Abstract
The antifungal activity of the plant defensin NaD1 involves specific interaction with the fungal cell wall, followed by permeabilization of the plasma membrane and entry of NaD1 into the cytoplasm. Prior to this study, the role of membrane permeabilization in the activity of NaD1, as well as the relevance of cell wall binding, had not been investigated. To address this, the permeabilization of Fusarium oxysporum f. sp. vasinfectum hyphae by NaD1 was investigated and compared with that by other antimicrobial peptides, including the cecropin-melittin hybrid peptide CP-29, the bovine peptide BMAP-28, and the human peptide LL-37, which are believed to act largely through membrane disruption. NaD1 appeared to permeabilize cells via a novel mechanism that required the presence of the fungal cell wall. NaD1 and Bac2A, a linear variant of the bovine peptide bactenecin, were able to enter the cytoplasm of treated hyphae, indicating that cell death is accelerated by interaction with intracellular targets.
Keywords: Antimicrobial Peptides, Cell Permeabilization, Cell Wall, Fluorescence, Fungi, Antifungal Peptides, NaD1, Plant Defensin
Introduction
Peptides with the ability to kill microbes are found ubiquitously throughout nature. The Antimicrobial Peptide Database (1) currently lists 1528 peptides from a diverse range of sources, including bacteria, fungi, fish, amphibians, spiders, mammals, and plants. Despite a number of common characteristics such as their small size and net positive charge, the mechanism of action for these peptides varies significantly. One common feature is their ability to permeabilize the plasma membrane of target cells. Initially, this permeabilization event was believed to be the sole event responsible for cell killing. Although this may be true for some peptides, it is now clear that other peptides may permeabilize membranes to access the cytoplasm and interact with intracellular targets (reviewed in Ref. 2).
Plants produce a number of cysteine-rich cationic peptides for protection against infection by potential microbial pathogens. Defensins are one such group of peptides and are one of the largest families of antimicrobial peptides in plants. Plant defensins are small (45–54 amino acids) basic proteins with four to five disulfide bonds (3). They share structural and functional similarities with defensins from insects (4), mammals (5), and fungi (6). A variety of functions have been attributed to plant defensins, including antibacterial activity (7, 8), protein synthesis inhibition (9), and inhibition of α-amylases and proteases (10–12). A large proportion of plant defensins display antifungal activity. NaD1 (Nicotiana alata defensin 1) is expressed at high concentrations in the flowers of the ornamental tobacco N. alata (13). It targets filamentous fungi and appears to act via a three-step process that begins with interaction with the fungal cell wall, followed by permeabilization of the plasma membrane and subsequent entry of the defensin into the cytoplasm (14).
Two groups of models have been proposed to explain how peptides interact with and disrupt membranes. The first, known as the barrel-stave model, involves the formation of discrete oligomeric pores that allow ions and other molecules to cross the membrane. Variations of this are the “toroidal pore” model, in which the pore is composed of both peptide monomers and lipid head groups, and the aggregate model, in which the peptides and lipids form informal aggregates within the membrane that permit ion leakage or peptide translocation across the membrane. The second type of model is termed the “carpet” model, in which at low concentrations, peptides lie across the surface of the membrane, and once a critical concentration is reached, the peptides insert in a detergent-like manner, causing the formation of micelles and ultimately the disintegration of the membrane (reviewed in Refs. 2, 15, and 16). Peptides that act via each of these mechanisms have been studied, and the models, or variations of them, probably hold true for many antimicrobial peptides. However, it is now well known that at their minimum effective concentrations, a number of peptides do not kill cells by permeabilizing the membrane in this manner.
In this study, we investigated permeabilization of the plasma membrane of the plant fungal pathogen Fusarium oxysporum f. sp. vasinfectum by the plant defensin NaD1 and compared the activity of this peptide with the antifungal activity of four other peptides. These peptides included the human and bovine cathelicidins LL-37 and BMAP-28; CP-29, a variant of the cecropin-melittin hybrid CEME (17); and Bac2A, a linear variant of the bovine peptide bactenecin (17). NaD1 seems to permeabilize the fungal membrane via a novel mechanism that requires the presence of the fungal cell wall.
EXPERIMENTAL PROCEDURES
Purification of NaD1 from Flowers
NaD1 was isolated from whole N. alata flowers as described previously (14).
Antifungal Activity
Antifungal activity against F. oxysporum f. sp. vasinfectum (Australian isolate VCG01111 isolated from cotton; a gift from Wayne O'Neill, Farming Systems Institute, Department of Primary Industries, Queensland, Australia) and Fusarium graminearum (wheat isolate CS3005; Commonwealth Scientific and Industrial Research Organization, University of Queensland) was assessed as described previously (14). Spores were isolated from cultures growing in half-strength potato dextrose broth (PDB).4 Spore concentrations were adjusted to 5 × 104 spores/ml in half-strength PDB, and 80 μl was added to the wells of sterile 96-well flat-bottomed microtiter plates along with 20 μl of filter-sterilized NaD1, CP-29, LL-37, BMAP-28, Bac2A, or water to give final protein concentrations of 0–10 μm. The plates were incubated in the dark at 25 °C for 24 h before hyphal growth was determined by measuring absorbance at 595 nm using a microtiter plate reader (SpectraMax Pro M5e, Molecular Devices). Each test was performed in quadruplicate.
Cell Viability Assay
F. oxysporum f. sp. vasinfectum hyphae were grown in half-strength PDB from a starting concentration of 5 × 104 spores/ml for 18 h at 25 °C (A595 = 0.38). Hyphal suspension (90 μl) was then transferred to 96-well microtiter plates along with 10 μl of filter-sterilized NaD1, CP-29, LL-37, BMAP-28, Bac2A, or water to give final protein concentrations of 0–10 μm. Plates were incubated for 2 h before the addition of 10 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; 5 mg/ml; Sigma). Plates were then incubated for 16 h at room temperature, followed by the addition of 100 μl of MTT solvent (0.1 n HCl in anhydrous isopropyl alcohol). The presence of MTT/formazan was monitored spectrophotometrically by measuring absorbance at 570 nm and subtracting background absorbance at 690 nm using a SpectraMax Pro M5e microtiter plate reader.
