Abstract
Bone morphogenetic proteins (BMPs) are critically involved in early development and cell differentiation. In humans, dysfunction of the bone morphogenetic protein type II receptor (BMPR-II) is associated with pulmonary arterial hypertension (PAH) and neoplasia. The ability of Kaposi sarcoma-associated herpesvirus (KSHV), the etiologic agent of Kaposi sarcoma and primary effusion lymphoma, to down-regulate cell surface receptor expression is well documented. Here we show that KSHV infection reduces cell surface BMPR-II. We propose that this occurs through the expression of the viral lytic gene, K5, a ubiquitin E3 ligase. Ectopic expression of K5 leads to BMPR-II ubiquitination and lysosomal degradation with a consequent decrease in BMP signaling. The down-regulation by K5 is dependent on both its RING domain and a membrane-proximal lysine in the cytoplasmic domain of BMPR-II. We demonstrate that expression of BMPR-II protein is constitutively regulated by lysosomal degradation in vascular cells and provide preliminary evidence for the involvement of the mammalian E3 ligase, Itch, in the constitutive degradation of BMPR-II. Disruption of BMP signaling may therefore play a role in the pathobiology of diseases caused by KSHV infection, as well as KSHV-associated tumorigenesis and vascular disease.
Keywords: Bone Morphogenetic Protein (BMP), E3 Ubiquitin Ligase, Herpesvirus, Lysosomes, Receptor Regulation, Trafficking, Transforming Growth Factor Beta (TGFbeta), Ubiquitin
Introduction
Bone morphogenetic proteins are multifunctional cytokines regulating growth and differentiation (1). BMPs,3 similar to other members of the transforming growth factor-β superfamily, signal by ligating heterodimeric transmembrane serine-threonine kinase receptors. Association of the constitutively active type II receptor leads to activation of the type I receptor and phosphorylation of signaling Smad proteins (1). One of the main type II receptors for BMPs in vascular cells is BMPR-II. Complete loss of BMPR-II in mice leads to fetal death prior to gastrulation (2), whereas postnatal loss leads to failure of vessel maturation (3). In humans and rodents, partial loss or mutation in BMPR-II predisposes to severe pulmonary arterial hypertension (PAH) (4–7). Viral infections, including HIV and Kaposi sarcoma-associated herpesvirus (KSHV), have been implicated as etiological agents in the development of PAH (8), although the evidence for the involvement of KSHV is controversial (9, 10). Nevertheless, KSHV is the etiologic agent of Kaposi sarcoma and primary effusion lymphoma (11) and may contribute to the pathogenesis of the plasmablast form of multicentric Castleman disease (12). Severe PAH has also been reported as a complication of Castleman disease (13).
KSHV preferentially infects endothelial cells. It is dysfunctional endothelial cell growth that gives rise to both the plexiform lesion in PAH and the spindle cell in Kaposi sarcoma (14, 15). Interestingly, KSHV expresses a lytic gene, K5, which unlike other lytic genes is expressed for a prolonged period of time (16). K5 is a membrane-associated RING E3 viral ubiquitin ligase (17). K5 is known to target a number of host cell receptors for ubiquitination and degradation, including the immunoreceptors MHC Class I, ICAM-1, and B7.3 (17–20) and the endothelial cell receptors platelet-endothelial cell adhesion molecule and vascular endothelial cadherin (21, 22). Because down-regulation of BMPR-II is associated with the pathology of PAH (23, 24) and also the development of some forms of neoplasia (25–27), we investigated the regulation of BMPR-II in vitro in the context of KSHV infection and ectopic K5 expression.
Our findings demonstrate that KSHV infection down-regulates cell surface BMPR-II. We propose that this is caused through the action of K5. We show that K5 ubiquitinates a membrane-proximal lysine residue in the cytoplasmic domain of BMPR-II. K5-mediated ubiquitination of BMPR-II leads to its lysosomal degradation. Importantly, through these studies with K5, we found a previously unreported endogenous mechanism of BMPR-II regulation in mammalian cells: constitutive lysosomal degradation of BMPR-II. This may be mediated partly by the E3 ligase, Itch, or atrophin-1-interacting protein 4 (AIP4).
