERK upon MT depolymerization in mutant htt but not in parental or WT htt cells (Fig. 4, B and C). Finally, we found that MT-associated Rho activator, GEF-H1, links MT assembly to RhoA-ROCK signaling and cell survival (Fig. 7). GEF-H1 is uniquely positioned to transduce signals upon MT depolymerization; it is bound to MTs and is released and activated upon MT depolymerization (54). These data have elucidated a pathway that links MT assembly to cell survival specifically in the presence of mutant htt (Fig. 8).
This study raises several points. First, the selective rescue of cell death in mutant htt cells is intriguing because MT depolymerization causes cell death in various cell types, and is the rationale for using these agents for cancer therapy (55). However, exceptions to the generalized cytotoxicity of these agents exist; MT depolymerizing agents enhance survival in cardiac myocytes (56, 57), suppress Fas-mediated death of hepatocytes (58) and are protective in an in vivo model of hereditary spastic paraplegia, a neurodegenerative disorder (59). Additionally, resistance to MT depolymerizing agents is a frequent problem in cancer chemotherapy. Several mechanisms including tubulin mutations that alter tubulin binding to MT depolymerizing agents or stabilize MT, overexpression of tubulin isoforms, and drug efflux transporters are implicated in resistance to MT depolymerizers (60). Because MT depolymerizing agents clearly destabilized MT in mutant htt cells at the reported EC50 for these compounds (Fig. 1, D and E), these known mechanisms of resistance are unlikely explanations for the resistance observed in mutant htt cells. However, these mechanisms explain resistance in relatively few resistant cell lines and tumors. In the vast majority of cases, the molecular mechanisms of resistance to MT depolymerizing drugs remain obscure, although altered cell signaling and cell survival pathways are implicated (60). Our results show that protection from toxicity is caused, at least in part, by specific activation of ERK survival signaling in mutant htt cells upon MT depolymerization (Fig. 4C). This is dependent on increased RhoA levels in mutant htt relative to parental cells, because RhoA overexpression in parental cells activates ERK and confers resistance to MT depolymerizer toxicity (Fig. 6F). Because GEF-H1 and RhoA are frequently increased in cancers (54, 61), it would be interesting to test if altered GEF-H1-RhoA signaling contributes to resistance to MT depolymerizer toxicity in other cellular systems.
Our findings also reveal how distinct cellular fates upon MT depolymerization may be explained by differential cell signaling. MT depolymerization can have distinct effects on cell signaling depending upon cell type (41), including activation of both cell survival (NFκB) and cell death signaling (p38 MAPK) pathways (62, 63). Although MT depolymerization did not affect NFκB or p38 MAPK signaling in mutant htt cells, we observed selective pro-survival ERK activation in mutant htt but not in ST14A and WT htt cells (Fig. 4, B and C). ERK activation in mutant htt cells was dependent upon the RhoA-ROCK pathway (Figs. 5B and 6E) with elevated RhoA protein in mutant htt-expressing cells contributing to the selective cell survival by up-regulating CTGF and activating ERK. CTGF is a transcriptional target of RhoA (64) and our results are consistent with this mechanism; RhoA inhibition, using siRNA or pharmacological inhibitors, and ROCK inhibition using structurally diverse ROCK inhibitors suppressed CTGF induction, ERK activation, and rescue upon MT depolymerization, whereas RhoA overexpression induced CTGF and activated ERK (Figs. 5 and 6, E and F). Although CTGF induction correlated with rescue by Pdx, and preventing CTGF induction using siRNA suppressed the rescue upon Pdx treatment (Fig. 3F), it is possible that additional mediators contribute to ERK activation and cell survival (Fig. 8).
It is notable that MT depolymerizers have recently been shown to determine other cell phenotypes by impacting transcription. For example, MT depolymerizing agents increase bone growth in vivo by inducing Gli2 expression (65) and enhance oxidative phosphorylation by inducing PGC-1α (66) in muscle cells, indicating a wider role for MT depolymerization induced transcriptional changes in regulating cellular phenotypes.