Kinetics of Membrane Permeabilization
F. oxysporum f. sp. vasinfectum hyphae were grown in half-strength PDB from a starting concentration of 5 × 104 spores/ml for 18 h at 25 °C. Hyphal suspension (90 μl) was then transferred to 96-well microtiter plates and incubated with SYTOX Green (0.5 μm) for 10 min prior to the addition of 10 μl of peptide solution to give a final peptide concentration of 2.5, 5, 10, 20, or 40 μm. SYTOX Green uptake was quantified by measuring fluorescence using a SpectraMax M5e microtiter plate reader with excitation and emission wavelengths of 488 and 538 nm, respectively. Readings were taken every 2 min for 2 h.
Liposome Preparation Using Purified Lipids
Calcein-entrapped small unilamellar liposomes (SUVs) were made from the lipids l-α-phosphatidylcholine (PC; egg), l-α-phosphatidyl-dl-glycerol (PG; chicken egg), l-α-phosphatidylethanolamine (PE; chicken egg), l-α-phosphatidylinositol (PI; bovine liver), and l-α-phosphatidylserine (PS; porcine brain) (all from Avanti Polar Lipids Inc.) and ergosterol (Sigma) as described by Zhang et al. (18). Lipid mixtures (3:1 PC/PG, 5:3:1:1 PC/PE/PI/PS, and 5:3:1:1:0.1 PC/PE/PI/PS/ergosterol) were dissolved in chloroform (10 mg/ml lipid) and then dried under a stream of nitrogen gas, followed by vacuum drying for 2 h in a 10 × 75-mm glass test tube. Lipid films were rehydrated with 1 ml of 5 mm HEPES (pH 7.4) containing 100 mm calcein (Sigma) for 2 h and freeze-thawed five times with liquid nitrogen. The solution was then sonicated for 15 min using a MicrosonTM XL sonicator (Misonix) to produce small unilamellar liposomes. Free calcein was removed by passing the liposome suspension through a Sephadex G-50 column (0.5 × 10 cm; Amersham Biosciences) and eluting in buffer containing 20 mm HEPES (pH 7.4), 150 mm NaCl, and 1 mm EDTA. Calcein-free liposomes of the same lipid composition were also made in 20 mm HEPES (pH 7.4), 150 mm NaCl, and 1 mm EDTA.
Calcein Release Assay
Calcein-entrapped liposomes (1 μl) and calcein-free liposomes (4 μl) were added to 1 ml of buffer (20 mm HEPES (pH 7.4), 150 mm NaCl, and 1 mm EDTA) in a four-sided quartz cuvette (PerkinElmer Life Sciences). Fluorescence was monitored using a SpectraMax M5e microtiter plate reader with excitation and emission wavelengths of 490 and 520 nm, respectively. Fluorescence was monitored before and after the addition of either Triton X-100 (final concentration of 0.1%) or test peptides (20 μm).
Fluorescent Labeling of Peptides with the Fluorophore BODIPY
Peptides were fluorescently labeled via carboxyl groups (NaD1 and CP-29) or reactive amines (Bac2A). For labeling via carboxyl groups, lyophilized peptide was dissolved in 0.1 m MES (pH 5.0) to a final concentration of 2 mm. The fluorescent tag BODIPY-FL-EDA (Molecular Probes) was added to a final concentration of 10 mm along with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (final concentration of 2 mm). For labeling via reactive amines, lyophilized peptide was dissolved in 0.1 m HEPES (pH 8.0), and the fluorescent tag BODIPY-FL-SE (Molecular Probes) was added to a final concentration of 2 mm. The reactions were incubated at room temperature for 2 h with gentle stirring before centrifugation (13,000 rpm, 10 min) to remove any precipitated protein. A Vivaspin 500 spin column (molecular weight cutoff of 3000 Da; Sartorius) was used to remove salts, unbound BODIPY, and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide. The BODIPY-labeled peptides were resuspended in water, and the protein concentration was determined using the BCA protein assay (Pierce).
Confocal Microscopy of BODIPY-labeled Peptides
Hyphae grown for 18 h in half-strength PDB from a starting concentration of 5 × 104 spores/ml were treated with BODIPY-labeled NaD1, CP-29, or Bac2A (5 μm) for 1 h. Hyphae were then visualized by confocal microscopy using a Leica TCS SP2 confocal microscope with an HCX APO 63×/W Corr/0.17 CS objective. Samples were excited at 488 nm with an argon laser while using an RP500 cutoff filter. BODIPY fluorescence was monitored at 509 nm. Images were captured using confocal LCS 3D software (Leica) and processed using Adobe Photoshop.
Effect of Cell Wall Modification on NaD1-induced Permeabilization
F. oxysporum f. sp. vasinfectum hyphae were grown for 18 h in half-strength PDB from a starting concentration of 5 × 104 spores/ml, and 1-ml samples were treated in one of the following ways prior to incubation with NaD1. (a) Hyphae were treated with proteinase K (1 mg/ml; Sigma) for 20 min at room temperature, followed by washing (3 × 10 min) with half-strength PDB. (b) Hyphae were collected by centrifugation (10,000 × g, 10 min) and resuspended in 50 mm NaOH containing 10 mm DTT. After a 20-min incubation at room temperature, hyphae were washed (3 × 10 min) with half-strength PDB. (c) Hyphae were treated with β-glucanase (2 mg/ml; Sigma) for 1 h at 30 °C, followed by washing (3 × 10 min) with half-strength PDB. (d) Hyphae were incubated with concanavalin A (100 μg/ml; Sigma) for 1 h at room temperature, followed by washing (3 × 10 min) with half-strength PDB. Samples were then incubated with NaD1 (2 μm) for 1 h prior to the addition of SYTOX Green (final concentration of 1 μm) and visualization by fluorescence microscopy using an Olympus BX51 fluorescence microscope. SYTOX Green fluorescence was detected using an MWIB filter (excitation wavelength of 460–490 nm). Images were captured using a SPOT RT 3CCD digital camera (Diagnostic Instruments) and processed using Adobe Photoshop.