EXPERIMENTAL PROCEDURES
Tissue Culture
HeLa cell lines were grown in DMEM (Invitrogen), and BC3 cells and Sultan cells were grown in RPMI 1640 (Invitrogen). All medium was supplemented with 10% fetal calf serum (FCS) (PAA Laboratories) and 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (Sigma-Aldrich). Cells were incubated at 37 °C with 5% CO2. The K5 stable cell line was created by viral transduction of a HeLa parent cell line with pMSCV-puro-FLAG-K5 and selected with puromycin (1 μg/ml) (Sigma-Aldrich). Human pulmonary artery endothelial cells and pulmonary artery smooth muscle cells were purchased from Lonza Wokingham. Cells were propagated according to the instructions supplied. Human umbilical vein endothelial cells (HUVECs) were isolated and maintained in Medium 199 (M199; Invitrogen) containing 20% fetal calf serum, 28 μg/ml gentamycin, 2.5 μg/ml amphotericin B, 1 ng/ml epidermal growth factor, and 1 μg/ml hydrocortisone (Sigma-Aldrich) until confluent. Primary cultures were dissociated with trypsin/EDTA (Sigma-Aldrich) and passaged into tissue culture multiwell plates (Falcon; BD Biosciences). Seeding density yielded confluent monolayers within 24 h. HUVECs were cultured in M199 as above for 18–24 h before infection with rKSHV.219.
KSHV Production
rKSHV.219 was produced from the latently infected Vero line, which expresses the red fluorescent protein (RFP) from the KSHV lytic PAN promoter and the green fluorescent protein (GFP) from the EF-1α promoter, with the gene for puromycin resistance as a selectable marker. Briefly, Vero cells were maintained in Eagle's minimal essential medium, 2.2 g/liter NaHCO3, 10% FCS, penicillin and streptomycin, and 5 μg/ml puromycin (all Sigma-Aldrich). Vero cells were infected with BacK50 and treated with 1.25 mm sodium butyrate (Sigma-Aldrich) to induce lytic replication. After 48 h, supernatants were removed, centrifuged at 500 × g for 15 min to remove cell debris, and then ultracentrifuged at 15,000 rpm for 4 h. The resultant pellet was resuspended overnight in EBM2 medium (Lonza). Infectious units were quantified by infection of 293 cells and quantification of GFP-positive cells.
Infection of HUVECs and HeLa Cells with KSHV
KSHV was diluted to the appropriate concentration in EBM2 and then added to the confluent monolayers at a multiplicity of infection of 1 (HeLa) or 10 (HUVEC) before centrifugation at 450 × g for 30 min (uninfected samples were treated in an identical manner, except that there was no addition of rKSHV.219). HUVECs were then incubated for a further 90 min at 37 °C with 5% CO2 before the KSHV-containing medium was removed and replaced with M199/DMEM including supplements as described above.
Stimulation of BC3 Cells
Viral replication in BC3 cells was induced by treating 2 × 107 cells (1 × 106/ml) with 2 mm sodium butyrate (Sigma-Aldrich) for 24 h.
Flow Cytometry and Cell Sorting
Cells were lifted from plates/flasks with 0.1 mm EDTA in PBS, stained with primary antibody, and incubated at 4 °C for 1 h. Cells were then washed three times with FACS solution (3% FCS in PBS) and incubated with secondary, fluorophore-labeled antibody for 1 h at 4 °C and washed with FACS solution prior to being resuspended in 500 μl of PBS for analysis. Analysis was carried out on a FACSCalibur flow cytometer (BD Biosciences). Cell sorting was carried out on a MoFlo cell sorter (Beckman-Coulter). Antibodies used are: rabbit anti-BMPR-II H-300 (Santa Cruz Biotechnology); secondary antibody goat anti-rabbit IgG allophycocyanin (R & D Systems); and allophycocyanin-conjugated mouse anti-ICAM-1 (CD54) (BD Biosciences).