These findings raise questions regarding HD pathophysiology. We observed resistance to, or enhanced survival, upon MT depolymerization in three independent models (Figs. 1C, 2, A–C, and supplemental Fig. 5), using neuronal cell lines expressing physiological levels of mutant htt (Fig. 1A) and primary neurons from HD mice. This suggests that our results are based on a well conserved pathophysiological mechanism in HD. Previous studies implicate multiple mechanisms, including altered MT function in HD. Htt is highly associated with MT and evidence exists for mutant htt decreasing MT-based axonal transport (34, 67, 68). Because MT disruption would exacerbate rather than reverse the reported transport defects, this excluded an increase in MT-based transport as a mechanism for rescue in our model. One study reported that overexpression of mutant htt destabilized MT and caused toxicity that was alleviated by the MT stabilizing agent Taxol (69). However, in that study, mutant htt toxicity was suppressed by a narrow concentration range (5–20 nm) of Taxol, even though Taxol stabilized MT without toxicity up to 200 nm, raising the possibility that a subtle change in MT dynamics or possible off-target effects of Taxol (70) could mediate the rescue. We tested Taxol over a wide concentration range, including the protective concentrations reported, but found that Taxol was not protective in the ST14A model but slightly enhanced toxicity (supplemental Fig. S1, A and B). This excluded altered MT dynamics per se in rescue in the ST14A model. Other studies suggest that not only can mutant htt affect MT function, but also that altered MT dynamics can impact mutant htt toxicity. MT depolymerization using nocodazole can inhibit aggregate formation and enhance toxicity of overexpressed polyglutamine-containing proteins (37, 38), although the toxicity of nocodazole in this regard may be explained by inhibition of autophagy (71). However, we observed little role for aggregation in our model (supplemental Fig. S8). This is likely due to physiological expression levels of a relatively large N-terminal 548-amino acid fragment of htt; such large htt fragments have a low propensity for aggregation (39). Thus the reasons for differences in our results and previous reports may include different levels of expression (physiological in the ST14A model versus overexpression in other models), differences in levels of aggregation, and overt toxicity because of physiological expression levels of mutant htt, and differences in the cell systems used such as yeast (37) and HEK293 cells (38) in some previous studies, versus striatal neuronal cell lines and primary neurons from HD mice in this study.
Although full-length htt is the physiological construct, our results are largely based on cell lines expressing an N-terminal 548-amino acid fragment of mutant htt. Evidence supports that N-terminal htt fragments containing the expanded polyglutamine stretch are the toxic species in HD. First, full-length htt is processed to N-terminal fragments in cell culture models and such fragments are detected in brain tissue of HD patients and animal models (72, 73). Transgenic mice expressing N-terminal mutant htt fragments develop a rapid HD phenotype (74), whereas those expressing full-length mutant htt develop a slowly progressing, mild and often variable phenotype; this delay is thought to be due to the time needed to process and accumulate short N-terminal htt fragments (74). Finally, preventing mutant htt cleavage to fragments smaller than the N-terminal Asn568 htt fragment completely abrogates HD phenotype in transgenic mice (75). These data strongly indicate that htt fragments smaller than the Asn568 htt fragment are the toxic species in HD, and guided our choice of the N548 mutant htt fragment expressing cells for the majority of our study.
Previous studies have suggested a multifaceted interplay between MT and mutant htt. Our results indicate a novel role for mutant htt in regulating MT-based signaling events. Although the role for MT in cell signaling is well established (41), a role for alterations in such signaling in HD is not well documented. We found that mutant htt and parental cells showed differential transcriptional response and ERK activation upon MT depolymerization (supplemental Table S1 and Fig. 4C), suggesting that mutant htt could affect MT-based signaling events. We also observed increased RhoA protein in mutant htt expressing cells. Mutant htt could contribute to the increase in RhoA via several potential mechanisms, including altered transcription because mutant htt causes widespread transcriptional alteration (6). Additionally, mutant htt can impact protein degradation specifically as in the case of β-catenin where mutant htt binds and inhibits the destructive complex and thus increase β-catenin levels (76), or by more widespread alterations in the ubiquitin-proteasomal system reported in HD (77). Although it remains to be determined how the RhoA increase contributes to disease, several observations suggest a potential role in HD pathophysiology. RhoA profoundly affects neuronal function by negatively regulating dendrite formation, and mutations in RhoA regulators can cause neurological disorders (78). Furthermore, decreased dendrite density and spine formation are noted in HD mouse models and patient brain tissue (79, 80). We speculate that increased RhoA levels, while enhancing survival upon MT depolymerization, paradoxically induce neuronal dysfunction, an early event in HD (81). A similar paradox is reported in several HD mouse models, where mutant htt expression induces neurodegeneration, but causes resistance to excitotoxins (9). We suspect that mutant htt causes multiple changes, including altered MT-based signaling, which make the cells resistant to certain perturbations, while at the same time contributing to neuronal dysfunction.