Effect of Cell Wall Modification on NaD1-induced Cell Death
Hyphae were pretreated with either proteinase K or β-glucanase and then incubated for 2 h with NaD1, CP-29, or Bac2A (5 μm). Cell viability was monitored using the MTT assay as described above.
RESULTS
Antifungal Activity
The amino acid sequences and characteristics of the peptides used in this study are listed in Table 1. The IC50 of each peptide against the fungal pathogens F. oxysporum f. sp. vasinfectum and F. graminearum is provided in Table 2. NaD1 and Bac2A were the most efficient, inhibiting the hyphal growth of both pathogens at 1 μm or less.
TABLE 1.
Amino acid sequences of antimicrobial peptides used in this study
| Peptide | Type | Sequence | Charge |
|---|---|---|---|
| NaD1 | Mixed αβ | RECKTESNTFPGICITKPPCRKACISEKFTDGHCSKILRRCLCTKPC | +7 |
| CP-29 | α-Helical | KWKSFIKKLTTAVKKVLTTGLPALIS | +6 |
| LL-37 | α-Helical | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | +5 |
| BMAP-28 | α-Helical | GGLRSLGRKILRAWKKYGPIIVPIIRIG | +6 |
| Bac2A | Unknown | RLARIVVIRVAR-NH2 | +4 |
TABLE 2.
IC50 values for peptides against fungal pathogens
| Fungal strain | IC50 |
||||
|---|---|---|---|---|---|
| NaD1 | CP-29 | LL-37 | Bac2A | BMAP-28 | |
| μm | |||||
| F. oxysporum f. sp. vasinfectum | 1 | 3.5 | 2.5 | 1 | 3 |
| F. graminearum | 0.5 | 1.25 | 1 | 1 | 2 |
Effect of NaD1 on Cell Viability
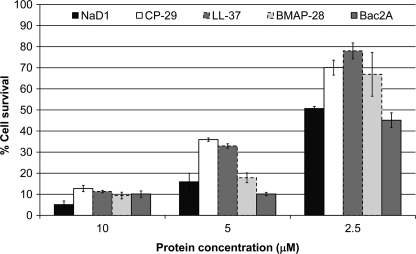
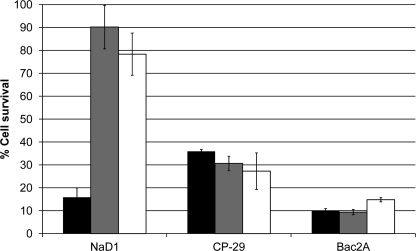
Because peptides that inhibit the growth of fungal pathogens can be either fungicidal or fungistatic, the effect of NaD1 on the viability of hyphal cells was examined. Cell viability was monitored using an MTT-based assay that measures production of MTT/formazan crystals by mitochondrial dehydrogenases in living cells. After treatment with 2.5 μm NaD1, only 50% of the cells remained viable (Fig. 1). This number decreased to 16 and 5% in the presence of 5 and 10 μm NaD1, respectively. The peptides CP-29, LL-37, BMAP-28, and Bac2A also killed hyphal cells. Consistent with results from growth inhibition assays, Bac2A killed cells as efficiently as NaD1, whereas CP-29, LL-37, and BMAP-28 were slightly less effective, leading to 30, 32, and 21% cell death, respectively, at a concentration of 2.5 μm.
FIGURE 1.
Cell viability of hyphal cells after treatment with antifungal peptides. The viability of fungal cells after treatment with NaD1 (black bars), CP-29 (white bars), LL-37 (dark gray bars, dashed edge), BMAP-28 (light gray bars), and Bac2A (dark gray bars, solid edge) was monitored using an MTT-based assay. All peptides were able to kill fungal cells, with NaD1 and Bac2A being the most effective. Error bars represent S.E. (n = 4).
Kinetics of Membrane Permeabilization
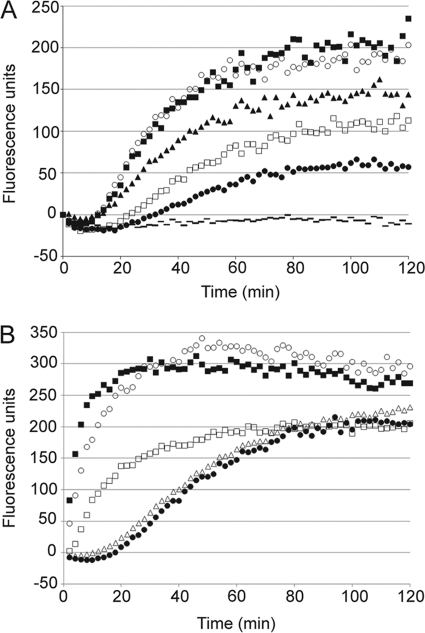
The rate at which NaD1 permeabilized F. oxysporum f. sp. vasinfectum hyphal membranes was monitored over time by measuring the uptake of the normally impermeable nucleic acid-binding molecule SYTOX Green. At all concentrations, permeabilization was observed only after a lag time of 15 min, and fluorescence began to plateau after 80 min (Fig. 2A). The rate of permeabilization was partially concentration-dependent and increased with increasing concentrations of NaD1 up to 20 μm. At concentrations above 20 μm, there was no difference in the kinetics of SYTOX Green uptake, suggesting that permeabilization was saturable. The rate of permeabilization was also examined for four other cationic antimicrobial peptides, CP-29, LL-37, Bac2A, and BMAP-28. At 5 μm, CP-29 and BMAP-28 permeabilized cells very rapidly, with >80% of the total SYTOX Green uptake occurring within 20 min (Fig. 2B). For LL-37, uptake demonstrated different kinetics but still reached 75% within 20 min. Fluorescence began to plateau after 40 min. Bac2A permeabilized hyphae with very similar kinetics to NaD1.
FIGURE 2.