Transient Transfection
Transfections were performed using the Mirus Bio TransIT-HeLaMONSTER® transfection kit according to the manufacturer's instructions. Transgene expression was assayed 48 h after transfection. Plasmids used are: pEGFP-N1-BMPR-II-3′GFP-tagged, pcDNA3-5′-Myc-tagged BMPR-II, pcDNA3-5′-Myc-tagged BMPR-II-R899X, MSCV-1-puro-AU1-tagged K5, MSCV-1-puro-AU1-tagged K5W/I, and MSCV-1-puro-AU1-tagged K3.
Short Interfering RNA Transfection
HeLa cells were plated out in 6-well plates in DMEM (with 10% FCS, no antibiotics) to be 50% confluent the next day. Human pulmonary artery endothelial cells were plated into 60-mm culture dishes in complete EGM2 to be 70% confluent after 48 h. DharmaFECT 1 transfection reagent was used as per the manufacturer's instructions. 60 h after transfection, the cells were lysed in 1% Triton X-100 in TBS (Complete protease inhibitor (Roche Applied Science)) at 1 × 107cells/ml lysis buffer; incubated on ice for 30 min; and then centrifuged at 13,500 rpm for 10 min. The supernatant was then assayed by immunoblot. The sequence of the short interfering RNA molecule (siRNA) oligonucleotides purchased from Dharmacon is: Itch/AIP4, 5′-guugggaacugcugcauua-3′. Sequences of the SMARTpool duplex for BMPR2 are: sense, 5′-gaaccuguguuauuagugauu-3′, antisense, 5′-pucacuaauaacacagguucuu-3′; sense, 5′-caacauugcccgcuuuauauu-3′, antisense, 5′-puauaaagcgggcaauguuguu-3′; sense, 5′-gaaaggauggcugaacuuauu-3′, antisense, 5′-puaaguucagccauccuuucuu-3′; sense, 5′-cauguaugcucuuggacuauu-3′, antisense 5′-puaguccaagagcauacauguu-3′.
Lentiviral Transduction
Packaging, envelope, and vector plasmid, pDG268 His6-WT ubiquitin-GFP, were cotransfected into 293T cells. Viral supernatants harvested at 48 and 72 h were concentrated by ultracentrifugation. The high titer virus was incubated with HeLa and HeLa-K5 cells for 1 h at 37 °C for transduction.
Immunoprecipitation and Immunoblotting
Cells were lysed in 1% Triton X-100 TBS with Roche Applied Science Complete protease inhibitors for 30 min in ice. Lysates were then centrifuged at 13,500 relative centrifugal force for 10 min at 4 °C. For immunoblots, the supernatant was heated in SDS sample buffer, separated by SDS-PAGE, transferred to PVDF membranes (Millipore), and probed with primary and secondary antibody. Reactive bands were detected by AmershamTM ECL (GE Healthcare). For immunoprecipitation, the supernatant was precleared with CL-4B-Sepharose and protein G. Then the sample was immunoprecipitated with antibody and protein G, eluted in SDS reducing sample buffer, and immunoblotted and then probed with primary and secondary antibodies and developed using ECL reagent. Antibodies used are: mouse anti-BMPR-II (BD Transduction Laboratories), mouse anti-ubiquitin P4D1 (Cell Signaling), mouse anti-His (Qiagen), mouse anti-Myc 9B11 (Cell Signaling), rabbit anti-phospho-Smad 1/5 (Cell Signaling), rabbit anti-phospho-Smad 2 (Cell Signaling), mouse anti-Itch (BD Transduction Laboratories), and mouse anti-α-tubulin (Sigma).
Immunofluorescence Microscopy
HeLa cells were grown on collagen coated glass coverslips and transfected 24 h later. 48 h after transfection, the coverslips were washed with PBS and fixed with 3.8% formaldehyde. Cells were permeabilized with 0.1% Triton X-100 and blocked with 10% fetal calf serum in PBS. Finally, the coverslips were stained with primary and secondary antibodies before being mounted in glycerol/PBS solution DAPI (VECTASHIELD)). Cells were viewed and photographed using an ultraviolet confocal microscope (TCS Leica), and images were captured using ImagePro Plus 4.1 software. Antibodies used are: mouse anti-Myc clone 4A6 (Upstate Biotech Millipore) and mouse anti-LAMP-1 (Dako).
