Abstract
Vacuolar H+-ATPases (V-ATPases) are highly expressed in ruffled borders of bone-resorbing osteoclasts, where they play a crucial role in skeletal remodeling. To discover protein-protein interactions with the a subunit in mammalian V-ATPases, a GAL4 activation domain fusion library was constructed from an in vitro osteoclast model, receptor activator of NF-κB ligand-differentiated RAW 264.7 cells. This library was screened with a bait construct consisting of a GAL4 binding domain fused to the N-terminal domain of V-ATPase a3 subunit (NTa3), the a subunit isoform that is highly expressed in osteoclasts (a1 and a2 are also expressed, to a lesser degree, whereas a4 is kidney-specific). One of the prey proteins identified was the V-ATPase B2 subunit, which is also highly expressed in osteoclasts (B1 is not expressed). Further characterization, using pulldown and solid-phase binding assays, revealed an interaction between NTa3 and the C-terminal domains of both B1 and B2 subunits. Dual B binding domains of equal affinity were observed in NTa, suggesting a possible model for interaction between these subunits in the V-ATPase complex. Furthermore, the a3-B2 interaction appeared to be moderately favored over a1, a2, and a4 interactions with B2, suggesting a mechanism for the specific subunit assembly of plasma membrane V-ATPase in osteoclasts. Solid-phase binding assays were subsequently used to screen a chemical library for inhibitors of the a3-B2 interaction. A small molecule benzohydrazide derivative was found to inhibit osteoclast resorption with an IC50 of ∼1.2 μm on both synthetic hydroxyapatite surfaces and dentin slices, without significantly affecting RAW 264.7 cell viability or receptor activator of NF-κB ligand-mediated osteoclast differentiation. Further understanding of these interactions and inhibitors may contribute to the design of novel therapeutics for bone loss disorders, such as osteoporosis and rheumatoid arthritis.
Keywords: Bone, Drug Design, Membrane Proteins, Osteoclast Acidification, Protein-Protein Interactions, Vacuolar ATPase, Osteoporosis
Introduction
V-ATPases2 are proton pumps ubiquitous in eukaryotic cells, where they acidify numerous intracellular membrane compartments, including Golgi, endosomes, lysosomes, clathrin-coated vesicles, chromaffin granules, and insulin secretory granules (reviewed in Refs. 1–11). V-ATPases also pump protons across the plasma membrane into the extracellular space in a variety of specialized cells, including renal duct intercalated cells, clear cells of the epididymis, and osteoclasts. Here they are involved in functions, including pH homeostasis, sperm maturation, and bone resorption and remodeling. Mutations in V-ATPase subunits lead to diseases, such as renal tubular acidosis and osteopetrosis. Furthermore, their inappropriate activity can contribute to osteoporosis and tumor metastasis (12–15).
V-ATPases are multisubunit molecular motors, structurally analogous to the F1F0-ATP synthases (F-ATPases), but working “in reverse” (16–20). Thus, V-ATPases create proton gradients across membranes by utilizing the energy of ATP hydrolysis, rather than utilizing the potential energy of proton gradients to synthesize ATP.
The mammalian V-ATPase complex includes at least 14 different subunits, some of which have multiple isoforms (13). The complex can be divided into two sectors, the cytoplasmic V1 sector and the integral membrane V0 sector. The V1 sector consists of subunits A–H, with a likely stoichiometry of A3B3CDE3FG3H (18). The mammalian V0 sector consists of subunits a, c, c″, d, e, and Ac45, and the stoichiometry appears to be ac5c″deAc45, whereas in yeast one of the c subunits is replaced with a homologous c′ subunit. Regulation of V-ATPase function can occur through dissociation of the complex into its separately inactive V1 and V0 sectors, with the reversible loss of the C subunit from the complex (21, 22).
Much of the architecture of the V-ATPases, and their molecular motor function, has been inferred from the structure-function analogy with F-ATPases, which, unlike the V-ATPase complexes, have had their structures largely resolved by x-ray crystallography. Crude confirmation of this analogy has been obtained, thus far, largely from single particle electron microscopic analysis of V-ATPases and chemical cross-linking experiments (5, 18, 23, 24). Many of the more precise details of intra-complex interactions of V-ATPase subunits, or their extra complex associations with non-V-ATPase polypeptides, remain to be characterized.
Landolt-Marticorena et al. (25) have shown previously, using yeast two-hybrid analysis, that the a subunit of the V0 sector interacts with the A and H subunits of the V1 sector, interactions that are thought to contribute to the stator complex that is required for the function of the V-ATPase molecular motor (26, 27). The work presented here approaches the problem of further characterizing V-ATPase structure and function by implementing new yeast two-hybrid studies using a cDNA library derived from the murine RAW 264.7 cell line, which is capable of differentiating into osteoclasts in the presence of the cytokine, RANKL (28–30). This system is of particular interest because of its high level expression of plasma membrane V-ATPase, which is responsible for proton secretion into the extracellular resorption lacunae of osteoclasts through their specialized ruffled border. This function is required to dissolve bone (31, 32), and its disruption in mammals results in the sclerosing bone disorder, osteopetrosis (33, 34), although excessive activity results in pathological bone loss, as in osteoporosis (13). Further understanding of the structure and function of V-ATPases will aid the development of targeted therapeutics for the treatment of bone loss diseases, such as osteoporosis and rheumatoid arthritis (13).
EXPERIMENTAL PROCEDURES
Materials and Reagents
PBS was 0.2 g/liter KCl, 0.2 g/liter KH2PO4, 8.0 g/liter NaCl, 1.15 g/liter Na2HPO4, pH 7.4. TBST was 20 mm Tris-HCl, 0.9% (w/v) NaCl, 0.05% (w/v) Triton X-100 (Sigma, catalog no. T8787), pH 7.40. All buffer pH values were adjusted at 25 °C. Gelatin for blocking in pulldown assays and ELISA was EIA grade (Bio-Rad, catalog no. 170-6537). Glutathione-Sepharose 4B was from GE Healthcare (catalog no. 17-0756-01). Ni-NTA-agarose was from Qiagen (catalog no. 30210). Anti-GST antibody was rabbit polyclonal GST(Z-5) from Santa Cruz Biotechnology (catalog no. sc-459). Anti-His tag antibody was His-Probe (H-3), from Santa Cruz Biotechnology (catalog no. sc-8036); horseradish peroxidase (HRP)-conjugated anti-rabbit second antibody (HRP-GAR) was polyclonal goat anti-rabbit IgG from Santa Cruz Biotechnology (catalog no. sc-2004); HRP-conjugated anti-mouse second antibody (HRP-GAM) was polyclonal goat anti-mouse IgG from Santa Cruz Biotechnology (catalog no. sc-2005).
Cell Culture
The Saccharomyces cerevisiae strain YRG-2 (Stratagene) was maintained on YPAD agar plates at 30 °C. Transformed yeast in two-hybrid experiments were grown on SD agar plates, or in SD liquid media, with amino acid dropouts appropriate to the experiment, according to supplier formulations and recommendations (Stratagene). Cloning and plasmid production were done in the Escherichia coli strain DH5α, pET vector protein expression in the strain BL21(DE3) Rosetta (Novagen), and pGEX vector protein expression in the strain K12 KS1000 (New England Biolabs). Murine macrophage RAW 264.7 cells (ATCC, catalog no. TIB-71)were grown in complete DMEM, consisting of Dulbecco's modified Eagle's medium (DMEM) with high glucose (4.5 g/liter), l-glutamine, sodium bicarbonate, and without sodium pyruvate (Sigma, catalog no. D5796), supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin (Sigma antibiotic mix, catalog no. 15140), and 10% (v/v) fetal bovine serum (Invitrogen; catalog no. 10437-028, lot 1233072). Differentiation of RAW 264.7 cells to osteoclasts was performed by exposure to 100 ng/ml soluble recombinant RANKL in complete α-MEM, consisting of α-MEM (Invitrogen, catalog no. 12571-063) containing FBS and antibiotics as for complete DMEM, for 5 days. This resulted in a mixture of cells, containing 1–20 nuclei, >90% positive for tartrate-resistant acid phosphatase (TRAP) staining. All mammalian cells were cultured in a humidified 5% CO2 atmosphere at 37 °C.
Yeast Two-hybrid Assays
The commercial HybriZap 2.1 system (Stratagene) was used throughout. Initial screening was done with a cDNA library constructed according to the supplier's recommendation, using mRNA (oligo(dT) magnetic bead-purified; Absolutely mRNA purification kit, Stratagene) from the mouse osteoclast cell culture model, RANKL-differentiated RAW 264.7 cells. After reverse transcription of mRNA, second strand synthesis, ligation of linkers, and restriction enzyme digestion (as per supplier's instructions), size-selected cDNA was inserted between EcoRI and XhoI sites in the HybriZap 2.1 λphage vector. The primary λphage library was mass-excised to yield a pAD-GAL4–2.1 cDNA phagemid library. The library was screened in YRG-2 host yeast with a bait construct consisting of the N-terminal, cytoplasmic domain of the murine V-ATPase subunit a3 (NTa3; amino acids 1–393) inserted between EcoRI and SalI sites in the pBD-GAL4 Cam phagemid vector. Additionally, a 5′-ggtggt-3′ linker (Gly-Gly) was constructed in-frame between the EcoRI site and the a3 subunit start codon. Sequences for a3 were obtained by PCR using the a3-containing vector pcDNA3.1-a3 as template, a kind gift of Dr. Beth S. Lee (Department of Physiology and Cell Biology, Ohio State University).
Constructs
Full-length mouse B1 subunit was cloned by PCR using as template the B1-containing vector pEF6/V5-His-TOPO-B1, also a gift from Dr. Beth S. Lee. The oligonucleotide primers used are described in Table 1. The PCR product was agarose gel-purified (GFX PCR DNA and gel band purification kit; GE Healthcare), digested with EcoRI and SalI, and cloned into EcoRI/SalI-digested pAD-GAL4 2.1 (Stratagene). To facilitate further cloning, the internal EcoRI site at nucleotide position 319–324 (with respect to start “a” of B1 ORF sequence) was knocked out with the silent mutation 321A→G (QuikChange site-directed mutagenesis kit; Stratagene). Constructs for other V-ATPase subunits followed the same strategy, but PCR templates were from varied sources, and inserts were cloned into EcoRI/SalI sites in pBD GAL4 Cam, pET-32a(+), or pGEX-4T-1, as elaborated in Table 1. All expression clones contained sequences for a Gly-Gly flexible coupler (G2) between the N and C termini of the fusion protein partner (either E. coli thioredoxin (TRX) or Schistosoma japonicum GST, respectively). As noted, full-length B1 and B2 constructs also had a C-terminal His tag, His6, with flanking Gly-Gly (G2H6G2), to facilitate full-length purification. Clones that were produced by RT-PCR from total RNA extracts of mouse tissues (see Table 1) were obtained by extraction with TRIzol reagent (Invitrogen) and reverse transcription using the RevertAid H Minus First Strand cDNA synthesis Kit (Fermentas, catalog no. K1632).
TABLE 1.
Primers, plasmid constructs, and protein expression products
a All a subunits were cloned partially, as their hydrophilic N-terminal (NT) cytoplasmic domains; V-ATPase subunits were of mouse origin; all PCR primers had a small, random 5′ extension (not shown) to improve restriction enzyme cleavage.
b pET constructs were in pET32a(+), pGEX constructs in pGEX-4T-1; PCR products were EcoRI/SalI-digested and ligated into EcoRI/SalI-digested vectors; all constructs were verified by full-length sequencing of inserts and junctions.
c Expressed protein domain organization is indicated as follows: TRX and GST are N-terminal E. coli thioredoxin (with His tag and S tag domains) and S. japonicum glutathione S-transferase fusions, respectively; G2 is a Gly-Gly coupler; and G2H6G2 is a C-terminal His tag extension flanked by Gly-Gly sequences; the superscript numbers in parentheses are the amino acid (a.a.) ranges of the expressed subunits (homologous target V-ATPase subunit sequence only, numbered with respect to the natural subunit N-terminal methionine).