In summary, we have identified a pathway that links MT disassembly to cell survival and demonstrated how a genetic alteration can affect cell fate in response to a drug. These findings provide insight into disease mechanisms and may be a basis for developing disease-specific therapies.
Acknowledgments
We thank Dr. Elena Cattaneo (University of Milan, Italy) and Dr. Marcy MacDonald (Massachusetts General Hospital) for providing the ST14A and the STHdhQ7 and STHdhQ111 cell lines, respectively, and Dr. Akiko Mammoto (Harvard Medical School) for providing lentiviral vectors.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R01GM085081 (to B. R .S.), RO1NS050199 (to A. Y.), R01NS055247, and Training Grant T32 AG000266-10 (to M. R. S.), grants from the Parkinson's Disease Foundation (to A. Y.), the High Q Foundation (to H. V.), a Beckman Young Investigator Award from the Arnold and Mabel Beckman Foundation, Cure Huntington Disease Initiative, Inc., Hereditary Disease Foundation, a Burroughs Wellcome Fund Career Award at the Scientific Interface (to B. R. S.), and the Buck Trust (to R. E. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
- HD
- Huntington disease
- CTGF
- connective tissue growth factor
- EGF-1
- epidermal growth factor-1
- GEF-H1
- guanine nucleotide exchange factor-H1
- Htt
- huntingtin
- MT
- microtubules
- Pdx
- podophyllotoxin
- ROCK
- Rho associated kinase
- SDM
- serum deprived medium
- BDNF
- brain derived neurotrophic factor
- DMSO
- dimethyl sulfoxide
- fmk
- fluoromethyl ketone.
REFERENCES
- 1. Evans W. E., Relling M. V. (2004) Nature 429, 464–468 [DOI] [PubMed] [Google Scholar]
- 2. Makaryus J. N., Catanzaro J. N., Katona K. C. (2007) Hematology 12, 349–352 [DOI] [PubMed] [Google Scholar]
- 3. Picard C., Casanova J. L., Abel L. (2006) Curr. Opin. Immunol. 18, 383–390 [DOI] [PubMed] [Google Scholar]
- 4. Ogawa M., Miyakawa T., Nakamura K., Kitano J., Furushima K., Kiyonari H., Nakayama R., Nakao K., Moriyoshi K., Nakanishi S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bell J. (2004) Nature 429, 453–456 [DOI] [PubMed] [Google Scholar]
- 6. Ross C. A. (2004) Cell 118, 4–7 [DOI] [PubMed] [Google Scholar]
- 7. Brouillet E., Condé F., Beal M. F., Hantraye P. (1999) Prog. Neurobiol. 59, 427–468 [DOI] [PubMed] [Google Scholar]
- 8. Cha J. H. (2007) Prog. Neurobiol. 83, 228–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zuchner T., Brundin P. (2008) Cell Death Differ. 15, 435–442 [DOI] [PubMed] [Google Scholar]
- 10. Gines S., Ivanova E., Seong I. S., Saura C. A., MacDonald M. E. (2003) J. Biol. Chem. 278, 50514–50522 [DOI] [PubMed] [Google Scholar]
- 11. Rigamonti D., Bauer J. H., De-Fraja C., Conti L., Sipione S., Sciorati C., Clementi E., Hackam A., Hayden M. R., Li Y., Cooper J. K., Ross C. A., Govoni S., Vincenz C., Cattaneo E. (2000) J. Neurosci. 20, 3705–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martín-Aparicio E., Yamamoto A., Hernández F., Hen R., Avila J., Lucas J. J. (2001) J. Neurosci. 21, 8772–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petersén A., Larsen K. E., Behr G. G., Romero N., Przedborski S., Brundin P., Sulzer D. (2001) Hum. Mol. Genet. 10, 1243–1254 [DOI] [PubMed] [Google Scholar]