Kinetics of hyphal membrane permeabilization. Permeabilization of F. oxysporum f. sp. vasinfectum hyphae induced by varying concentrations of NaD1 (40 μm (○), 20 μm (■), 10 μm (▴), 5 μm (□), 2.5 μm (●), and no NaD1 ( )) (A) or a range of antifungal peptides (B) was monitored using a SYTOX Green uptake assay. A, after a lag time of 15 min, NaD1 caused a concentration-dependent increase in SYTOX Green uptake up to 80 min. At concentrations over 20 μm, there was no increase in the rate of permeabilization. B, at 5 μm, CP-29 (■) and BMAP-28 (○) caused rapid SYTOX Green uptake with maximum fluorescence reached within 20 min. LL-37 (□) was slightly slower, taking 40 min to reach the maximum. NaD1 (●) and Bac2A (△) had very similar kinetics, with a slight lag prior to induction of SYTOX Green uptake, followed by a linear increase in fluorescence up to 80 min. Data are from a single experiment representative of three individual replicates.
)) (A) or a range of antifungal peptides (B) was monitored using a SYTOX Green uptake assay. A, after a lag time of 15 min, NaD1 caused a concentration-dependent increase in SYTOX Green uptake up to 80 min. At concentrations over 20 μm, there was no increase in the rate of permeabilization. B, at 5 μm, CP-29 (■) and BMAP-28 (○) caused rapid SYTOX Green uptake with maximum fluorescence reached within 20 min. LL-37 (□) was slightly slower, taking 40 min to reach the maximum. NaD1 (●) and Bac2A (△) had very similar kinetics, with a slight lag prior to induction of SYTOX Green uptake, followed by a linear increase in fluorescence up to 80 min. Data are from a single experiment representative of three individual replicates.
Effect of Peptides on Artificial Liposomes
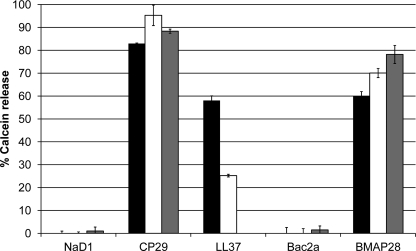
The peptides (20 μm, 1:10 peptide/lipid ratio) were tested for their ability to disrupt artificial liposomes using a calcein release assay. Calcein-entrapped liposomes were prepared with either PC/PG at a ratio of 3:1 (microbe-like membranes) or PC/PE/PS/PI at a ratio of 5:3:1:1 with or without ergosterol (fungus-like membranes), respectively. CP-29, LL-37, and BMAP-28 induced calcein release from both PC/PG and PC/PE/PS/PI liposomes. CP-29 was the most efficient, causing 83 and 95% calcein release from PC/PG or PC/PE/PS/PI liposomes, respectively, whereas BMAP-28 caused 60 and 70%, respectively (Fig. 3). The presence of ergosterol did not affect the peptides' ability to permeabilize the liposomes. LL-37 was similarly effective against PC/PG liposomes (58% calcein release) but caused only 25% calcein release from PC/PE/PS/PI liposomes. This peptide was not tested against liposomes containing ergosterol. NaD1 and Bac2A did not disrupt any of the liposomes (Fig. 3), even at 40 μm (1:5 peptide/lipid ratio) (data not shown).
FIGURE 3.
Release of calcein from artificial liposomes. The ability of peptides to disrupt artificial liposomes was measured using a calcein release assay. NaD1 and Bac2A failed to release calcein from liposomes composed of PC/PG (3:1; black bars), PC/PE/PS/PI (5:3:1:1; white bars), or PC/PE/PS/PI/ergosterol (5:3:1:1:0.1; gray bars). CP-29 and BMAP-28 induced substantial calcein release from all three liposomes, whereas LL-37 was more effective against PC/PG liposomes but was still able to disrupt PC/PE/PS/PI liposomes. Percent calcein release is relative to Triton-X-100 (0.1%), and error bars represent S.E. (n = 4). All peptides were used at 20 μm.
Confocal Microscopy of BODIPY-labeled Peptides
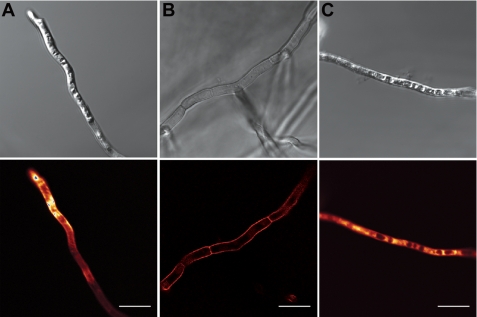
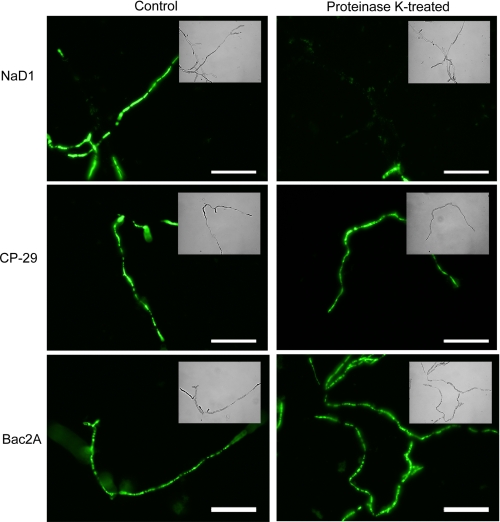
The location of fluorescently labeled peptides in and on treated hyphae was monitored by confocal microscopy. The small uncharged fluorescent label BODIPY was used to label peptides via their carboxyl groups (NaD1 and CP-29) or N-terminal reactive amines (Bac2A). All labeled peptides retained full antifungal activity (data not shown). NaD1 accumulated to high concentrations in the cytoplasm of hyphae, and after 1 h, cells that had taken up NaD1 had a very granular appearance (Fig. 4A). Like NaD1, Bac2A entered hyphae and was abundant in the cytoplasm, which appeared granular (Fig. 4C). In contrast to NaD1 and Bac2A, CP-29 was located predominantly along the cell surface, and the cytoplasm appeared relatively normal (Fig. 4B).
FIGURE 4.