Reverse Transcription-PCR and Quantitative PCR
RNA was prepared using either the Qiagen RNeasy mini kit or TRIzol (Invitrogen) and converted to cDNA using SuperScript III first-strand synthesis supermix (Invitrogen) as described in the manufacturer's instructions with PCR settings as follows: 44 °C for 90 min, 90 °C for 10 min, and then 0 °C for 5 min. A PCR reaction was then performed using Go Taq® Flexi DNA polymerase (Promega) according to the manufacturer's instructions with PCR conditions as follows: 94 °C for 2 min and then 30 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 1 min followed by 72 °C for 10 min and 4 °C for 10 min. The products were then run on a 1% agarose gel. Specific primers used are: K5 sense, 5′-ggatccaccatggcgtccaaggacgtagaag-3′, K5 antisense, 5′-atgcggccgctcaaccgttgttttttggatgattt-3′, GAPDH sense, 5′-tgccagccccagcgtcaaag-3′, GAPDH antisense, 5′-gcaggggggagccaaaagggy-3′, β-actin, sense, 5′-catcaccattggcaatgagc-3′, and antisense, 5′-cgatccacacggagtacttg-3′. For quantitative PCR reactions, 45 ng of cDNA was used in a PCR reaction according to the manufacturer's instructions using the SYBR® Green JumpstartTMTaqReadymixTM (Sigma-Aldrich) containing 200 nm of the relevant sense and antisense primers and 10 nm fluorescein (Invitrogen). Specific primers used are: BMPR2 sense, 5′-caaatctgtgagcccaacagtcaa-3′, BMPR2 antisense, 5′-gaggaagaataatctggataaggaccaat-3′. The QuantiTect primer for β2-microglobulin was purchased from Qiagen.
Competition Radiolabeled Ligand Binding Studies
Cells were grown to confluence in 24-well plates, pre-equilibrated in binding buffer (DMEM/0.5% BSA containing 25 mm HEPES) for 60 min at 4 °C, and then incubated at 4 °C for 3 h with binding buffer containing either 125I-BMP4 (∼6 pm or 0.22 ng/ml) in the absence or presence of unlabeled BMP4 (300 ng/ml) or 125I-TGF-β (∼25 pm) in the presence of absence of unlabeled TGF-β (100 ng/ml). Cells were then washed three times with ice-cold binding buffer and solubilized in lysis buffer (20 mm HEPES, pH 7.4, containing 10% (v/v) glycerol, 1% (v/v) Triton X-100 and 0.05% (w/v) BSA). Each point was determined in triplicate for each experiment. Lysates were transferred to polystyrene tubes, and radioactivity was measured with a Packard γ counter.
BMP4/6 and TGF-β Stimulation
Cells plated into 6-well plates were quiesced in DMEM supplemented with 0.1% FCS overnight. Cells were stimulated by adding DMEM supplemented with 0.1% FCS plus 10 ng/ml BMP4, 10 ng/ml TGF-β, or 1 ng/ml TGF-β for 60, 30, 15, and 0 min. At the end of stimulation, the medium was removed, and the cells were snap-frozen at −20 °C in 350 μl of lysis buffer (10% Tris-HCL, 20% SDS, 20% glycerol and Complete protease inhibitors (Roche Applied Science)). The lysates were sonicated and analyzed by immunoblot.
Concanamycin/Lactacystin Treatment
Concanamycin A was dissolved in 100% ethanol and used at a final concentration of 50 nm for 24 h. Lactacystin was used at 10 μm for up to 5 h (both Sigma-Aldrich). After treatment, the cells were lysed for immunoblotting.