Purification of GST Fusion Proteins
Bacteria transfected with expression constructs that were growing exponentially at 37 °C, in LB medium with selective antibiotics, were chilled on ice, and isopropyl β-d-1-thiogalactopyranoside was added to 0.2 mm. Cells were incubated further with shaking at 16 °C for 16 h. Cells were harvested by centrifugation at 4 °C, and pellets were resuspended in 4 ml of ice-cold PBS, containing 0.2 mg/ml lysozyme per 200 ml of original culture volume. The suspension was incubated on ice for 30 min and then mixed with 2.5 volumes of ice-cold 0.2% (w/v) Triton X-100. This was sonicated on ice four times for 15 s with 30 s of cooling between bursts. DNase and RNase were added to 5 μg/ml each, from 10 mg/ml stocks, and the lysate was incubated a further 10 min on ice and then centrifuged at 20,000 × g for 15 min. The supernatant was mixed with 10 ml/liter of starting culture of a 50% slurry of glutathione-Sepharose 4B in PBS. After 1 h of incubation at 4 °C with rocking, beads were washed three times for 5 min with PBS and collected by centrifugation at 500 × g for 5 min. The beads were transferred to a Poly-Prep chromatography column (Bio-Rad) and washed twice with 20 ml of PBS and eluted with 10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0. Fractions were collected and analyzed by SDS-PAGE. GST-B1(GH) and GST-B2(GH) were repurified on Ni-NTA-agarose beads, as described for the TRX fusion protein purification.
Purification of TRX Fusion Proteins
This procedure was similar to purification of GST fusion proteins, with the following exceptions. Bacterial pellets were resuspended in ice-cold lysis buffer consisting of 50 mm NaH2PO4, 300 mm NaCl, and 10 mm imidazole, pH 8.0, at a ratio of 4 ml per 60 ml of original culture volume. After lysis, cleared supernatants were mixed with 2 ml of a 50% slurry of Ni-NTA-agarose beads in lysis buffer at 4 °C for 1 h with rocking. Beads were washed three times for 5 min with a 10-ml wash buffer containing 50 mm NaH2PO4, 300 mm NaCl, and 20 mm imidazole, pH 8.0, and packed in a Poly-Prep chromatography column. The column was washed twice with 20 ml of wash buffer and eluted with 50 mm NaH2PO4, 300 mm NaCl, 50 mm imidazole, pH 8.0. A second elution was performed at 150 mm imidazole concentration. Fractions were analyzed by SDS-PAGE.
Pulldown Assays
Glutathione-Sepharose 4B beads (0.5-ml packed volume) were combined with 4 ml of cleared bacterial lysate containing 0.35 mg/ml total protein and PMSF to 2 mm (from 0.2 m stock in anhydrous ethanol) from either GST-B1 or GST-B2 expression cultures. After coating at 4 °C for 1 h with rocking, the beads were washed five times with 10 ml of PBS, and 125 μl of coated beads was mixed with 400 μl of cleared supernatants of TRX-NTa1–4 expression cultures containing 0.13 mg/ml protein. After a 1-h incubation at 4 °C with rocking, beads were washed five times with 1 ml of ice-cold PBS. Pelleted beads were eluted with 100 μl of SDS sample buffer, and 10-μl aliquots were analyzed by SDS-PAGE.
ELISA Solutions
Protein Coating buffer was 10 mm sodium phosphate, pH 7.0; Tris-buffered saline (TBS) was 20 mm Tris-HCl, 0.9% (w/v) NaCl, pH 7.40; Blocking Buffer was 1% (w/v) gelatin in TBS containing 0.05% (w/v) Triton X-100 and 0.1% (w/v) phenol, pH 7.40; Wash Buffer was TBST; 3,3′,5,5′-tetramethylbenzidine stock was 4 mg/ml 3,3′,5,5′-tetramethylbenzidine (Sigma) in DMSO/ethanol (1:9), Developer was freshly prepared 80 μg/ml 3,3′,5,5′-tetramethylbenzidine (from stock), 0.01% hydrogen peroxide, 0.1 m sodium acetate, pH 6.0; Stop Solution was 1 n sulfuric acid. All antibodies were diluted in Blocking Buffer from commercial stock as follows: anti-GST antibody at a 1:4000 dilution of GST(Z-5); anti-His tag antibody at a 1:3000 dilution of His probe (H-3); HRP-GAR second antibody at a 1:4000 dilution; HRP-GAM second antibody at a 1:3000 dilution.
In Vitro ELISA-based Solid-phase Binding Assays
Volumes are per well. All washes were done with 100 μl of Wash Buffer (see under “ELISA Solutions”). High protein-binding polystyrene 384-well plates (Greiner Microlon 781097; Sigma catalog no. M6936) were coated with ligand protein (20 μl of Protein Coating Buffer containing 10 μg/ml TRX-NTa3 or 2 μg/ml GST-CTB2, as indicated), overnight at 4 °C. All further steps were at 25 °C. Coated plates were washed twice, blocked with 30 μl of Blocking Buffer for 1 h, and again washed twice. For binding experiments, wells were then incubated with analyte protein (20 μl of Blocking Buffer with 5 mm MgCl2, containing GST-B2(GH), concentrations as described) for 1 h. Plates were washed three times, followed by incubation with 20 μl of anti-GST antibody in Blocking Buffer for 30 min. Plates were again washed three times and incubated with 20 μl of HRP-GAR in Blocking Buffer for 30 min (in some assays, where the ligand was GST-CTB2 or GSTB1 anti-His tag antibody was used with HRP-GAM second antibody). Plates were then washed five times, followed by addition of 30 μl of Developer. After 15 min of incubation, the reaction was stopped with 30 μl of Stop Solution. Absorbance was quantified using a Envision Multilabel Reader (PerkinElmer Life Sciences) at 450 nm, with subtraction of an optical reference absorbance at 600 nm.
High Throughput Screening
The above protocol was adapted to automated liquid handling and quantification. Briefly, high throughput screening of a 10,000 compound DIVERSet synthetic organic chemical library (ChemBridge) was executed on a robotics platform where a CRS articulated arm controlled by Polara software (Thermo Electron) moved on a 3-m rail to utilize a Biomek FX liquid handler (Beckman Coulter), equipped with a 96-channel disposable tip pipetting head (pipetting in staggered quadrants into 384-well microplates) and attached stacker carousel for sourcing plates and tip boxes. Assay plates were sourced from room temperature hotels. A Multimek 384-channel automated pipettor (Beckman) with an accessory 200-nl 384-pin floating pin tool (V&P Scientific) was used to dispense library compounds; an EMBLA washer (Molecular Devices) was used for 384-well microplate washing; and a 2102 EnVision Xcite multilabel reader was used to perform absorbance measurements, using 450 and 600 nm (optical control) filters. Typically, 10–14 384-well plates were screened simultaneously, with 320 compounds per plate, and two positive (no library compound, vehicle only) and two negative (no ligand) control columns at left (columns 1 and 2) and right (columns 23 and 24) ends of the plate, respectively. Library compounds were dispensed into test wells using a 200-nl 384-pin tool, each compound being at 1 mm concentration in DMSO (5 μm final concentration in assay). Hits were scored after analyzing data using B-score statistics (35) and ranking the inhibitory compounds according to negative deviation from the mean. Selected hits were confirmed by repeating the primary assay and by comparing dose responses in the primary assay; reproducible hits were then tested in secondary cellular assays.
Model Compound
The hit described in detail in this work is referred to as KM91104 and has the chemical structure 3,4-dihydroxy-N′-(2-hydroxybenzylidene)benzohydrazide. KM91104 was purchased as powder (ChemBridge 5266986) and was diluted for use in secondary assays from 10 mm stock in DMSO. Vehicle controls were equivalent volumes of neat DMSO. Some experiments included an inactive analog of KM91104, which is referred to as KM91201. KM91201 has the structure N′,N″-(1,4-phenylenedimethylylidene)bis(3,4-dihydroxybenzohydrazide) and was purchased as a powder (ChemBridge 5318527). KM91201 was treated exactly as KM91104 in experiments.
Cell Growth Protein Assay
To assess the effect of primary screening hits on growth of RAW264.7 cells, the cells were seeded in complete DMEM in 96-well plates at 5 × 103 cells/well (200 μl/well), with varying concentrations of test compounds, and were allowed to grow for 5 days with a change of complete medium on day 3. Wells were then washed with PBS, and cells were lysed with Lysis Buffer, containing 90 mm trisodium citrate, 10 mm NaCl, (adjusted to pH 4.8 with HCl), and 0.1% Triton X-100 (added prior to use from 10% stock). 10-μl aliquots were withdrawn and mixed with 200 μl of Bio-Rad protein assay reagent (Bio-Rad, catalog no. 500-0002) in a microplate and incubated at 25 °C for 5 min. Absorbances were then read at 595 nm.
Cell Growth Cytotoxicity Assay
Proliferation and metabolic activity of cell cultures in the presence of primary hit compounds was determined using a modified tetrazolium dye assay (MTS reagent; Promega CellTiter 96 Aqueous One Solution cell proliferation assay, catalog no. G3582). Cells were grown in 96-well plates as for the cell growth protein assay; then 20 μl of MTS reagent was added to each well, and cells were further incubated overnight at 37 °C. Absorbances were then read at 490 nm.
Total Solubilized TRAP Assay
Cells grown in 96-well plates, as for the cell growth protein assay, were washed twice with PBS, and wells were aspirated, and then 200 μl/well of Lysis Buffer (see under “Cell Growth Protein Assay”) was added to the plates. After lysis, 20-μl samples were withdrawn and added to microplate wells on ice, containing 50 μl each of ice-cold Substrate Solution and Tartaric Acid Buffer. Substrate Solution was prepared as 0.1 g of disodium p-nitrophenyl phosphate (Sigma catalog no. 104 phosphatase substrate, Sigma; catalog no. 104-0) in 25 ml of distilled water. Tartaric Acid Buffer was 40 mm l(+)-tartaric acid added to Citrate Buffer (final pH ∼4.0). The assay plate was mixed and then incubated for 30 min at 37 °C. The reaction was stopped with 80 μl of 2 n NaOH. Absorbances were measured at 405 nm and compared with 20-μl standards consisting of 1–100 μm p-nitrophenol.
TRAP Staining of Fixed Cells
Cells were treated according to the BD Biosciences TRAP staining protocol no. 445. Briefly, TRAP Buffer, pH 5.0, was prepared fresh for use by mixing 50 ml of Acetate Buffer (35.2 ml of 0.2 m sodium acetate and 14.8 ml of 0.2 m acetic acid), 10 ml of 0.3 m sodium tartrate, 1 ml of 10 mg/ml naphthol AS-MX phosphate disodium salt (Sigma, catalog no. N-5000), 0.10 ml of Triton X-100, and 38.9 ml of distilled water. TRAP Stain was prepared fresh for use by dissolving 0.3 mg of Fast Red Violet LB salt (Sigma, catalog no. F-3381) per ml of TRAP Buffer at 37 °C. For TRAP staining, medium was aspirated from cells, and cells were washed with PBS. Cells were fixed with 200 μl/well formalin (Sigma; catalog no. HT501128) for 15 min at 37 °C and then washed three times with PBS at 37 °C. Cells were incubated in TRAP stain for 5–10 min at 37 °C. TRAP stain was aspirated, and cells were washed with Ca2+/Mg2+-free PBS and were stored in the same buffer at 4 °C. Photomicrography was done using phase contrast or bright field illumination after warming cells to ambient temperature in the storage buffer.