Location of fluorescently labeled peptides in treated hyphae. The location of fluorescently labeled NaD1 (A), CP-29 (B), and Bac2A (C) was examined by confocal microscopy. NaD1 and Bac2A were located inside the cytoplasm of treated hyphae, whereas CP-29 was located at the cell surface. NaD1- and Bac2A-treated hyphae exhibited significant granulation of the cytoplasm. Scale bars = 10 μm.
Effect of Cell Wall Modifications on Permeabilization
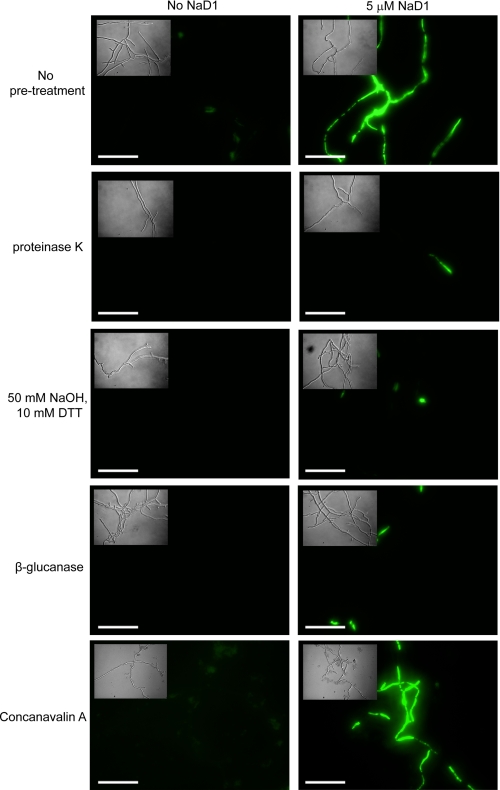
Fungal hyphae are surrounded by a cell wall that antifungal peptides must pass through before they reach the plasma membrane. We thus investigated whether modifications to the cell wall affected membrane permeabilization by antifungal peptides. After removal of the outer layer of glycosylated proteins with proteinase K, NaD1 failed to permeabilize the plasma membrane (Fig. 5). Removal of the (1–3)-β-glucan layer by treatment with either sodium hydroxide and DTT or β-glucanase also prevented NaD1 from permeabilizing the membrane (Fig. 5). Incubation of hyphae with concanavalin A, a lectin that binds to glucose and mannose residues, did not affect the ability of NaD1 to permeabilize the membrane. CP-29 and Bac2A were both able to permeabilize proteinase K-treated hyphae (Fig. 6).
FIGURE 5.
Permeabilization of hyphae after cell wall removal. SYTOX Green uptake was used to monitor NaD1-induced permeabilization of hyphal membranes after removal of either the outer layer of glycosylated proteins (proteinase K) or the (1–3)-β-glucan layer (50 mm NaOH and 10 mm DTT or β-glucanase) in the hyphal cell wall or after incubation of hyphae with concanavalin A. NaD1 (5 μm) failed to permeabilize hyphae when these layers had been removed. Scale bars = 50 μm.
FIGURE 6.
Permeabilization of hyphae after proteinase K treatment. The ability of peptides to permeabilize the plasma membrane of hyphae after removal of the outer layer of glycosylated proteins was monitored by SYTOX Green uptake. NaD1 (5 μm) was unable to permeabilize fungal hyphae following proteinase K treatment. In contrast, CP-29 and Bac2A (both at 5 μm) permeabilized proteinase K-treated hyphae as efficiently as the untreated control. Scale bars = 50 μm.
Effect of Cell Wall Modifications on NaD1-induced Cell Death
The ability of NaD1 to kill fungal cells that had been treated with proteinase K or β-glucanase was also investigated. Treatment with either of these enzymes prevented NaD1-induced killing of hyphal cells (Fig. 7). In contrast, the activities of CP-29 and Bac2A were unaffected.
FIGURE 7.
Killing of hyphal cells after cell wall treatments. The death of untreated fungal hyphae (black bars) was compared with that of hyphae that had been treated with either proteinase K (gray bars) or β-glucanase (white bars). Both proteinase K and β-glucanase treatments prevented NaD1-induced cell death. In contrast, CP-29 and Bac2A killed treated hyphae as efficiently as the untreated control. All peptides were used at 5 μm, and error bars represent S.E. (n = 4).
DISCUSSION
Plant defensins with potent antifungal activity have the potential to be used in both transgenic plant and human pharmaceutical applications. The antifungal activity of NaD1, a defensin from the flowers of N. alata, involves specific interaction with the fungal cell wall, followed by permeabilization of the plasma membrane and entry of NaD1 into the cytoplasm (14). To investigate the role of membrane permeabilization in NaD1-induced cell death, permeabilization of F. oxysporum f. sp. vasinfectum hyphae by NaD1 was compared with that by antimicrobial peptides that are believed to act solely through membrane disruption.