- 14. Apostol B. L., Illes K., Pallos J., Bodai L., Wu J., Strand A., Schweitzer E. S., Olson J. M., Kazantsev A., Marsh J. L., Thompson L. M. (2006) Hum. Mol. Genet. 15, 273–285 [DOI] [PubMed] [Google Scholar]
- 15. Varani K., Bachoud-Lévi A. C., Mariotti C., Tarditi A., Abbracchio M. P., Gasperi V., Borea P. A., Dolbeau G., Gellera C., Solari A., Rosser A., Naji J., Handley O., Maccarrone M., Peschanski M., DiDonato S., Cattaneo E. (2007) Neurobiol. Dis. 27, 36–43 [DOI] [PubMed] [Google Scholar]
- 16. Apostol B. L., Simmons D. A., Zuccato C., Illes K., Pallos J., Casale M., Conforti P., Ramos C., Roarke M., Kathuria S., Cattaneo E., Marsh J. L., Thompson L. M. (2008) Mol. Cell. Neurosci. 39, 8–20 [DOI] [PubMed] [Google Scholar]
- 17. Varma H., Voisine C., DeMarco C. T., Cattaneo E., Lo D. C., Hart A. C., Stockwell B. R. (2007) Nat. Chem. Biol. 3, 99–100 [DOI] [PubMed] [Google Scholar]
- 18. Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V. C., Sharp A. H., Persichetti F., Cattaneo E., MacDonald M. E. (2000) Hum. Mol. Genet. 9, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 19. Yamamoto A., Lucas J. J., Hen R. (2000) Cell 101, 57–66 [DOI] [PubMed] [Google Scholar]
- 20. Shimizu A., Mammoto A., Italiano J. E., Jr., Pravda E., Dudley A. C., Ingber D. E., Klagsbrun M. (2008) J. Biol. Chem. 283, 27230–27238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varma H., Cheng R., Voisine C., Hart A. C., Stockwell B. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14525–14530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cattaneo E., Conti L. (1998) J. Neurosci. Res. 53, 223–234 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Leavitt B. R., van Raamsdonk J. M., Dragatsis I., Goldowitz D., MacDonald M. E., Hayden M. R., Friedlander R. M. (2006) EMBO J. 25, 5896–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jordan M. A., Wilson L. (1998) Methods Enzymol. 298, 252–276 [DOI] [PubMed] [Google Scholar]
- 25. Stähelin H. F., von Wartburg A. (1991) Cancer Res. 51, 5–15 [PubMed] [Google Scholar]
- 26. Morris R. L., Hollenbeck P. J. (1995) J. Cell. Biol. 131, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldberg Y. P., Nicholson D. W., Rasper D. M., Kalchman M. A., Koide H. B., Graham R. K., Bromm M., Kazemi-Esfarjani P., Thornberry N. A., Vaillancourt J. P., Hayden M. R. (1996) Nat. Genet. 13, 442–449 [DOI] [PubMed] [Google Scholar]
- 28. Wellington C. L., Singaraja R., Ellerby L., Savill J., Roy S., Leavitt B., Cattaneo E., Hackam A., Sharp A., Thornberry N., Nicholson D. W., Bredesen D. E., Hayden M. R. (2000) J. Biol. Chem. 275, 19831–19838 [DOI] [PubMed] [Google Scholar]
- 29. Rigamonti D., Sipione S., Goffredo D., Zuccato C., Fossale E., Cattaneo E. (2001) J. Biol. Chem. 276, 14545–14548 [DOI] [PubMed] [Google Scholar]
- 30. Schiff P. B., Horwitz S. B. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jordan M. A., Toso R. J., Thrower D., Wilson L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9552–9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gutekunst C. A., Levey A. I., Heilman C. J., Whaley W. L., Yi H., Nash N. R., Rees H. D., Madden J. J., Hersch S. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8710–8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karsenti E., Vernos I. (2001) Science 294, 543–547 [DOI] [PubMed] [Google Scholar]