BMPR-II Constructs, BMPR-II Tail Truncations
A forward primer was designed to amplify 5′-Myc-tagged BMPR-II from the BamHI restriction enzyme cleavage site of pcDNA3-5′-Myc-tagged BMPR-II-R899X. Reverse primers introduced a premature stop codon and a new XhoI cleavage site. Another reverse primer introduced a point mutation at amino acid 180, mutating the lysine to an arginine. The surrounding wild type amino acid sequence from the start of the BMPR-II cytoplasmic domain (amino acids 171) is YRMLTGDRKQGLHSMNMME. After amplification, the products were subcloned into pCR4-TOPO cloning vector. The inserts were then cloned back into pcDNA3, to give pcDNA3-BMPR-II-Y172X, pcDNA3-BMPR-II-S185X, and pcDNA3-BMPR-II-K180R/S185X. Site-directed mutagenesis was performed on the full-length 5′-Myc-tagged BMPR-II to give pcDNA3-BMPR-II-K180R. Oligonucleotides used are: forward primer for truncated constructs, 5′-tcggatccggccagggatgactt-3′, and reverse primers, BMPR-II-Y172X, 5′-ctcgagttatccaaagcataag-3′; BMPR-II-S185X, 5′ctcgagttagtgaagaccttgtttacgctctcctgt-3′; BMPR-II-K180R/S185X, 5′-ctcgagttagtgaagaccttgtctacgctctcctgt-3′. The forward primer for BMPR-II K180R is 5′-aatgttgaccggtgaccgtagacaaggtcttcacagtatg-3′, and the reverse primer is 5′-catactgtgaagaccttgtctacggtcaccggtcaacatt-3′.
Statistics
A Student's t test was used for analysis of two normally distributed variables. For non-parametric data, a Kruskall-Wallis test was used with post hoc Dunn's test with significance accepted as p < 0.05.
RESULTS
Induction of Lytic KSHV Genes Reduces BMPR-II Protein Expression in BC3 Cells
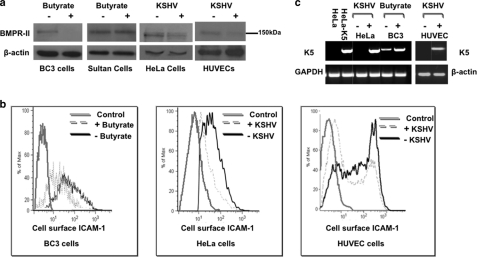
First we studied the primary effusion lymphoma cell line, BC3, which is naturally latently infected with KSHV. Untreated BC3 cells express only a subset of latent viral genes, but lytic gene expression can be induced by treatment with sodium butyrate (28). Treatment of BC3 cells and Sultan cells (a Burkitt lymphoma cell line that lacks KSHV infection) with 2 mm sodium butyrate for 24 h substantially reduced BMPR-II protein levels in BC3 cells, but not in control Sultan cells (Fig. 1a). Next we assessed whether de novo KSHV infection down-regulates BMPR-II in HeLa cells and in endothelial cells. Primary cultured human umbilical vein endothelial cells (HUVECs) or HeLa cells were infected with KSHV and cultured for 7 or 2 days, respectively. KSHV infection of both HUVEC and HeLa cells also reduced BMPR-II protein levels (Fig. 1a).
FIGURE 1.
KSHV infection causes a reduction in BMPR-II protein. a, immunoblot for BMPR-II protein in BC3 cells and control Sultan cells before and after treatment with 2 mm sodium butyrate for 24 h. BMPR-II levels in HeLa cells 2 days after KSHV infection and also in HUVECS 7 days after KSHV infection when compared with uninfected control cells are shown. b, flow cytometry for cell surface ICAM-1, a known target of K5, in BC3 cells after induction of lytic KSHV and in HeLa cells infected with KSHV after 2 days and in HUVECs 7 days after infection. Controls in these experiments were uninduced BC3 cells and uninfected HeLa and HUVEC cells stained with secondary antibody only. c, RT-PCR analysis for K5 mRNA from KSHV-infected HeLa cells when compared with HeLa controls and BC3 cells before and after induction of lytic KSHV infection with butyrate and in control and KSHV-infected HUVECs. Control HeLa cells and stably transfected HeLa-K5 cells served as negative and positive controls for the PCR, respectively. Representative images of three experiments are shown.