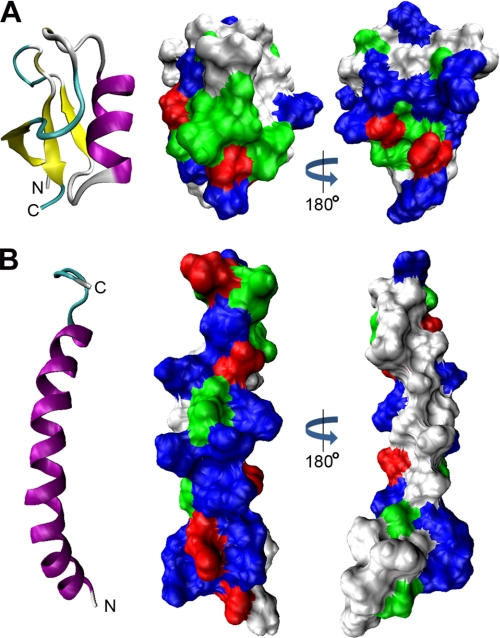
Antimicrobial peptides can be categorized based on their structure into α-helical peptides, β-sheet peptides, mixed α-helical β-sheet peptides, and peptides with an extended conformation. They can be further categorized into linear peptides and peptides containing disulfide bonds. Many linear antimicrobial peptides, including LL-37 and CP-29, are unstructured in an aqueous environment and adopt an α-helical structure when they come into contact with membranes (19). This α-helical structure is usually amphipathic (Fig. 8), allowing the peptide to interact with phospholipid bilayers, which is key to membrane disruption and translocation across membranes. The extent to which they do this is highly variable, with some α-helical peptides such as bee melittin being very membrane lytic even for eukaryotic (less charged) membranes as well as anionic bacterial membranes, whereas others such as toad buforin translocate into cells and act on cytoplasmic targets. Despite significant sequence variation, plant defensins form a well conserved structure consisting of an α-helix tethered to a triple-stranded β-sheet by three disulfide bonds (3, 20). A fourth disulfide joins the N and C termini. The resulting structure is a highly stable cystine-stabilized αβ-motif (CSαβ) (Fig. 8). Although some defensins display an amphipathic distribution of charged and hydrophobic resides on the surface, NaD1 does not. This suggests that it is unlikely that NaD1 interacts directly with the phospholipid membrane via a classical model. Permeabilization of the fungal hyphae by NaD1 was compared with that by four other cationic antimicrobial peptides, three of which have been postulated to exert their antibacterial activity via direct permeabilization of the plasma membrane.
FIGURE 8.
Surface charge distribution of NaD1 and LL-37. A and B, comparison of the three-dimensional structures and surface charge distributions of NaD1 (Protein Data Bank code 1MR4) and LL-37 (code 2K6O), respectively. Ribbon diagrams show β-strands in yellow and α-helices in purple. Space-filling models show basic residues in blue, acidic residues in red, polar residues in green, and hydrophobic residues in white. A, NaD1 contains a conserved triple-stranded antiparallel β-sheet tethered to an α-helix. The molecule is hydrophilic and lacks a large hydrophobic face. B, LL-37 forms a single amphipathic α-helix.
LL-37 is the only human member of the cathelicidin family of proteins. It inhibits the growth of bacteria (21) and yeast (22), although its physiological role is probably more related to its modulation of the innate immune system (23, 24). BMAP-28 is a bovine cathelicidin that has antibacterial and antifungal activities (25). The N-terminal region of this peptide forms an amphipathic α-helix, whereas the C terminus forms a hydrophobic tail. CP-29 is a synthetic peptide based on the cecropin-melittin hybrid CEME (19). It was designed to have an increased amphipathic α-helical content, has improved activity on Gram-negative bacteria, and is believed to act via membrane perturbation (19, 26). Bac2A is a linear variant of bactenecin, a peptide produced in bovine neutrophils (17). The two cysteine residues that form the disulfide bond in bactenecin have been replaced with alanines, and the C terminus has been amidated to increase the charge of the protein. The antifungal activity of LL-37 and BMAP-28 has been reported previously (22, 27). The activity of all five peptides was examined against two agronomically important fungal pathogens, F. oxysporum f. sp. vasinfectum and F. graminearum. All five peptides were efficient inhibitors of fungal growth, although NaD1 and Bac2A were the most effective. The peptides all appear to act via a fungicidal mechanism, as measured by an MTT-based cell viability assay.
To compare the relative membrane-permeabilizing activities of these peptides, the rate of fungal membrane disruption was monitored using a SYTOX Green uptake assay. CP-29, BMAP-28, and LL-37 all caused rapid permeabilization, consistent with reports of LL-37 activity against Candida albicans (22) and CP-29 and BMAP-28 activity against bacteria (19, 25). This supports the hypothesis that these peptides act primarily by direct interaction with and disruption of the plasma membrane. In contrast, SYTOX Green uptake in the presence of NaD1 was much slower, with no significant permeabilization observed within the first 10 min. This lag time might have represented the time required for NaD1 to concentrate on and cross the fungal cell wall, to interact with a receptor, or to assemble in the membrane. Maximum permeabilization was achieved only after 80 min. Interestingly, Bac2A displayed very similar SYTOX Green uptake kinetics to NaD1, suggesting that these two peptides may have similar mechanisms of permeabilization. Given their substantial sequence and structural differences, it seems unlikely that this would involve a common receptor. This is consistent with observations that the permeabilization of Gram-positive bacteria by Bac2A is significantly slower than that by CP-29 (28). The effect of NaD1 concentration on SYTOX Green entry was also investigated. At all concentrations tested, NaD1 started permeabilizing only after 10 min and took 80 min to reach maximum SYTOX Green uptake. At concentrations above 20 μm, there was no change in the kinetics of SYTOX Green uptake, indicating that the process was saturable. This may indicate the requirement for a receptor. In contrast to mammalian defensins, NaD1 requires its disulfide bonds for activity, indicating that its three-dimensional structure is essential (14). This is also consistent with the proposal that interaction with a receptor may be involved. Alternatively, the slow permeabilization kinetics of NaD1 and Bac2A may represent the time taken to interact with intracellular targets and cause cell death, which would subsequently result in disintegration of the membrane.
Most antimicrobial peptides that exhibit their effects through membrane permeabilization also cause the disruption of artificial liposomes and bilayers at low concentrations, and these provide valuable tools for assaying the interaction of these peptides with membranes. The effect of NaD1 on SUVs composed of combinations of purified lipids representative of bacterial (PC/PG) and fungal (PC/PE/PS/PI with or without ergosterol) membranes was examined. In both cases, even at the very high peptide/lipid ratio of 1:5, NaD1 failed to cause release of the encapsulated fluorescent molecule calcein, indicating that it was unable to permeabilize the SUVs. This supports the idea that NaD1-induced permeabilization requires the presence of a receptor. This finding is also consistent with the finding that the plant defensin RsAFP2 is unable to permeabilize liposomes containing glucosylceramide, a known binding partner of the peptide (29). In contrast, LL-37, CP-29, and BMAP-28 caused rapid calcein release due to disruption of the SUVs. Like NaD1, Bac2A was also unable to disrupt SUVs. This is consistent with the fact that the two proteins have similar permeabilization kinetics on intact fungal hyphae. Studies on the Gram-positive bacterium Staphylococcus aureus indicated that Bac2A does not destroy membrane potential at its minimum effective concentrations and instead causes a range of physiological defects (10). Consistent with this, in this study, it did not affect the stability of the fungal membrane under the same conditions that led to destabilization of the membrane by CP-29.