Isolation of Bone Marrow Mononuclear (BMM) Cells
BMM cells were isolated from tibias and femurs of normal C57BL/6 mice and seeded into 24-well plates at 1.0 × 105 cells/well in 1 ml of complete α-MEM containing 15% FBS. The cells were incubated for 2 days in the presence of 50 ng/ml M-CSF. To induce osteoclast differentiation, the medium was changed and included, additionally, 200 ng/ml RANKL. Test compounds were also added at this time. The cells were allowed to grow and differentiate for 5 days with no further change of medium.
Hydroxyapatite Resorption Assay
Early passage (P3 to P6 after thawing) RAW 264.7 cells were plated at 5 × 103 cells per well in 100 μl of complete α-MEM containing 100 ng/ml soluble recombinant RANKL in Corning Osteo-Assay Surface 96-well plates (Corning Life Sciences, catalog no. 3988XX1). Plates were incubated for 2 h at 37 °C in a humidified 5% CO2 incubator to allow cell attachment. A further 100 μl of complete α-MEM containing RANKL and test compound was then added to each well. The latter complete medium was changed after 3 days, and cells were incubated for a further 2 days prior to processing for von Kossa staining.
Modified von Kossa Staining
Plates from hydroxyapatite resorption experiments were stripped with 1.2% sodium hypochlorite solution for 5 min to remove cells, rinsed with distilled water, and air-dried until further use. For staining, plates were treated in darkness at ambient temperature with 100 μl/well 5% (w/v) silver nitrate solution for 10 min. Wells were then aspirated and washed for 5 min in distilled water. Wells were again aspirated, and 100 μl/well 5% (w/v) sodium carbonate in 10% formalin was added. After a 4-min incubation at ambient temperature, wells were washed twice with PBS and three times with distilled water. The plates were then aspirated and air-dried prior to imaging.
Dentin Resorption Assay
Elephant ivory (donated by the Toronto Zoo, Toronto, Canada) was sliced using a Buehler Isomet slow speed saw with a 4-inch diameter diamond impregnated wafering blade with a 300 μm kerf. Slices were end-cut 150 μm thick and washed in distilled water, and discs were punched using a 0.25-inch diameter paper hole-punch. Discs were stored in 20% (v/v) ethanol solution. For tissue culture use, discs were soaked in complete medium overnight and then transferred to fresh medium in wells of a 96-well microplate. RAW 264.7 cells were plated in 100-mm culture dishes containing 20 ml of complete α-MEM with 100 ng/ml soluble recombinant RANKL. Plates were incubated for 4 days to allow cell differentiation. Mature osteoclasts were harvested by scraping into 2 ml of medium and were pelleted at 1,000 × g for 5 min at 4 °C. Cells were resuspended in complete medium with RANKL and appropriate concentrations of test compound, then were plated onto dentin slices. Further growth was allowed for 3 days prior to processing for Picro-Sirius Red staining.
Picro-Sirius Red Staining
Discs from dentin resorption experiments were washed in distilled water and ethanol. The discs were then stained with Picro-Sirius Red stain (0.1% Sirius Red F3B (C.I. 35782) in saturated aqueous picric acid) overnight. Discs were destained for 1 h in 0.5% acetic acid solution, then washed in distilled water, and mounted on slides for fluorescence imaging (36).
Imaging
Photomicrography was done using a Leica DM IRE2 microscope with OpenLab software (Leica Microsystems). Image analysis was carried out with NIH ImageJ software. For resorption assays in Corning plates, all microplate wells were imaged with five fields/well (20× objective) using an automated stage (Applied Scientific Instrumentation MS-2000). Dentin slices were imaged using epifluorescence with GFP filters.
Statistical Analysis
Repetition of experiments is given in the figure legends. Determination of IC50 values for dose-response curves or half-maximal binding in protein interaction assays was done using GraphPad Prism (version 4.02) curve-fitting software. In some experiments, where saturation was not achieved, automated curve fitting failed, and manual estimates were made by interpolation in the log linear portion of the curve. This is noted in the figure legends. Standard deviations were calculated and are shown as ±1 S.D. wherever values are quoted or in error bars in histograms and graphs. Unpaired two-tailed t tests were used to test significance of differences, as appropriate.
RESULTS
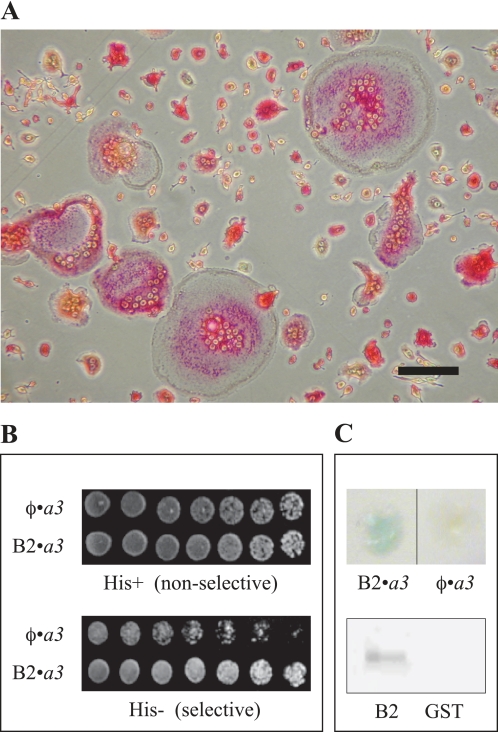
The initial intention of this work was to determine the set of protein binding partners that interact specifically with the a3 subunit that is highly expressed as part of the plasma membrane V-ATPase of osteoclasts. Crucial interactions with it could potentially be exploited as molecular targets for the treatment of osteolytic disease (12, 13). Therefore, a mouse cDNA library was constructed from RANKL-differentiated RAW 264.7 cells (29, 30) representing a range of osteoclasts from single TRAP-positive cells to cells containing over 20 nuclei (Fig. 1A) in the HybriZap 2.1 phagemid vector system (37). The cDNA library was probed in YRG-2 yeast, using a construct of NTa3 (the N-terminal domain of a3, amino acids 1–393; see Table 1) in the yeast two-hybrid GAL4 binding domain vector pBD GAL4 Cam. Although many hits were obtained, the outcome of this screening was difficult to interpret because of unresolved issues with a high background of self-activation with all a subunit bait constructs attempted (data not shown except for NTa3); however, a strong interaction that was consistently above background was found with the full-length B2 subunit of V-ATPase (Fig. 1B). This interaction was confirmed using the second reporter gene, lacZ, and by affinity pulldowns of NTa3 with B2-coated glutathione-Sepharose 4B beads (Fig. 1C).
FIGURE 1.
Initial evidence that the NTa3 domain interacts with the B2 subunit. A, TRAP staining of RANKL-differentiated RAW 264.7 osteoclasts. A representative distribution of small, intermediate, and large osteoclasts in the cell culture used to construct a HybriZAP-2.1 yeast two-hybrid cDNA library is shown. Red staining indicates expression of the osteoclast differentiation marker enzyme TRAP. Cells were grown for 3 days in complete DMEM with 100 ng/ml soluble recombinant RANKL prior to RNA extraction. Scale bar, lower right, 100 μm. B, upper plate, YRG-2 yeast co-transformed with empty pAD vector and pBD-NTa3 (φ·a3, top row) or pAD-B2 and pBD-NTa3 (φ·a3, bottom row) were spotted onto complete SD agar in 2-fold serial dilutions starting at 1.0 × 104 cells (left to right); the lower plate is the same as the upper plate, but cells were grown on SD medium lacking histidine. All a subunit constructs in pAD showed autoactivation (e.g. top-most row); nevertheless, cells simultaneously expressing NTa3 and B2 fusions had a growth advantage due to activation of the HIS3 reporter gene (bottom-most row). C, upper plate shows independent verification of NTa3 and B2 interaction using the second reporter gene of the HybriZAP yeast two-hybrid system lacZ. Blue staining with X-gal indicates activation of lacZ (φ·a3, left half). Only background level staining is seen with empty vector pAD and pBD-NTa3 co-transformation (φ·a3, right half). Lower plate shows pulldown assays using B2 and NTa3 as GST and TRX fusion proteins, respectively. Glutathione beads coated with GST-B2 from whole bacterial lysates were washed and exposed to lysates of bacteria expressing TRX-NTa3 (see “Experimental Procedures”). Beads were washed, eluted, and subjected to SDS-PAGE. Staining with anti-His tag antibody revealed the TRX-NTa3 band for pulldowns using beads coated with GST-B2 (B2, left lane) but not with control empty-vector GST (GST, right lane).
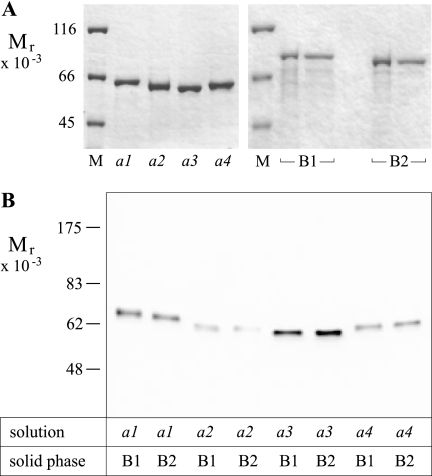
It was of interest to determine whether there were differences among the four murine a subunit isoforms (a1-4) in terms of their interactions with the two murine B isoforms (B1 and B2). TRX fusion proteins (with His tag), NTa1, NTa2, NTa3, and NTa4, were expressed in E. coli and purified using standard Ni(II) affinity chromatography. GST fusion proteins, B1 and B2, were similarly prepared but using glutathione affinity chromatography (Fig. 2A; see also “Experimental Procedures” and Table 1). Affinity pulldowns, as shown in Fig. 2B, were performed, showing that a1, a2, a3, and a4 were efficiently pulled down with both B1 and B2, with somewhat less a2 and strikingly more a3 being pulled down; however, differences between the comparative abilities of B1 and B2 to pull down any of the NTa isoforms were not discernible in replicate experiments.
FIGURE 2.