The Viral Ubiquitin Ligase, K5, Is Induced by Lytic KSHV Infection
We next sought to determine the mechanism by which lytic KSHV down-regulates BMPR-II. K5 is a KSHV-encoded lytic gene that remains expressed for a prolonged period of time after infection (16). K5 ubiquitinates and down-regulates a number of host immunoreceptors, including ICAM-1 (19, 20). To determine whether K5 is functional in our in vitro system, we measured the cell surface expression of ICAM-1 in BC3 cells by flow cytometry, before and after the lytic reactivation of KSHV with sodium butyrate. A reduction in ICAM-1 staining in BC3 cells after treatment with sodium butyrate is seen at 24 h. Infection of HeLa and HUVECS with a KSHV virus engineered to express GFP upon infection allowed gating on the infected cell population. Again, a reduction in cell surface ICAM-1 expression is seen in both HeLa cells and HUVECs following de novo infection (Fig. 1b). To investigate whether this reduction in ICAM-1 could be due to K5 expression, we measured K5 mRNA levels in HeLa and HUVEC following their infection with KSHV and in BC-3 cells in which lytic replication was induced. We observed a substantial increase in the relative expression levels of K5 in all three cell types after lytic infection with KSHV (Fig. 1c). Furthermore, the expression of K5 in HUVECs was maintained for at least 7 days after infection.
Ectopic Expression of K5 Reduces Cell Surface BMPR-II Expression
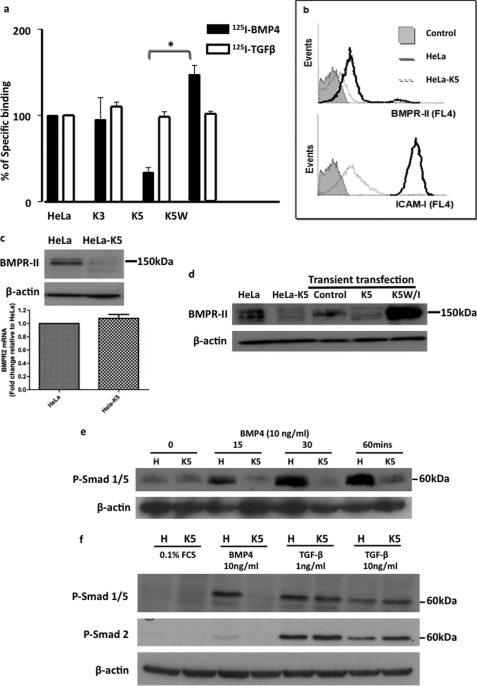
To determine whether BMPR-II is a novel target for K5, HeLa cell lines stably expressing ectopic K5 or K3 (another E3 ubiquitin ligase expressed by KSHV, which shares 40% homology with K5) were evaluated (HeLa-K5 and HeLa-K3) (29). Initially, we employed radioligand binding studies with 125I-BMP4, the natural ligand for BMPR-II, and 125I-TGFβ1 as a control. We observed a ≥50% reduction in cell surface 125I-BMP4 binding in HeLa-K5 cells when compared with HeLa-K3 cells (Fig. 2a). 125I-TGFβ binding was unaffected. These data suggested that in the presence of K5, there is a specific reduction in the number of BMP4 binding sites at the cell membrane. Furthermore, in HeLa cells expressing an E3 ligase-inactive form of K5, K5W- (containing a single mutation tryptophan alanine 47 in the RING domain) (30) specific cell surface binding was increased above baseline levels (Fig. 2a). By flow cytometry, we confirmed a reduction in cell surface BMPR-II in HeLa-K5 cells when compared with HeLa cells. In addition, ICAM-1 expression was also reduced, indicating that K5 was active in HeLa-K5 cells (Fig. 2b).
FIGURE 2.