- 34. Gunawardena S., Her L. S., Brusch R. G., Laymon R. A., Niesman I. R., Gordesky-Gold B., Sintasath L., Bonini N. M., Goldstein L. S. (2003) Neuron 40, 25–40 [DOI] [PubMed] [Google Scholar]
- 35. Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. (1999) Science 286, 971–974 [DOI] [PubMed] [Google Scholar]
- 36. Tobey R. A., Crissman H. A. (1972) Exp. Cell Res. 75, 460–464 [DOI] [PubMed] [Google Scholar]
- 37. Muchowski P. J., Ning K., D'Souza-Schorey C., Fields S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. (2003) Hum. Mol. Genet. 12, 749–757 [DOI] [PubMed] [Google Scholar]
- 39. Gutekunst C. A., Li S. H., Yi H., Mulroy J. S., Kuemmerle S., Jones R., Rye D., Ferrante R. J., Hersch S. M., Li X. J. (1999) J. Neurosci. 19, 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hackam A. S., Singaraja R., Wellington C. L., Metzler M., McCutcheon K., Zhang T., Kalchman M., Hayden M. R. (1998) J. Cell. Biol. 141, 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gundersen G. G., Cook T. A. (1999) Curr. Opin. Cell Biol. 11, 81–94 [DOI] [PubMed] [Google Scholar]
- 42. Leask A., Abraham D. J. (2006) J. Cell Sci. 119, 4803–4810 [DOI] [PubMed] [Google Scholar]
- 43. Croci S., Landuzzi L., Astolfi A., Nicoletti G., Rosolen A., Sartori F., Follo M. Y., Oliver N., De Giovanni C., Nanni P., Lollini P. L. (2004) Cancer Res. 64, 1730–1736 [DOI] [PubMed] [Google Scholar]
- 44. Song M. R., Ghosh A. (2004) Nat. Neurosci. 7, 229–235 [DOI] [PubMed] [Google Scholar]
- 45. Arévalo J. C., Waite J., Rajagopal R., Beyna M., Chen Z. Y., Lee F. S., Chao M. V. (2006) Neuron 50, 549–559 [DOI] [PubMed] [Google Scholar]
- 46. Cho S. R., Benraiss A., Chmielnicki E., Samdani A., Economides A., Goldman S. A. (2007) J. Clin. Investig. 117, 2889–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mueller B. K., Mack H., Teusch N. (2005) Nat. Rev. Drug Discov. 4, 387–398 [DOI] [PubMed] [Google Scholar]
- 48. Cicha I., Goppelt-Struebe M., Muehlich S., Yilmaz A., Raaz D., Daniel W. G., Garlichs C. D. (2008) Atherosclerosis 196, 136–145 [DOI] [PubMed] [Google Scholar]
- 49. Narumiya S., Ishizaki T., Uehata M. (2000) Methods Enzymol. 325, 273–284 [DOI] [PubMed] [Google Scholar]
- 50. Nakamura K., Nishimura J., Hirano K., Ibayashi S., Fujishima M., Kanaide H. (2001) J. Cereb. Blood Flow Metab. 21, 876–885 [DOI] [PubMed] [Google Scholar]
- 51. Schmandke A., Schmandke A., Strittmatter S. M. (2007) Neuroscientist 13, 454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang G., Beier F. (2005) J. Bone Miner. Res. 20, 1022–1031 [DOI] [PubMed] [Google Scholar]
- 53. Aktories K. (1997) Trends Microbiol. 5, 282–288 [DOI] [PubMed] [Google Scholar]
- 54. Birkenfeld J., Nalbant P., Yoon S. H., Bokoch G. M. (2008) Trends Cell. Biol. 18, 210–219 [DOI] [PubMed] [Google Scholar]
- 55. Mollinedo F., Gajate C. (2003) Apoptosis 8, 413–450 [DOI] [PubMed] [Google Scholar]
- 56. Chatterjee K., Zhang J., Honbo N., Simonis U., Shaw R., Karliner J. S. (2007) J. Mol. Cell. Cardiol. 43, 327–336 [DOI] [PubMed] [Google Scholar]
- 57. Saji K., Fukumoto Y., Suzuki J., Fukui S., Nawata J., Shimokawa H. (2007) Tohoku J. Exp. Med. 213, 139–148 [DOI] [PubMed] [Google Scholar]