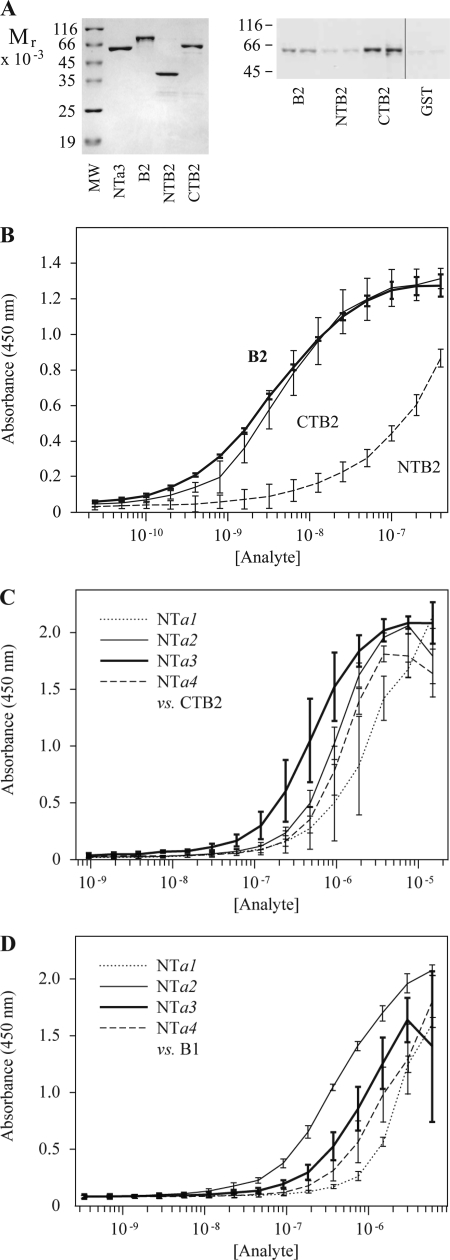
Expression of NTa and B subunits and pulldown assays of subunit pairs. A, affinity-purified fusion proteins (2 μg, each) separated on 8% SDS-PAGE and stained with Coomassie Blue R-250. Left panel shows proteins expressed from pET32a(+) vector; M, molecular weight standards; followed by (2 μg each) a1, TRX-NTa1 fusion protein; a2, TRX-NTa2; a3, TRX-NTa3; a4, TRX-NTa4. Right panel shows proteins expressed from pGEX-4T-1 vector; B1, GST-B1(GH) (left lane, affinity-purified on GST beads; right lane, repurified on Ni(II) beads); B2, GST-B2(GH) (left lane, affinity-purified on GST beads; right lane, repurified on Ni(II) beads). Photographs are of dried gels. B, pulldowns of TRX fusions of all N-terminal a subunit domains with GST fusions of either B1 or B2 subunits. Glutathione beads were coated with bacterial lysates from cells expressing either GST-B1 or GST-B2 fusion proteins (1 μg/ml total protein), washed, and then exposed to lysates containing TRX-NTa fusions derived from a1, a2, a3, and a4 subunits (1 μg/ml total protein). Washed beads were eluted and run on 8% SDS-PAGE, blotted, and probed with anti-GST antibody. Image was developed using chemiluminescence.
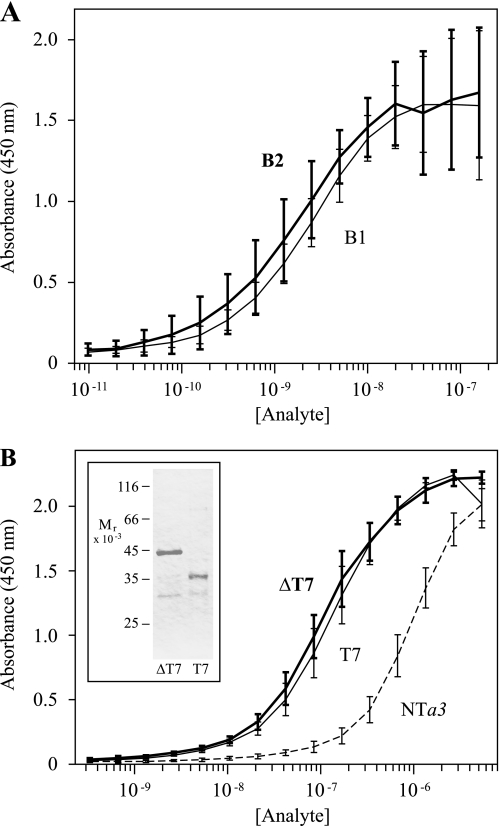
To make more quantifiable determinations of relative binding, an ELISA-based solid-phase binding assay was designed to compare saturation curves for the binding of NTa3, the N-terminal domain of the osteoclast specific isoform, with B1 and B2. Saturation curves (with log abscissae), shown in Fig. 3A, again indicated that differences between NTa3-B1 binding and NTa3-B2 binding were not significant at p ≤ 0.05. Half-maximal binding values were 2.7 ± 0.5 nm for NTa3-B1 and 1.9 ± 0.4 nm for NTa3-B2.
FIGURE 3.
ELISA-based binding assays of NTa3 interactions with B1 and B2 subunits. A shows ELISA saturation curves of B1 (heavy line and error bars) and B2 (light line and error bars) binding to NTa3. ELISA plates were coated with TRX-NTa3 ligand and probed with 2-fold serial dilutions of analyte (GST-B1 or GST-B2 fusion proteins, 9.8 pm to 160 nm). Binding of GST alone was negligible, as were the signals obtained without ligand or either analyte protein (data not shown). Absorbance at 450 nm was determined after staining with an anti-GST-HRP sandwich and color development with 3,3′,5,5′-tetramethylbenzidine (see “Experimental Procedures”). Absorbance at the reference wavelength of 600 nm was subtracted to normalize optical variance among wells (A600 was typically <3% of the maximum A450 signal). Each curve shows means ± S.D. bars (n = 3, in duplicate). Differences between B1 and B2 interacting with NTa3 were not significant. B shows ELISA saturation curves of NTa3ΔT7 (heavy line and error bars), NTa3T7 (light line and error bars), and NTa3(dashed line) binding to CTB2. ELISA plates were coated with GST-CTB2 ligand and probed with 2-fold serial dilutions of analyte (TRX-NTa3ΔT7, TRX-NTa3T7, or TRX-NTa3 fusion proteins, 25 pm to 400 nm). Shown also is purification of TRX-NTa3ΔT7 and TRX-NTa3T7 (SDS-PAGE, inset panel). TRX alone showed negligible binding (data not shown). Each curve shows means ± S.D. bars (n = 3, in duplicate).
Because a3 and B2 subunit isoforms both are highly expressed in the plasma membranes of active osteoclasts (38, 39), attention was focused on that pair, and experiments were done to further delineate the sites of binding. A splice variant of the a3 subunit, referred to as Tirc7, with an N-terminal truncation of 217 amino acids, is expressed in T cells (40). This variant may delineate a natural domain boundary between the deleted, non-Tirc7, N-terminal half of NTa3 (referred to here as ΔT7) and the Tirc7-specific, C-terminal half of NTa3 (referred to here as T7). We prepared NTa3ΔT7 and NTa3T7 fusion constructs (Fig. 3B, inset, and see Table 1) and, as shown in Fig. 3B, used these to probe B2 ligand, in a reverse of the binding experiment shown in Fig. 3A. NTa3 binding to immobilized B2 was observed at a much lower apparent affinity than B2 binding to immobilized NTa3 (950 ± 310 nm, half-maximal; a 500-fold lower apparent affinity). Surprisingly, the fragmented NTa3 not only bound with a significantly higher apparent affinity (p = 0.01) than the intact moiety, but both halves bound equally well. The half-maximal binding concentrations were 121 ± 84 nm for NTa3ΔT7 and 134 ± 81 nm for NTa3T7, which are not significantly different. Possible explanations for these apparent anomalies are addressed under “Discussion.”
Holliday et al. (41) split B2 into an N-terminal domain (amino acids 1–117 in the mouse ortholog) and a C-terminal domain (amino acids 118–511) and showed that actin binding was confined to the N-terminal domain. In this study, similar constructs were prepared (Table 1) to express GST fusions of N- and C-terminal domains of B2 (Fig. 4A), and solid-phase binding assays were performed (Fig. 4B). In contrast to actin binding, a3 binding to the B2 subunit appeared to be confined to the C-terminal domain, CTB2 (Fig. 4B). Binding of CTB2 to NTa3 was half-maximal at 4.8 ± 1.2 nm, which was not significantly different from binding to the full-length B2 subunit (half-maximal at 3.6 ± 0.8 nm), whereas binding of the N-terminal domain (NTB2) to NTa3 was reduced 50-fold, being half-maximal at 240 ± 30 nm. This difference was highly significant (p = 0.0002).
FIGURE 4.
Determining the domains of interaction between the a and B subunits. A, gel on left shows purified proteins used in pulldown assays and ELISA binding analysis. MW, molecular weight standards; followed by (2 μg each) NTa3, TRX-NTa3; B2, full-length B2 fusion protein, GST-B2(GH); NTB2, GST-NTB2 (N-terminal domain); CTB2, GST-CTB2 (C-terminal domain). Blot on right shows pulldowns of TRX-NTa3 with GST-B2(GH), GST-NTB2, and GST-CTB2, as described. Right-most lane shows control with GST alone (identical blot and exposure with intervening lanes removed). Note that NTB2 pull down is not greatly different from GST control. B, ELISA, as in Fig. 3, but comparing binding of B2 (full-length B2 subunit, GST-B2(GH), heavy line) with that of CTB2 (C-terminal domain of B2, GST-CTB2, light line) and NTB2 (the N-terminal domain of B2, GST-NTB2, dashed line). Fusion protein analytes were 2-fold serially diluted from 400 nm to 24 pm. Each curve shows means ± S.D. bars (n = 3, in duplicate). C shows ELISA-based binding assay using plates coated with GST-CTB2. Analytes were TRX-NTa1–4 (a1, dotted line; a2, light line; a3, heavy line; a4, dashed line) 2-fold serially diluted from 15.6 μm to 0.95 nm. Each curve shows means ± S.D. bars (n = 3, in duplicate). D, similar to C, but using plates coated with GST-B1. Analytes were TRX-NTa1–4 (a1, dotted line; a2, light line; a3, heavy line; a4, dashed line) 2-fold serially diluted from 6.0 μm to 0.15 nm. Each curve shows means ± S.D. bars (n = 2, in duplicate). Automated curve fitting failed for the NTB2 curve because it does not approach saturation. A manually estimated interpolation was derived on the assumption that the data converge in a manner similar to the convergence for B2 and CTB2.
It was of interest to compare the binding of a1–4 subunit isoforms to B2. To do this, ELISA plates were coated with CTB2 ligand and probed with a1–4 analytes (Fig. 4C). Interestingly, NTa3 showed a significantly higher (>2-fold) apparent affinity (half-maximal at 390 ± 32 nm) for binding CTB2 than was seen for NTa2-CTB2 (p = 0.006) and NTa4-CTB2 (p = 0.02) binding, which were statistically indistinguishable at (half-maximal values of 830 ± 140 nm and 1.1 ± 0.3 μm, respectively). The NTa1-CTB2 interaction seemed to have the lowest apparent affinity, 6-fold less than that of the NTa3-CTB2 interaction, with a half-maximal value of 2.5 ± 1.3 μm, which could be distinguished from the NTa2-B2 and NTa4-B2 interactions (p = 0.01). The data suggest that a3-B2 interaction may have a privileged affinity, in comparison with the other a-B subunit interactions.
This latter observation begs the question whether binding of a1–4 subunit isoforms to B1 follows a similar distribution. Fig. 4D shows that this is not the case. Apparent affinities are as follows: NTa1-B1, 2.3 ± 0.4 μm; NTa2-B1, 460 ± 57 nm; NTa3-B1, 740 ± 240 nm; NTa4-B1, 1.24 ± 0.04 μm. It is of interest that although the apparent affinities of binding are nearly identical for a1 and a4 with B1 and B2, a2 and a3 have switched relative positions, i.e. a3 paired with B2 seems to be favored in the B2-binding series, whereas a2 paired with B1 seems to be favored in the B1-binding series. Statistical tests, however, indicate that, for binding to B1, a2 and a4 are different (p = 0.001) and a1 binding differs from the rest of the subunits (p ≤ 0.05), but a2 and a3 are not significantly different (p = 0.25).
One of the proposed functions of the a subunit in the V-ATPase complex is to act as a stator to prevent futile rotation of the A3B3 catalytic headpiece. It is presumed to accomplish this by anchoring to the membrane through its C-terminal domain, as part of the V0 complex, and binding to the V1 complex through its N-terminal domain, by interactions with the catalytic head group and peripheral stalk components (12, 27). We speculated that small molecule inhibitors of a3-B2 interaction might disrupt stator organization and/or destabilize the V1V0 complex and provide a means of regulating V-ATPase activity, possibly specifically targeting the plasma membrane V-ATPase that contains the a3 and B2 subunit isoforms. This could have therapeutic potential for alleviating bone loss disease (13) and limiting the metastatic potential of some tumors (15).