K5 specifically reduces BMPR-II, a process that is dependent on the RING domain of K5. a, cell surface binding of radiolabeled BMP4 in HeLa cells stably expressing K5 (HeLa-K5) when compared with control HeLa cells and HeLa cells stably expressing K3 (a homolog of K5) (HeLa-K3) and in HeLa cells expressing a mutated, non-functioning K5 (K5W). Radiolabeled TGF-β1 binding is also shown. (*, p < 0.01). Error bars indicate S.E. b, flow cytometry for cell surface BMPR-II and ICAM-1 expression in HeLa and HeLa-K5 cells. Controls were incubated with secondary antibody alone. Error bars indicate S.E. c, immunoblot for BMPR-II protein in HeLa and HeLa-K5 cells showing reduced BMPR-II protein in HeLa-K5 cells. Graph shows similar levels of BMPR-II mRNA transcripts in both cell lines. d, immunoblot for BMPR-II protein levels in control HeLa cells, stably expressing HeLa-K5 cells and HeLa cells transiently transfected with K5 or a non-functioning RING mutant of K5 (K5W/I). e, immunoblot showing the level of Smad 1 phosphorylation (P-Smad 1/5) following BMP4 stimulation in HeLa and HeLa-K5 cells. f, immunoblot of the TGF-β downstream signaling molecule, phospho-Smad 2 (P-Smad 2), in HeLa and HeLa-K5 cells following TGF-β1 stimulation. Studies shown are representative of 3 independent experiments.
This K5-dependent decrease in BMPR-II levels was confirmed by immunoblotting for BMPR-II. The predicted 150-kDa doublet seen in control cells was reduced in abundance in HeLa-K5 cells (Fig. 2c). Importantly, we confirmed that there was no difference in BMPR-II mRNA transcript levels in HeLa-K5 cells when compared with HeLa cells (Fig. 2c). To ensure that this effect of K5 was not restricted to clonal effects of K5-expressing stable cell lines, we confirmed the reduction in BMPR-II protein expression in HeLa cells transiently transfected with K5 (Fig. 2d). We provided further evidence that the effect of K5 on BMPR-II was dependent on the RING domain of K5 using an additional ligase-defective K5 construct (K5W/I), which harbors two mutations in the K5 RING domain (tryptophan 47 and isoleucine 17 to alanines). These mutations prevent recruitment of the cognate cellular E2 (31), and K5W/I is therefore unable to catalyze the transfer of ubiquitin to substrate. When K5W/I was transiently expressed in HeLa cells, we observed a marked increase in BMPR-II protein when compared with control HeLa cells (Fig. 2d). This observation was consistent with the increased 125I-BMP4 binding in HeLa cells expressing K5W, where cell surface BMP4 binding is higher than in control HeLa cells (Fig. 2a). Taken together, these findings suggested that K5W/I is effectively “trapping” BMPR-II at the cell surface and inhibiting endogenous degradation of BMPR-II, analogous to the related K3 (W41A) mutant that associates with, but is unable to ubiquitinate, MHC I molecules (16).
K5 Expression Inhibits BMP-mediated Signaling via Smad 1/5
To confirm the functional relevance of the decrease in BMPR-II expression, we investigated the effect of K5 on BMP signaling. Usually, activation of the type I BMP receptor by the constitutively active BMPR-II leads to phosphorylation of Smads 1, 5, and 8 (32). By immunoblotting, we found that phosphorylation of Smad 1 was markedly reduced in HeLa-K5 cells when compared with HeLa cells following BMP4 stimulation (Fig. 2e). Furthermore, this effect was specific to the BMP signaling pathway as there was no difference in the levels of phospho-Smad 2 following TGF-β stimulation (Fig. 2f). Interestingly, there was no difference in phospho-Smad 1/5 levels following TGF-β stimulation, suggesting that there was no alteration in the cell surface levels of TGF-β type I receptors mediating Smad 1/5 phosphorylation (e.g. ALK1 or ALK5) in HeLa and HeLa-K5 cells.