- 58. Feng G., Kaplowitz N. (2000) J. Clin. Investig. 105, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Orso G., Martinuzzi A., Rossetto M. G., Sartori E., Feany M., Daga A. (2005) J. Clin. Investig. 115, 3026–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kavallaris M. (2010) Nat. Rev. Cancer 10, 194–204 [DOI] [PubMed] [Google Scholar]
- 61. Sahai E., Marshall C. J. (2002) Nat. Rev. Cancer 2, 133–142 [DOI] [PubMed] [Google Scholar]
- 62. Deacon K., Mistry P., Chernoff J., Blank J. L., Patel R. (2003) Mol. Biol. Cell 14, 2071–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mistry P., Deacon K., Mistry S., Blank J., Patel R. (2004) J. Biol. Chem. 279, 1482–1490 [DOI] [PubMed] [Google Scholar]
- 64. Chowdhury I., Chaqour B. (2004) Eur. J. Biochem. 271, 4436–4450 [DOI] [PubMed] [Google Scholar]
- 65. Zhao M., Ko S. Y., Liu J. H., Chen D., Zhang J., Wang B., Harris S. E., Oyajobi B. O., Mundy G. R. (2009) Mol. Cell. Biol. 29, 1291–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Arany Z., Wagner B. K., Ma Y., Chinsomboon J., Laznik D., Spiegelman B. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4721–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Szebenyi G., Morfini G. A., Babcock A., Gould M., Selkoe K., Stenoien D. L., Young M., Faber P. W., MacDonald M. E., McPhaul M. J., Brady S. T. (2003) Neuron 40, 41–52 [DOI] [PubMed] [Google Scholar]
- 68. Morfini G. A., You Y. M., Pollema S. L., Kaminska A., Liu K., Yoshioka K., Björkblom B., Coffey E. T., Bagnato C., Han D., Huang C. F., Banker G., Pigino G., Brady S. T. (2009) Nat. Neurosci. 12, 864–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Trushina E., Heldebrant M. P., Perez-Terzic C. M., Bortolon R., Kovtun I. V., Badger J. D., 2nd, Terzic A., Estévez A., Windebank A. J., Dyer R. B., Yao J., McMurray C. T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12171–12176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang J., Lou P., Lesniewski R., Henkin J. (2003) Anticancer Drugs 14, 13–19 [DOI] [PubMed] [Google Scholar]
- 71. Webb J. L., Ravikumar B., Rubinsztein D. C. (2004) Int. J. Biochem. Cell Biol. 36, 2541–2550 [DOI] [PubMed] [Google Scholar]
- 72. Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., Qin Z. H., Aronin N., DiFiglia M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang C. E., Tydlacka S., Orr A. L., Yang S. H., Graham R. K., Hayden M. R., Li S., Chan A. W., Li X. J. (2008) Hum. Mol. Genet. 17, 2738–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ferrante R. J. (2009) Biochim. Biophys. Acta 1792, 506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., Hayden M. R. (2006) Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 76. Godin J. D., Poizat G., Hickey M. A., Maschat F., Humbert S. (2010) EMBO J. 29, 2433–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 78. Linseman D. A., Loucks F. A. (2008) Front. Biosci. 13, 657–676 [DOI] [PubMed] [Google Scholar]
- 79. Klapstein G. J., Fisher R. S., Zanjani H., Cepeda C., Jokel E. S., Chesselet M. F., Levine M. S. (2001) J. Neurophysiol. 86, 2667–2677 [DOI] [PubMed] [Google Scholar]
- 80. Ferrante R. J., Kowall N. W., Richardson E. P., Jr. (1991) J. Neurosci. 11, 3877–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tobin A. J., Signer E. R. (2000) Trends Cell Biol. 10, 531–536 [DOI] [PubMed] [Google Scholar]