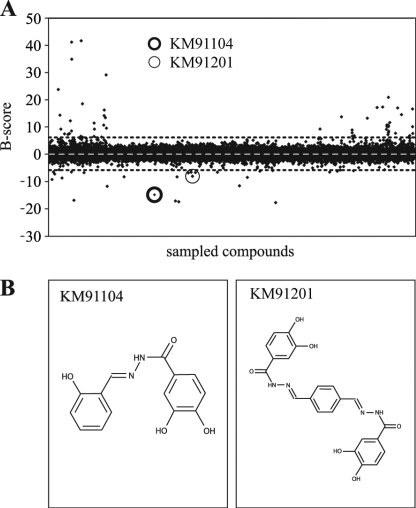
To screen for small molecule inhibitors, we adapted the above described ELISA-based solid-phase binding assay for use in a high throughput robotics platform (see under “Experimental Procedures”). Synthetic chemical screening yielded primary hits at a rate of ∼0.2%. Rescreening and evaluation of primary assay dose-response curves reduced this to 0.04% of compounds initially tested. B-scores (35), for all 10,241 compounds that were screened, are plotted in Fig. 5A.
FIGURE 5.
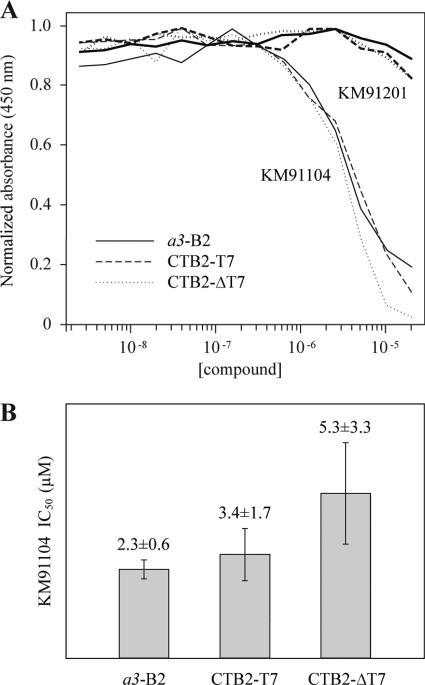
High throughput primary screening for inhibitors of a3-B2 interaction, using an ELISA-based binding assay. The Chembridge DIVERSet collection of 10,000 synthetic chemical compounds was screened using the ELISA-based binding assay described and executed on a robotics platform (see “Experimental Procedures”). A, B-score statistics are shown for the compounds, where ELISA absorbances were analyzed in the presence of test compounds. The B-score statistic smoothes systematic machine error and takes into account positive and negative controls. Small black diamonds indicate results for individual compounds. White dashed line indicates mean (B-score = 0), and dotted black lines indicate ± 3 S.D. from the mean. Outliers with a B-score less than ± 3 S.D. were regarded as hits. Hits were retested twice in the primary screen and ranked according to deviation from the mean. The highest ranking hits (with lowest B-scores) were picked for secondary screening. The hit discussed here, KM91104, is indicated with a heavy circle. The only analog of KM91104 in the library (KM91201) is indicated with a light circle. It was also identified as a hit in the primary screen but failed in downstream validation screens (unacceptably high IC50; see also Fig. 9). B shows the chemical structure of compound KM91104, 3,4-dihydroxy-N′-(2-hydroxybenzylidene)benzohydrazide (left panel). This compound was the best hit, overall, in terms of effectiveness in inhibiting osteoclast resorption without effect on osteoclast viability or differentiation, as described in subsequent figures. The inactive analog, KM91201 (N′,N″-(1,4-phenylenedimethylidene)bis(3,4-dihydroxybenzohydrazide)), is shown in the right panel.
The model compound discussed here, KM91104, indicated with a heavy circle, Fig. 5A, had a B-score of minus 14.8. Interestingly, the only analog of KM91104 in the 10,000-compound library (light circle, Fig. 5A, identified here as KM91201) was also identified as a primary hit, but it failed to reproducibly give a low B-score on rescreening. Other compounds, including those with a lower B-score than KM91104, were mostly eliminated in rescreening. Of the four compounds that reproducibly gave low B-scores, all except KM91104 were found to be overtly toxic to mammalian cells in growth and cytotoxicity assays (see “Experimental Procedures”). The structures of KM91104 and KM91201 are shown in Fig. 5B.
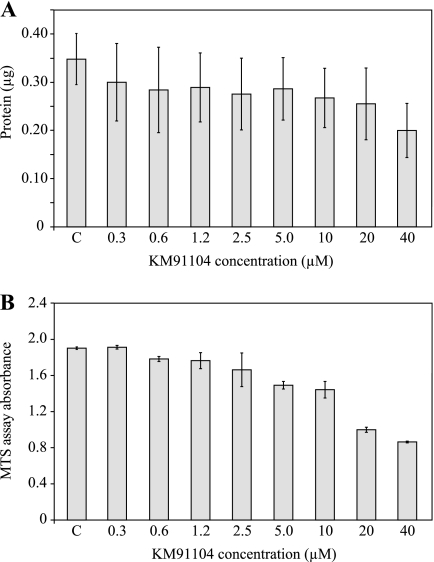
Secondary screening of compounds consisted of performing growth and cytotoxicity assays, testing effects on osteoclast differentiation and maturation, and testing whether osteoclast mineral resorption, which is dependent on V-ATPase-mediated acid secretion, could be effectively inhibited. Assays were developed using formats that would enable future large scale high throughput screening. Fig. 6A shows that 5-day growth of undifferentiated RAW 264.7 cells was negligibly affected by concentrations of KM91104 up to 20 μm, with an IC50 ≥40 μm. Fig. 6B shows that there was negligible cytotoxicity, according to the mitochondrial reductase-based MTS assay, up to 2.5 μm, with an IC50 of ∼20 μm. Thus, the potentially cytotoxic effects of KM91104 seemed to become manifest only at a >20 μm concentration.
FIGURE 6.
Secondary screening for cytotoxicity of primary hits in RAW 264.7 cells. Secondary cellular assays were used to screen primary hits to determine their effect on growth (total cellular protein) and metabolic status (MTS tetrazolium dye reduction) after 5 days of continuous exposure. The test cell line was undifferentiated RAW 264.7 cells, and results are shown for the subject compound KM91104. A, total cell protein was determined in cultures exposed to a concentration range of KM91104, from 0.3 to 40 μm, as indicated (n = 3, in duplicate; error bars are ± S.D.). C indicates control, vehicle (DMSO) only added. Concentration of KM91104 up to 20 μm had negligible effect on cell growth, with an IC50 >40 μm. B, as in A, but MTS reduction was determined to assess metabolic status. KM91104 had negligible effect up to 2.5 μm, with an IC50 of ∼20 μm (n = 3, in triplicate; error bars are ± S.D.).
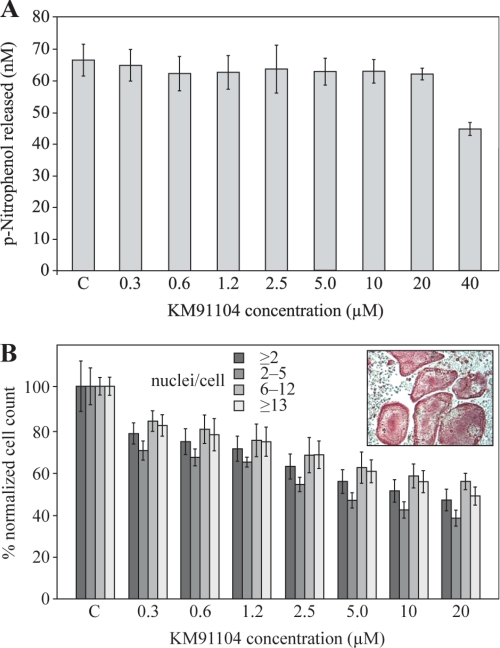
It was of interest also to determine effects on osteoclastogenesis. Fig. 7 shows the effects of KM91104 on RAW 264.7 cell differentiation in the presence of RANKL. In Fig. 7A, expression of the osteoclast differentiation marker enzyme, TRAP, was not significantly affected at up to 20 μm KM91104, with an IC50 >40 μm, reflecting the growth inhibition results of Fig. 6A. Fig. 7B shows a small, but significant, drop-off in formation of osteoclasts even at 0.3 μm KM91104 and a small, but steady, decline at higher concentrations up to 20 μm (IC50 ∼20 μm). Osteoclast subpopulation counts are shown (2–5, 6–12, and ≥13 nuclei) in comparison with total osteoclasts (≥2 nuclei). Histograms are shown normalized to control (actual mean counts for controls are given in the figure legend). This figure illustrates that up to 20 μm KM91104 does not significantly alter the distribution of small versus large osteoclasts, relative to total osteoclasts. The inset panel in Fig. 7B shows TRAP-positive (red) osteoclasts with >20 nuclei derived from mouse BMM cells in primary tissue culture exposed to 1.2 μm KM91104. There did not appear to be any inhibition of differentiation or maturation of these authentic osteoclasts compared with untreated cultures (data not shown).
FIGURE 7.
Secondary screening for effect on osteoclast differentiation by KM91104. A secondary cellular assay was used to determine the effect of KM91104 on osteoclast differentiation and fusion after 5 days of continuous exposure. TRAP is a marker enzyme for osteoclast differentiation that converts p-nitrophenyl phosphate to p-nitrophenol under acidic conditions. The test cell line was RAW 264.7 cells differentiated in the presence of 100 ng/ml soluble recombinant RANKL. A, total solubilized TRAP assay was used to determine the effect of compound KM91104 on RANKL-mediated differentiation of RAW 264.7 cells (see “Experimental Procedures”). The concentration range of compound tested was 0.3–40 μm, as indicated. C indicates control, vehicle only added. Amount of p-nitrophenol released indicates the degree of differentiation after 5 days of growth with RANKL. Differentiation was not significantly affected up to 20 μm KM91104 concentration (n = 3, in duplicate; error bars are ± S.D.). B shows quantitative analysis of TRAP-stained fixed cultures (see “Experimental Procedures”). Cell counts for TRAP-positive (red) multinucleated cells with ≥2 nuclei after treatment with compound KM91104 are plotted. Controls are normalized to 100% (n = 3, in duplicate; error bars are ± S.D.). Inset panel shows primary culture mouse BMM cells (TRAP-stained, red) differentiated with M-CSF and RANKL and exposed continuously to 1.2 μm KM91104 for 5 days. Ability of primary BMM cells to form large osteoclasts (>20 nuclei) has not been impaired at this concentration.