Interestingly, the two peptides that were least efficient in permeabilizing both the fungal membrane and artificial liposomes (NaD1 and Bac2A) had the highest antifungal activity. This could indicate that other (possibly intracellular) targets are involved in their activity and that these targets represent a more efficient method of killing cells. When the relative activities of CP-29 and Bac2A against S. aureus were measured, Bac2A had a 2-fold lower minimum inhibitory concentration but caused substantially less membrane permeabilization (26). We have reported previously that NaD1 is able to enter the fungal cytoplasm (14), but the location of Bac2A had not been investigated. The location of fluorescently labeled peptides on live hyphae was examined by confocal microscopy. NaD1 and Bac2A were both found at high concentrations in the cytoplasm of hyphae that had a granular appearance.
In contrast, CP-29 was restricted to the cell surface of the hyphae. This is consistent with the permeabilization kinetics of the peptides on fungal hyphae and their ability to disrupt artificial liposomes. It suggests that membrane permeabilization may be solely responsible for the activity of CP-29 but not NaD1 and Bac2A. The observation that Bac2A caused granulation of the hyphal cytoplasm and CP-29 did not is consistent with their reported activities on bacterial cells in that S. aureus cells treated with Bac2A show intracytoplasmic membrane inclusions and DNA condensation, whereas cells treated with CP-29 do not (26). Previous work has shown that LL-37 is located along the plasma membrane of C. albicans (30). In contrast, a truncated variant of the peptide, RK-31, is not restricted to the plasma membrane and enters the cytoplasm. This truncated variant has significantly better antifungal activity than LL-37, potentially due to interaction with an intracellular target (30). The data presented here show that LL-37 causes less membrane permeabilization but is more lethal to fungal hyphae compared with CP-29 and BMAP-28. It is possible that LL-37 acts predominantly by damaging the plasma membrane but that some LL-37 is able to enter the cytoplasm and interact with the same intracellular target as RK-31. Consistent with this concept, LL-37 enters eukaryotic cells freely (31).
Like bacteria, fungi are surrounded by a cell wall, although the composition of these cell walls is significantly different. The cell wall of Gram-positive bacteria is composed mainly of peptidoglycan with proteins and teichoic acids attached, whereas the cell wall of Gram-negative bacteria consists of a thin layer of peptidoglycan adjacent to the plasma membrane as well as an outer membrane (32). This outer membrane contains phospholipids in addition to proteins and large amounts of LPS. The fungal cell wall is composed of a chitin layer that sits adjacent to the plasma membrane topped by a (1–3)-β-glucan network that also contains (1–6)-β-glucosidic linkages (33). The outermost layer of the cell wall is composed of highly glycosylated proteins that are anchored via glycopeptide linkages to the (1–3)-β-glucan layer. For peptides that act directly with the plasma membrane, removal of the cell wall does not inhibit their activity (34). To examine the role of the cell wall in the activity of NaD1, hyphae were treated with proteinase K to degrade the outer layer of glycosylated proteins or with NaOH and DTT to degrade the β-glucan network. When the outer proteinaceous layer was removed, through either direct digestion with proteinase K or removal of the β-glucan network, NaD1 was unable to either permeabilize the plasma membrane or kill hyphal cells. This proteinaceous layer may therefore be the location of an NaD1 receptor. Proteins in the fungal cell wall are required for the activity of other antifungal peptides such as the tobacco protein osmotin and the human peptide histatin-5. Osmotin interacts with the carbohydrate groups attached to the proteins (35), whereas histatin-5 interacts with a specific protein known as Ssa1 (36). The inability of concanavalin A to block the activity of NaD1 suggests that NaD1 does not need to bind to the carbohydrate side chains of glycoproteins in the cell wall. CP-29 and Bac2A were also investigated for their ability to act on cells that had been treated with proteinase K or β-glucanase. Both peptides were able to permeabilize the membrane of hyphae that had been treated with proteinase K and kill hyphal cells that had been treated with either proteinase K or β-glucanase. This suggests that these peptides do not require cell wall proteins for their activity.
In summary, it appears that NaD1 permeabilizes the plasma membrane of fungal hyphae via a novel mechanism that may involve a receptor located in the proteinaceous layer of the cell wall. CP-29, BMAP-28, and LL-37 appear to kill fungal cells primarily via disruption of the plasma membrane, whereas NaD1 and Bac2A appear to enhance cell killing by interacting with intracellular targets. The correlation of membrane permeabilization kinetics with the location of peptides suggests that this method could be used to predict whether peptides require interaction with intracellular targets for their activity.
Acknowledgments
We thank Barbara Barbeta for assistance with the confocal microscopy and Melissa Elliot for assistance with the calcein release assays.
This work was supported in part by an Australian Research Council grant (to M. A. A.).
- PDB
- potato dextrose broth
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- SUV
- small unilamellar liposome
- PC
- l-α-phosphatidylcholine
- PG
- l-α-phosphatidyl-dl-glycerol
- PE
- l-α-phosphatidylethanolamine
- PI
- l-α-phosphatidylinositol
- PS
- l-α-phosphatidylserine.