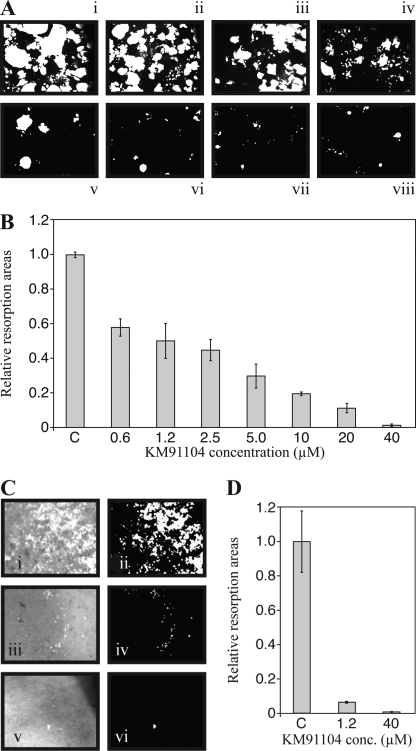
Finally, it was of greatest interest to determine whether KM91104 is able to inhibit osteoclast resorptive activity. Fig. 8A shows photomicrographs of synthetic hydroxyapatite mineral surfaces (Corning Osteo-Assay Surface, post-stained black with metallic silver; images are ImageJ (National Institutes of Health)-processed as described under “Experimental Procedures”) where resorption has occurred (white areas) due to active acid secretion by osteoclasts differentiated in situ with RANKL from RAW 264.7 cells. Fig. 8B shows the quantified resorption areas from these experiments, indicating that KM91104 does inhibit osteoclast mineral resorptive activity, with an EC50 of approximately 1.2 μm. This suggests that there is an acceptable ratio (∼20-fold) between the concentration of KM91104 that is efficacious in reducing mineral resorption (∼1 μm) and the concentration that is toxic, as measured either by cytotoxicity assays or assays of cell differentiation (≥20 μm). Because these data were derived using an artificial mineral surface, we also tested KM91104 using dentin (150-μm-thick ivory slices from elephant), which is a means of assessing authentic bone resorption. Fluorescence images are shown in Fig. 8C, where type I collagen, exposed by osteoclast resorption, is stained by the fluorescent dye, Picro-Sirius Red. Quantified resorption areas are shown in Fig. 8D. These data suggest that the efficacy of KM91104 in inhibiting authentic bone resorption is greater than in inhibiting resorption of the artificial mineral surface. Likely there are differences in the way that osteoclasts mobilize hydrated mineral interspersed with extracellular matrix proteins, as in dentin, compared with dissolving a continuous microcrystalline surface, as is found in the commercial test plates.
FIGURE 8.
Secondary screening for inhibition of hydroxyapatite resorption by RANKL-differentiated RAW 264.7 cells. A, cells were differentiated in the presence of 100 ng/ml RANKL on synthetic mineralized surfaces of Corning Osteo-Assay Surface 96-well plates. Cells were cultured for 5 days with continuous exposure to RANKL and KM91104. The complete medium was changed on the 3rd day. On day 5, cells were stripped with 1.2% sodium hypochlorite solution, and the mineral surface was stained using a modified von Kossa method (see “Experimental Procedures”). Plates were air-dried and imaged using digital bright field photomicrography. Images shown are ImageJ-processed for quantification. Concentrations of KM91104 (μm) in medium were as follows: panel i, control, vehicle only; panel ii, 0.6; panel iii, 1.2; panel iv, 2.5; panel v, 5.0; panel vi, 10; panel vii, 20; panel viii, 40. B shows quantitative image analysis of resorption areas (white areas shown in A), using ImageJ software. The concentration range of KM91104 was from 0.6 to 40 μm, as indicated (n = 3, in duplicate, 5 fields per well imaged; error bars are ± S.D.). C indicates control, vehicle only added. Highly significant reduction in resorption (p < 0.0001) was observed even at the lowest concentration. Approximate IC50 value of KM91104 for resorption was 1.2 μm, a concentration that was not cytotoxic and did not significantly affect osteoclast differentiation or fusion to form large osteoclasts (see Figs. 6 and 7). C, osteoclasts derived from RAW 264.7 cells were seeded on dentin (elephant ivory) slices. After 3 days of resorption, slices were stained with Picro-Sirius Red and viewed by epifluorescence (GFP filters). Left column is representative of fluorescence image, and right column is ImageJ-processed to quantify fluorescence (white) on a black background. Panels i and ii, control (vehicle only); panels iii and iv, 1.25 μm KM91104; panels v and vi, 40 μm KM91104. D, quantified fluorescence from C (average of results from three independent slices; error bars are ± S.D.).
DISCUSSION
This study has demonstrated that mouse V-ATPase a and B subunits interact. This protein-protein interaction has not been described previously, but the observation is easily reconciled structurally with published data showing that a and A subunits interact (25), because A and B subunits are in close proximity in the V1 sector. Although true affinities are difficult to determine in solid-phase binding assays, half-maximal binding values derived from saturation curves provided a means of comparison of “apparent affinity” among subunit isoforms. The calculated values for NTa3-B1 and NTa3-B2 (2.7 and 1.9 nm, respectively) were not significantly different, and this lack of discrimination between B1 and B2 seemed to be confirmed for all four a subunits in affinity pulldown assays. Because solid-phase binding assays tend to underestimate binding affinities, these values likely represent minimum affinities (usually expressed as the inverse, i.e. dissociation constants, Kd), suggesting that NTa3-B binding is a high affinity interaction having biological significance. In comparison, the reported affinity for F-actin binding to the B2 subunit is nearly 30-fold lower, with a Kd of 55 nm (42), and the affinity between the a2 subunit and cytohesin-2 (ARNO) is 160-fold lower, with a reported Kd of 310 nm (43).
The sites of a3-B2 interaction were crudely localized to two apparently equivalent “half-domains” (Tirc7 and non-Tirc7 domains, NTa3T7 and NTa3ΔT7, respectively) of NTa3 and the C-terminal domains of B1 and B2 subunit isoforms (CTB1 and CTB2). The division of B2 was based on previously published work by Holliday et al. (41), and a3 was divided on the basis of a previously defined, naturally occurring truncation (40). Binding of NTa3 to the C-terminal domain of B2 was not significantly different from binding to the full-length B2 subunit, whereas binding to the N-terminal domain of B2 was ∼50-fold less, similar to background with the isolated GST fusion partner, suggesting that the interaction of the NTa subunit domain is with the C-terminal domain of the B subunit.
Perhaps the most surprising observation of this work was that when the N-terminal domain of a3 was divided into halves, both halves bound equally well to B2. Furthermore, the halves bound with significantly higher (8-fold) apparent affinity than that of the intact NTa3 domain (possible reasons why this might be the case are discussed further in the legend to Fig. 10B). To lend further support to the former observation, we asked whether the inhibitor of the a3-B2 interaction, KM91104, would similarly inhibit the NTaΔT7-B2 and NTa3T7-B2 interactions. Fig. 9 shows that this is the case, with values for IC50 being statistically indistinguishable. Furthermore, an inactive structural analog of KM91104, KM91201, did not inhibit any of the interactions (IC50 ≫80 μm). This argues that the observed interactions of NTa3ΔT7 and NTa3T7 with B2 are similar to the interaction of intact NTa3 with B2.
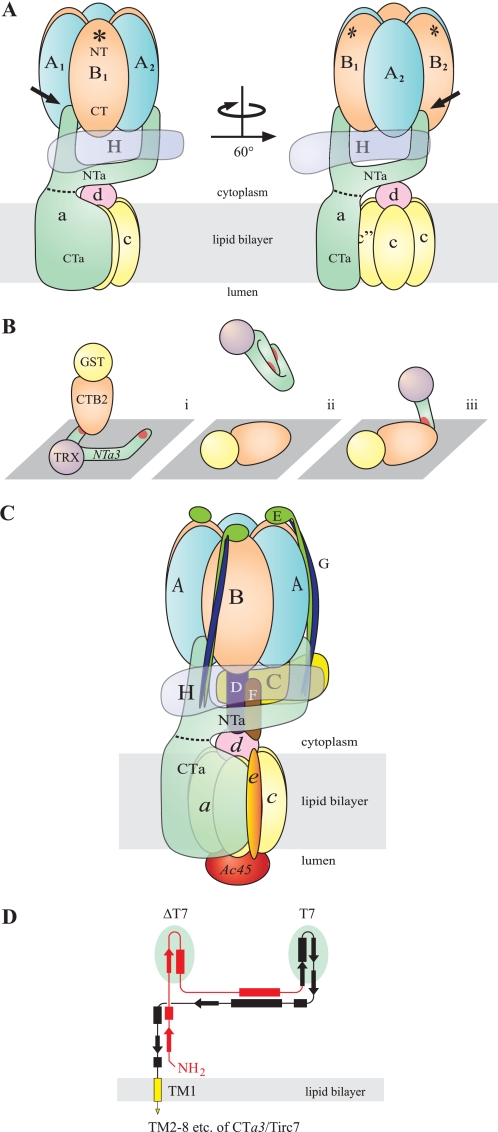
FIGURE 10.
Possible scenario for NTa interaction with the B subunit. A, for clarity, the C–G, e, and Ac45 subunits of the complex have been omitted (compare with C). Shown are the V1 sector, with catalytic head group comprising three each of alternating A and B subunits (blue and orange, respectively), and the H subunit (transparent purple). The V0 sector includes subunit a (green), with the cytoplasmic NTa and integral membrane CTa domains delimited (dashed line), the mammalian hexameric c5c″ proteolipid barrel (yellow), and subunit d (pink). Note that the B subunit is oriented with the N-terminal domain (NT) to the top (distal to the V1/V0 interface) and C-terminal domain (CT) to the bottom (see “Discussion”). Asterisks indicate the F-actin-binding site, which is localized to the NT domain in all B subunits. This diagram is patterned loosely after single particle EM reconstruction by Zhang et al. (18) and shows a bifurcated NTa with the left finger extended to the AB groove of the catalytic head group (left figure, black arrow). A 60° rotation, as indicated, shows the insertion of the right finger into the adjacent AB groove (right figure, black arrow). This takes into account interactions known to take place with both the A and B subunits. The “beam” that extends between the fingers is shown to cradle the H subunit and contact the d subunit, although this latter interaction is likely transient or manifest only when V1 and V0 are dissociated. These interactions have been demonstrated elsewhere (25, 59), but the precise assembly is speculative. Both ends of the beam, which traverses ∼120° around the rotor axis, are thought to support one each of three EG heterodimer peripheral stalks, and one end contacts the C subunit, which supports the third EG heterodimer at its distal end (see also C). Although the orientation shown here seems to be in reasonable agreement with the more detailed reconstruction images of Zhang et al. (18), the resolution of their images does not permit definitively choosing this scenario over one where the fingers of NTa are inserted into BA grooves, rather than an AB grooves, which would require only slight adjustment of the present model. In fact neither the reconstruction by Zhang et al. (18), nor similar work done by Muench et al. (24) resolves a potential “left finger” in contact with the catalytic head group in any orientation in their electron density maps. Better resolution has been obtained for T. thermophilus V-ATPase, which favors the scenario presented here (44). B, depicts an explanation for the data in Fig. 4B; panel i, the fusion protein ligand, TRX-NTa3, is splayed out, due to interaction with the hydrophobic plastic surface of the ELISA plate and is consequently accessible to the analyte, GST-CTB2, resulting in a high affinity interaction with either of the apparently equivalent binding sites (red) on the “fingers” of NTa3; panel ii,, in the reverse assay, the ligand is GST-CTB2 and the analyte is TRX-NTa3. This panel suggests that the unwieldy fingers of the intact NTa3 moiety fold on themselves while in free solution, unsupported by other subunits of the native V-ATPase. This results in sequestering its binding sites, resulting in significant reduction in binding affinity; panel iii, isolated NTa3ΔT7 or NTa3T7 domains (i.e. the isolated fingers of NTa3) are not self-sequestering and interact with immobilized CTB2 at higher affinity. C, this schematic shows the best-guess scenario of mammalian V-ATPase structure, taking into account this work and single-particle EM descriptions (18, 24, 44). It also suggests, speculatively, the co-alignment of the fingers of the a subunit with two of the three EG peripheral stalks, the third being supported by the C subunit. Recently, Lee et al. (60) have shown that the EG heterodimer forms a rare right-handed coiled-coil in T. thermophilus, also depicted here. Some subunits are depicted as semi-transparent for clarity (see also description for A). D shows a scale redrawing of the proposed secondary and tertiary structure of the NTa domain shown in Zhang et al. (18) to accommodate the present findings. It depicts the ΔT7 (red) and T7 (black) domains (with underlying green ovals indicating the two speculative fingers), which must have equivalent binding sites for the B subunit. No repetitive polypeptide sequences have been identified to suggest identical binding sites. The C-terminal end of the T7 domain continues as the CTa domain (yellow; truncated here), which includes eight predicted transmembrane α-helices (one shown) and a cytoplasmic C-terminal tail (61).
FIGURE 9.
Inhibition of binding of NTa3 and its N- and C-terminal halves to B2 by KM91104. A, plates were coated with either TRX-NTa3 (for B2 analyte) or GST-CTB2 (for NTa3ΔT7 or NTa3T7 analytes). TRX-NTa3, TRX-NTa3ΔT7, and TRX-NTa3T7 were used at 400, 47, and 158 nm constant concentration, respectively. Analytes were bound in the presence of compound KM91104 or KM91201, 2-fold serially diluted (2.4 nm to 20 μm). A450 ELISA absorbances were normalized to the highest value for the mean of each set of curves for comparison (n = 6, n = 4, and n = 3, in duplicate, respectively). B, values for IC50 were obtained by using GraphPad Prism nonlinear regression and curve-fitting. Only values for KM91104 are shown. Unpaired two-tailed t tests show that these values are not significantly different. Curves for KM91201 were essentially nonconvergent, with lowest of range having an IC50 >80 μm.
The above observations may correlate with EM observations suggesting that the NTa domain is bifurcated. Zhang et al. (18) have suggested a bifurcated cytoplasmic domain for the a subunit, based on secondary structure predictions and bulk fitting of subunits into single particle EM electron density volumes. Our present observations that NTa3ΔT7 and NTa3T7 both bind B2 equally well strongly support a model where both fingers of the bifurcated NTa domain contact B subunits of the catalytic head group. Based on the former predictions and present observations, one scenario for a structural configuration is that both of the two predicted finger-like projections of NTa lie in interfacial grooves between A and B subunits of the catalytic head group, making contact with both subunits simultaneously. Because of the distances involved, this must occur near the ends of the A and B subunits that are nearest the V1/V0 interface.
There are six interfacial grooves in the catalytic headpiece of V-ATPase, in two different conformations, three AB grooves alternating with three BA grooves. Presently, there are no data of sufficient resolution in eukaryotic systems to determine which of the two grooves is favored in the interaction with NTa. Clearly, higher resolution images of the V-ATPase complex would help greatly to resolve these scenarios. Recently, Lau and Rubinstein (44) have produced images of remarkably high resolution (16 Å), in eubacterial Thermus thermophilus V-ATPase, using cryo-electron microscopic single-particle analysis. The N-terminal domain of the I subunit (the a subunit ortholog) clearly is bifurcated, with two extensions toward the top of the V1 sector that must represent the EG heterodimeric peripheral stalks. It is impossible to resolve the junction between the I subunit and the EG heterodimers within the peripheral stalks in the latter work, but we speculate that the I subunit fingers co-align with the EG stalks as far as their contacts with the A and B subunits. A diagrammatic representation to summarize our speculative structure is shown in Fig. 10 (see detailed description in figure legend).
In the work of Lau and Rubinstein (44), the positioning of the peripheral stalks appears to favor contact with the AB interface, rather than the BA interface. In T. thermophilus V-ATPase, it has been shown that the BA interface is involved in ATP binding and catalysis, whereas the AB groove does not participate in either function (45). If the a-B interaction is purely structural, as in a stator function, one might argue that it is more likely to occur at the AB interface, so as to avoid interference with ATP binding and hydrolysis and the local conformational changes that ensue. This argument lends further support to the notion that NTa contacts the catalytic headpiece at AB interfaces.
This study has further demonstrated that the B subunit interaction with the a subunit is localized to a domain of the B subunit that is spatially distinct from the one that interacts with actin microfilaments. The B subunit can be crudely approximated by a prolate ellipsoid with its long axis perpendicular to the plane of the membrane bilayer. EM images suggest that actin binding to the N-terminal domain of the B subunit is distal to the membrane plane in the long axis of the B subunit (41, 42). The C terminus of the B subunit is thought to be at the opposite end, near the V1/V0 interface, proximal to the membrane plane. This orientation is also supported by analogy with the F-ATPase crystal structure (46). The model proposed here (Fig. 10) is consistent with these observations, taking into account the limited “reach” that the two fingers of NTa would likely have from the V1/V0 interface into the V1 sector.
A defect is observed in V-ATPase association with actin microfilaments in oc/oc mutant mice, which have an osteopetrotic phenotype (41, 47). Actin binds the V-ATPase B subunits, and lack of actin binding is thought to prevent V-ATPase from trafficking to the plasma membrane, where it needs to be for crucial acid secretion to take place in osteoclast bone resorption; however, oc/oc mice were shown to be truncation mutants in the V-ATPase a3 subunit gene (48). These observations beg the following question. How can a mutation in a3 affect the interaction between the B subunit and F-actin? We show here for the first time that there is a direct interaction between a and B subunits. Holliday et al. (41), who first posed this question, speculated that the a subunit might directly regulate access to the F-actin-binding site in the N-terminal domain of the B subunit. At the time, as now, structural models of V-ATPase lacked the resolution to prove a direct interaction between NTa and the F-actin-binding site near the N terminus of B. We have shown here that NTa does not bind to the N-terminal domain of the B subunit but rather to the C-terminal domain, which is not directly involved in actin binding. Furthermore, it is difficult to conceive how a single a subunit could simultaneously occlude the potential binding sites of the three available B subunits in V1 by physically blocking F-actin binding, even by an allosteric mechanism. It seems more likely that a single a subunit might exert some allosteric influence on the B subunit with which it makes contact to promote, rather than inhibit, F-actin binding. Actin binding would then presumably occur only in the B subunit(s) that interacts directly with the a3 subunit. In vitro binding experiments, however, do not require the a subunit to be present for F-actin to bind to the B subunit, so one needs to complicate this model with the notion that such regulation occurs only in native V-ATPase complexes. This explanation remains unsatisfying and suggests that there are as yet unknown factors involved. There is still much work to be done to fully understand this regulatory mechanism and to understand the functional consequences of a-B subunit interaction.
The functional significance of the NTa3-B2 interaction remains to be determined, but speculatively it may be important to the stator function of the a subunit, its role in docking of the V1 to the V0 sector, and possibly in intra-complex regulatory signal transduction mediated by the a subunit in response to pH or cellular energy status (1, 49–51). In this study, we have exploited the knowledge of the a3-B2 interaction to screen for inhibitors, using a simple in vitro a3-B2 binding model. It was largely speculative that such an inhibitor might also disrupt V-ATPase function in cells; however, one compound, KM91104, identified as an inhibitor in the primary screen has proved to inhibit bone resorption by osteoclasts, which is entirely dependent on V-ATPase-mediated acid secretion. The data of Fig. 8 lend support to the initial assumption that small molecule inhibitors of a3-B2 interaction might destabilize the V1/V0 association of the V-ATPase complex that is required for active proton translocation in vivo. It is also noteworthy that the IC50 for inhibition of osteoclast resorption (∼1.2 μm) is very similar to the IC50 seen for in vitro inhibition of a3-B2 protein interaction (2.3 μm).
In the RANKL-differentiated RAW 264.7 cell osteoclast model, V-ATPase a1, a2, and a3 subunits are expressed (in our hands, a2 is difficult to detect). The a3 subunit is found in late endosomes/lysosomes and the plasma membrane (the ruffled border of differentiated osteoclasts). The a1 and a2 subunits are found in Golgi, and a1 is also found in other nonlysosomal, non-Golgi organelles (52). The a4 subunit is thought to be kidney-specific, and the others are expressed fairly ubiquitously in many tissues. Nevertheless, mutations in a3 that cause malignant osteopetrosis do not cause any pathology that is not related to loss of osteoclast resorptive function. In many tissues, other subunits may be able to complement loss of a3 function, but in osteoclasts a3 function seems to be irreplaceable (38). Nyman and Väänänen (53) have recently argued this point, suggesting that a1 can compensate for a3 in lysosomes but not significantly in the plasma membrane of bone osteoclasts. They concluded that structure-independent mechanisms may cause bone resorption to be far more sensitive than lysosomal function to V-ATPase inhibition and that “submaximal inhibition of V-ATPase could have a dramatic impact on bone resorption with little impact on lysosomal function.” Thus, highly specific inhibitors of V-ATPase may not be required to specifically inhibit osteoclast resorptive function. A relatively nonspecific inhibitor, like KM91104, may therefore find some utility or at least point the way for further development of novel bone loss therapeutics.
Our data also suggest that the a3-B2 interaction may have a moderately higher affinity than other subunit pairs. This may drive a3-B2 formation in preference to other combinations when the two subunits are available. Whether this property can be exploited therapeutically is presently unclear.
The broader cell biological mechanism of action of K91104 inhibition of a3-B2 remains to be determined; it could be via disruption of catalytic function, disruption of proton translocation, interference with V-ATPase trafficking to the plasma membrane, interfering with actin ring formation, or other possible scenarios. Nevertheless, the data presented here suggest that development of a targeted inhibitor of a3-B2 interaction that exploits differences among the a subunit polypeptide sequences may be possible and might be useful in regulating osteoclast bone resorption activity at doses that preclude cytotoxicity or have an appreciable effect upon osteoclast differentiation or maturation.
Reducing bone resorption, while increasing bone formation, would be ideal to prevent pathological bone loss and, at the same time, restore bone that has been lost, yet clinical studies attempting this approach have failed. Previous or concomitant bisphosphonate therapy suppresses the efficacy of parathyroid hormone-related protein (teriparatide) therapy for increasing bone formation (54–57). It is speculated that bisphosphonates, by inducing apoptosis in osteoclasts (58), also eliminate osteoclast-osteoblast signaling essential for parathyroid hormone-stimulated bone formation. A therapeutic that inhibits osteoclast bone resorption, without affecting osteoclastogenesis, would be advantageous over bisphosphonates, which are the current gold standard for bone loss therapy. We show here, with the screening of a relatively small library, that selecting inhibitors of a3-B2 interaction is a viable approach. Although the compound we have selected (KM91104) may not show sufficient specificity, it provides proof-of-principle that a larger screen might provide the “magic bullet” specific to a3-B2 disruption.
In summary, we show here for the first time that the V-ATPase subunits a and B interact. A compound selected to inhibit this interaction reduced osteoclast resorption at concentrations that had no appreciable effect on osteoclastogenesis. A therapeutic that blocks resorption but not osteoclastogenesis would be advantageous over bisphosphonate treatment by preserving osteoclast-osteoblast signaling and thus could potentially be used concurrently and synergistically with osteoblast bone formation therapies. Our data also validate our approach for screening chemical libraries for potential osteoclast V-ATPase targeted bone-loss therapeutics and support the notion that the a3-B2 interaction is biologically important.
Acknowledgments
We thank Dr. Rhea Hudson (Hospital for Sick Children Research Institute, Toronto, Ontario, Canada) for helpful discussion on protein expression and purification and Drs. Beth S. Lee and Brooke McMichael (Department of Physiology and Cell Biology and Ohio State University College of Medicine, Ohio State University, Columbus) for providing cDNA clones for mouse a3 and B1 subunits and advice on osteoclast resorption assays. We also thank Thomas Sun and Frederick Vizeacoumar (Samuel Lunenfeld Research Institute SMART Robotics Facility, Mt. Sinai Hospital, Toronto, Canada) for help with high throughput screening and data analysis, Cristian Frusina for help with microscopy and imaging automation, and Feryal Sarraf and Nancy Valiquette (University of Toronto, Faculty of Dentistry) for help with ivory sectioning and staining. We thank the Toronto Zoo for the donation of elephant ivory from Tara, the former matriarch of the Toronto elephant herd, who died of natural causes at the age of 41 on November 30, 2009.