REFERENCES
- 1.Wang Z., Wang G. (2004) Nucleic Acids Res. 32, D590–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hale J. D., Hancock R. E. W. (2007) Expert Rev. Anti-infect. Ther. 5, 951–959 [DOI] [PubMed] [Google Scholar]
- 3.Lay F. T., Anderson M. A. (2005) Curr. Protein Pept. Sci. 6, 85–101 [DOI] [PubMed] [Google Scholar]
- 4.Bulet P., Stöcklin R. (2005) Protein Pept. Lett. 12, 3–11 [DOI] [PubMed] [Google Scholar]
- 5.Selsted M. E., Ouellette A. J. (2005) Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 6.Mygind P. H., Fischer R. L., Schnorr K. M., Hansen M. T., Sönksen C. P., Ludvigsen S., Raventós D., Buskov S., Christensen B., De Maria L., Taboureau O., Yaver D., Elvig-Jørgensen S. G., Sørensen M. V., Christensen B. E., Kjaerulff S., Frimodt-Moller N., Lehrer R. I., Zasloff M., Kristensen H. H. (2005) Nature 437, 975–980 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Lewis K. (1997) FEMS Microbiol. Lett. 149, 59–64 [DOI] [PubMed] [Google Scholar]
- 8.Chen G. H., Hsu M. P., Tan C. H., Sung H. Y., Kuo C. G., Fan M. J., Chen H. M., Chen S., Chen C. S. (2005) J. Agric. Food Chem. 53, 982–988 [DOI] [PubMed] [Google Scholar]
- 9.Mendez E., Moreno A., Colilla F., Pelaez F., Limas G. G., Mendez R., Soriano F., Salinas M., de Haro C. (1990) Eur. J. Biochem. 194, 533–539 [DOI] [PubMed] [Google Scholar]
- 10.Colilla F. J., Rocher A., Mendez E. (1990) FEBS Lett. 270, 191–194 [DOI] [PubMed] [Google Scholar]
- 11.Bloch C., Jr., Richardson M. (1991) FEBS Lett. 279, 101–104 [DOI] [PubMed] [Google Scholar]
- 12.Thomma B. P., Cammue B. P., Thevissen K. (2002) Planta 216, 193–202 [DOI] [PubMed] [Google Scholar]
- 13.Lay F. T., Brugliera F., Anderson M. A. (2003) Plant Physiol. 131, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Weerden N. L., Lay F. T., Anderson M. A. (2008) J. Biol. Chem. 283, 14445–14452 [DOI] [PubMed] [Google Scholar]
- 15.Brogden K. A. (2005) Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 16.Oren Z., Shai Y. (1998) Biopolymers 47, 451–463 [DOI] [PubMed] [Google Scholar]
- 17.Wu M., Hancock R. E. W. (1999) Antimicrob. Agents Chemother. 43, 1274–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., Rozek A., Hancock R. E. W. (2001) J. Biol. Chem. 276, 35714–35722 [DOI] [PubMed] [Google Scholar]
- 19.Friedrich C., Scott M. G., Karunaratne N., Yan H., Hancock R. E. (1999) Antimicrob. Agents Chemother. 43, 1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lay F. T., Schirra H. J., Scanlon M. J., Anderson M. A., Craik D. J. (2003) J. Mol. Biol. 325, 175–188 [DOI] [PubMed] [Google Scholar]
- 21.Larrick J. W., Hirata M., Zhong J., Wright S. C. (1995) Immunotechnology 1, 65–72 [DOI] [PubMed] [Google Scholar]
- 22.López-García B., Lee P. H., Yamasaki K., Gallo R. L. (2005) J. Invest. Dermatol. 125, 108–115 [DOI] [PubMed] [Google Scholar]
- 23.Mookherjee N., Brown K. L., Bowdish D. M., Doria S., Falsafi R., Hokamp K., Roche F. M., Mu R., Doho G. H., Pistolic J., Powers J. P., Bryan J., Brinkman F. S., Hancock R. E. W. (2006) J. Immunol. 176, 2455–2464 [DOI] [PubMed] [Google Scholar]
- 24.Bowdish D. M., Davidson D. J., Scott M. G., Hancock R. E. W. (2005) Antimicrob. Agents Chemother. 49, 1727–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skerlavaj B., Gennaro R., Bagella L., Merluzzi L., Risso A., Zanetti M. (1996) J. Biol. Chem. 271, 28375–28381 [DOI] [PubMed] [Google Scholar]
- 26.Friedrich C. L., Moyles D., Beveridge T. J., Hancock R. E. W. (2000) Antimicrob. Agents Chemother. 44, 2086–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benincasa M., Scocchi M., Pacor S., Tossi A., Nobili D., Basaglia G., Busetti M., Gennaro R. (2006) J. Antimicrob. Chemother. 58, 950–959 [DOI] [PubMed] [Google Scholar]
- 28.Wu M., Maier E., Benz R., Hancock R. E. W. (1999) Biochemistry 38, 7235–7242 [DOI] [PubMed] [Google Scholar]
- 29.Aerts A. M., François I. E., Meert E. M., Li Q. T., Cammue B. P., Thevissen K. (2007) J. Mol. Microbiol. Biotechnol. 13, 243–247 [DOI] [PubMed] [Google Scholar]
- 30.den Hertog A. L., van Marle J., Veerman E. C., Valentijn-Benz M., Nazmi K., Kalay H., Grün C. H., Van't Hof W., Bolscher J. G., Nieuw Amerongen A. V. (2006) Biol. Chem. 387, 1495–1502 [DOI] [PubMed] [Google Scholar]
- 31.Lau Y. E., Rozek A., Scott M. G., Goosney D. L., Davidson D. J., Hancock R. E. W. (2005) Infect. Immun. 73, 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabeen M. T., Jacobs-Wagner C. (2005) Nat. Rev. Microbiol. 3, 601–610 [DOI] [PubMed] [Google Scholar]
- 33.Bowman S. M., Free S. J. (2006) BioEssays 28, 799–808 [DOI] [PubMed] [Google Scholar]
- 34.Lee D. G., Kim D. H., Park Y., Kim H. K., Kim H. N., Shin Y. K., Choi C. H., Hahm K. S. (2001) Biochem. Biophys. Res. Commun. 282, 570–574 [DOI] [PubMed] [Google Scholar]
- 35.Ibeas J. I., Lee H., Damsz B., Prasad D. T., Pardo J. M., Hasegawa P. M., Bressan R. A., Narasimhan M. L. (2000) Plant J. 23, 375–383 [DOI] [PubMed] [Google Scholar]
- 36.Li X. S., Reddy M. S., Baev D., Edgerton M. (2003) J. Biol. Chem. 278, 28553–28561 [DOI] [PubMed] [Google Scholar]