This work was supported by Canadian Arthritis Network Grant 2008-02-DAP-02 and Canadian Institutes for Health Research Grant MOP-79322 and PDD-86132.
- V-ATPase
- vacuolar H+-ATPase
- BMM
- bone marrow mononuclear
- KM91104
- 3,4-dihydroxy-N′-(2-hydroxybenzylidene)benzohydrazide
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- RANKL
- receptor activator of NF-κB ligand
- TRAP
- tartrate-resistant acid phosphatase
- TRX
- thioredoxin
- Ni-NTA
- nickel-nitrilotriacetic acid
- α-MEM
- α-minimal essential medium
- M-CSF
- macrophage-colony stimulating factor.
REFERENCES
- 1.Marshansky V., Futai M. (2008) Curr. Opin. Cell Biol. 20, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kane P. M. (2006) Microbiol. Mol. Biol. Rev. 70, 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jefferies K. C., Cipriano D. J., Forgac M. (2008) Arch. Biochem. Biophys. 476, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cipriano D. J., Wang Y., Bond S., Hinton A., Jefferies K. C., Qi J., Forgac M. (2008) Biochim. Biophys. Acta 1777, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saroussi S., Nelson N. (2009) J. Exp. Biol. 212, 1604–1610 [DOI] [PubMed] [Google Scholar]
- 6.Gluck S. L., Underhill D. M., Iyori M., Holliday L. S., Kostrominova T. Y., Lee B. S. (1996) Annu. Rev. Physiol. 58, 427–445 [DOI] [PubMed] [Google Scholar]
- 7.Stevens T. H., Forgac M. (1997) Annu. Rev. Cell Dev. Biol. 13, 779–808 [DOI] [PubMed] [Google Scholar]
- 8.Nelson N., Harvey W. R. (1999) Physiol. Rev. 79, 361–385 [DOI] [PubMed] [Google Scholar]
- 9.Forgac M. (1999) J. Biol. Chem. 274, 12951–12954 [DOI] [PubMed] [Google Scholar]
- 10.Forgac M. (2000) J. Exp. Biol. 203, 71–80 [DOI] [PubMed] [Google Scholar]
- 11.Nishi T., Forgac M. (2002) Nat. Rev. Mol. Cell Biol. 3, 94–103 [DOI] [PubMed] [Google Scholar]
- 12.Forgac M. (2007) Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 13.Xu J., Cheng T., Feng H. T., Pavlos N. J., Zheng M. H. (2007) Histol. Histopathol. 22, 443–454 [DOI] [PubMed] [Google Scholar]
- 14.Sennoune S. R., Bakunts K., Martínez G. M., Chua-Tuan J. L., Kebir Y., Attaya M. N., Martínez-Zaguilán R. (2004) Am. J. Physiol. Cell Physiol. 286, C1443–C1452 [DOI] [PubMed] [Google Scholar]
- 15.Hinton A., Sennoune S. R., Bond S., Fang M., Reuveni M., Sahagian G. G., Jay D., Martinez-Zaguilan R., Forgac M. (2009) J. Biol. Chem. 284, 16400–16408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama K., Nakano M., Imamura H., Yoshida M., Tamakoshi M. (2003) J. Biol. Chem. 278, 24255–24258 [DOI] [PubMed] [Google Scholar]
- 17.Imamura H., Nakano M., Noji H., Muneyuki E., Ohkuma S., Yoshida M., Yokoyama K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2312–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Zheng Y., Mazon H., Milgrom E., Kitagawa N., Kish-Trier E., Heck A. J., Kane P. M., Wilkens S. (2008) J. Biol. Chem. 283, 35983–35995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drory O., Nelson N. (2006) Physiology 21, 317–325 [DOI] [PubMed] [Google Scholar]
- 20.Cross R. L., Müller V. (2004) FEBS Lett. 576, 1–4 [DOI] [PubMed] [Google Scholar]
- 21.Kane P. M. (1995) J. Biol. Chem. 270, 17025–17032 [PubMed] [Google Scholar]
- 22.Huss M., Wieczorek H. (2007) FEBS Lett. 581, 5566–5572 [DOI] [PubMed] [Google Scholar]
- 23.Grüber G., Wieczorek H., Harvey W. R., Müller V. (2001) J. Exp. Biol. 204, 2597–2605 [DOI] [PubMed] [Google Scholar]
- 24.Muench S. P., Huss M., Song C. F., Phillips C., Wieczorek H., Trinick J., Harrison M. A. (2009) J. Mol. Biol. 386, 989–999 [DOI] [PubMed] [Google Scholar]
- 25.Landolt-Marticorena C., Williams K. M., Correa J., Chen W., Manolson M. F. (2000) J. Biol. Chem. 275, 15449–15457 [DOI] [PubMed] [Google Scholar]
- 26.Féthière J., Venzke D., Diepholz M., Seybert A., Geerlof A., Gentzel M., Wilm M., Böttcher B. (2004) J. Biol. Chem. 279, 40670–40676 [DOI] [PubMed] [Google Scholar]
- 27.Diepholz M., Börsch M., Böttcher B. (2008) Biochem. Soc. Trans. 36, 1027–1031 [DOI] [PubMed] [Google Scholar]
- 28.Raschke W. C., Baird S., Ralph P., Nakoinz I. (1978) Cell 15, 261–267 [DOI] [PubMed] [Google Scholar]
- 29.Hsu H., Lacey D. L., Dunstan C. R., Solovyev I., Colombero A., Timms E., Tan H. L., Elliott G., Kelley M. J., Sarosi I., Wang L., Xia X. Z., Elliott R., Chiu L., Black T., Scully S., Capparelli C., Morony S., Shimamoto G., Bass M. B., Boyle W. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collin-Osdoby P., Yu X., Zheng H., Osdoby P. (2003) Methods Mol. Med. 80, 153–166 [DOI] [PubMed] [Google Scholar]
- 31.Blair H. C., Teitelbaum S. L., Ghiselli R., Gluck S. (1989) Science 245, 855–857 [DOI] [PubMed] [Google Scholar]
- 32.Väänänen H. K., Karhukorpi E. K., Sundquist K., Wallmark B., Roininen I., Hentunen T., Tuukkanen J., Lakkakorpi P. (1990) J. Cell Biol. 111, 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Fattore A., Cappariello A., Teti A. (2008) Bone 42, 19–29 [DOI] [PubMed] [Google Scholar]
- 34.Kane P. M. (2007) J. Bioenerg. Biomembr. 39, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brideau C., Gunter B., Pikounis B., Liaw A. (2003) J. Biomol. Screen. 8, 634–647 [DOI] [PubMed] [Google Scholar]
- 36.Dolber P. C., Spach M. S. (1993) J. Histochem. Cytochem. 41, 465–469 [DOI] [PubMed] [Google Scholar]
- 37.Fields S., Song O. (1989) Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 38.Manolson M. F., Yu H., Chen W., Yao Y., Li K., Lees R. L., Heersche J. N. (2003) J. Biol. Chem. 278, 49271–49278 [DOI] [PubMed] [Google Scholar]
- 39.Lee B. S., Holliday L. S., Ojikutu B., Krits I., Gluck S. L. (1996) Am. J. Physiol. 270, C382–C388 [DOI] [PubMed] [Google Scholar]
- 40.Heinemann T., Bulwin G. C., Randall J., Schnieders B., Sandhoff K., Volk H. D., Milford E., Gullans S. R., Utku N. (1999) Genomics 57, 398–406 [DOI] [PubMed] [Google Scholar]
- 41.Holliday L. S., Lu M., Lee B. S., Nelson R. D., Solivan S., Zhang L., Gluck S. L. (2000) J. Biol. Chem. 275, 32331–32337 [DOI] [PubMed] [Google Scholar]
- 42.Lee B. S., Gluck S. L., Holliday L. S. (1999) J. Biol. Chem. 274, 29164–29171 [DOI] [PubMed] [Google Scholar]
- 43.Merkulova M., Bakulina A., Thaker Y. R., Grüber G., Marshansky V. (2010) Biochim. Biophys. Acta 1797, 1398–1409 [DOI] [PubMed] [Google Scholar]
- 44.Lau W. C., Rubinstein J. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1367–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maher M. J., Akimoto S., Iwata M., Nagata K., Hori Y., Yoshida M., Yokoyama S., Iwata S., Yokoyama K. (2009) EMBO J. 28, 3771–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 47.Nakamura I., Takahashi N., Udagawa N., Moriyama Y., Kurokawa T., Jimi E., Sasaki T., Suda T. (1997) FEBS Lett. 401, 207–212 [DOI] [PubMed] [Google Scholar]
- 48.Scimeca J. C., Franchi A., Trojani C., Parrinello H., Grosgeorge J., Robert C., Jaillon O., Poirier C., Gaudray P., Carle G. F. (2000) Bone 26, 207–213 [DOI] [PubMed] [Google Scholar]
- 49.Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., Stevens T. H. (2001) J. Biol. Chem. 276, 47411–47420 [DOI] [PubMed] [Google Scholar]
- 50.Marshansky V. (2007) Biochem. Soc. Trans. 35, 1092–1099 [DOI] [PubMed] [Google Scholar]
- 51.Landolt-Marticorena C., Kahr W. H., Zawarinski P., Correa J., Manolson M. F. (1999) J. Biol. Chem. 274, 26057–26064 [DOI] [PubMed] [Google Scholar]
- 52.Toyomura T., Murata Y., Yamamoto A., Oka T., Sun-Wada G. H., Wada Y., Futai M. (2003) J. Biol. Chem. 278, 22023–22030 [DOI] [PubMed] [Google Scholar]
- 53.Nyman J. K., Väänänen H. K. (2010) Calcif. Tissue Int. 87, 273–283 [DOI] [PubMed] [Google Scholar]
- 54.Hodsman A. B., Bauer D. C., Dempster D. W., Dian L., Hanley D. A., Harris S. T., Kendler D. L., McClung M. R., Miller P. D., Olszynski W. P., Orwoll E., Yuen C. K. (2005) Endocr. Rev. 26, 688–703 [DOI] [PubMed] [Google Scholar]
- 55.Ettinger B., San Martin J., Crans G., Pavo I. (2004) J. Bone Miner. Res. 19, 745–751 [DOI] [PubMed] [Google Scholar]
- 56.Black D. M., Greenspan S. L., Ensrud K. E., Palermo L., McGowan J. A., Lang T. F., Garnero P., Bouxsein M. L., Bilezikian J. P., Rosen C. J.PaTH Study Investigators (2003) N. Engl. J. Med. 349, 1207–1215 [DOI] [PubMed] [Google Scholar]
- 57.Finkelstein J. S., Hayes A., Hunzelman J. L., Wyland J. J., Lee H., Neer R. M. (2003) N. Engl. J. Med. 349, 1216–1226 [DOI] [PubMed] [Google Scholar]
- 58.Karsdal M. A., Martin T. J., Bollerslev J., Christiansen C., Henriksen K. (2007) J. Bone Miner. Res. 22, 487–494 [DOI] [PubMed] [Google Scholar]
- 59.Ediger B., Melman S. D., Pappas D. L., Jr., Finch M., Applen J., Parra K. J. (2009) J. Biol. Chem. 284, 19522–19532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee L. K., Stewart A. G., Donohoe M., Bernal R. A., Stock D. (2010) Nat. Struct. Mol. Biol. 17, 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Toei M., Forgac M. (2008) J. Biol. Chem. 283, 20696–20702 [DOI] [PMC free article] [PubMed] [Google Scholar]