K5 Ubiquitinates BMPR-II for Lysosomal Degradation
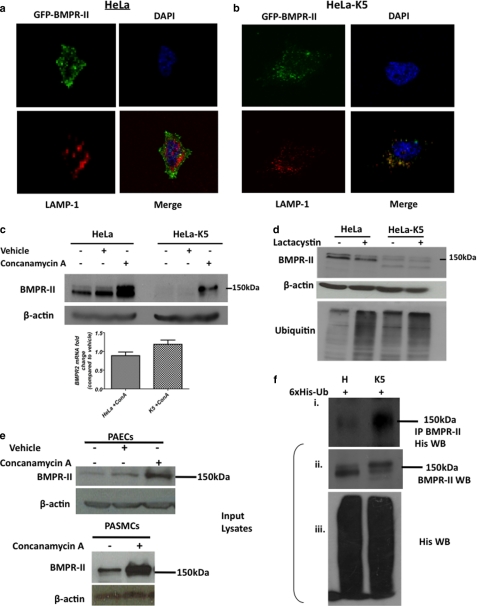
We next investigated the fate of BMPR-II when co-expressed with K5. We used confocal microscopy to visualize the location of a GFP-tagged BMPR-II expressed in both HeLa cells and HeLa-K5 cells. GFP-tagged BMPR-II was located mainly at the cell surface in HeLa cells (Fig. 3a). In contrast, in HeLa-K5 cells, GFP-tagged BMPR-II was internalized to discrete vesicles within the cytoplasm. These cytoplasmic locations completely co-localized with the lysosomal marker LAMP-1 (Fig. 3b). To confirm that K5 targets BMPR-II to the lysosomal pathway for degradation, we used pharmacological inhibitors to block either the proteasomal or the lysosomal pathway. When HeLa-K5 cells were treated with the lysosomal inhibitor, concanamycin A (a specific inhibitor that blocks lysosomal acidification through selective inhibition of the V-type ATPase), there was a striking reversal of the degradation of BMPR-II (Fig. 3c). Lactacystin inhibits the proteasomal pathway by inhibiting the 20 S proteasome directly. HeLa-K5 cells treated with lactacystin showed no reversal of the loss of BMPR-II despite clearly increasing the level of cellular ubiquitin (Fig. 3d). These data confirmed that K5 targets BMPR-II to be degraded via the lysosomal pathway and not the proteasome. Interestingly, in control HeLa cells treated with concanamycin A, there was also an increase in BMPR-II protein by Western blotting. This observation suggests that BMPR-II is constitutively turned over through the lysosomal pathway by endogenous E3 ligases (Fig. 3c). We confirmed this observation in primary cultured human pulmonary artery endothelial cells and pulmonary artery smooth muscle cells (Fig. 3e).
FIGURE 3.
K5 causes redistribution of BMPR-II from the cell surface to a lysosomal compartment. a and b, confocal microscopy showing the localization of GFP-tagged BMPR-II (green), the lysosomal marker, LAMP-1 (red), nuclear DAPI stain (blue), and merged images in HeLa (a) and HeLa-K5 (b) cells. c, immunoblot showing BMPR-II protein levels before and after treatment of HeLa and HeLa-K5 cells with concanamycin A (lysosomal inhibitor) (vehicle is ethanol) and graph of BMPR-II mRNA transcript levels under the same conditions. Error bars indicate S.E. d, immunoblots for BMPR-II and endogenous ubiquitin levels in the presence and absence of lactacystin (proteasomal inhibitor). e, immunoblots for BMPR-II protein in pulmonary arterial endothelial cells (PAECs) and pulmonary arterial smooth muscle cells (PASMCs) before and after treatment with concanamycin A. f, panel i, viral transduction of HeLa and HeLa-K5 cells with a His6-tagged ubiquitin (6xHis-Ub) followed by a BMPR-II immunoprecipitation (IP) and His Western blot. f, panels ii and iii, Western blots (WB) show the presence of endogenous BMPR-II in input lysates (panel ii) and also the presence of transduced His6 ubiquitin (panel iii). Studies shown are representative of 3 independent experiments.
We next sought to demonstrate whether BMPR-II was indeed ubiquitinated in the presence of K5. A His6-ubiquitin construct was transduced into HeLa and HeLa-K5 cells, and after treatment with concanamycin A, BMPR-II was immunoprecipitated and the associated His-tagged ubiquitinated proteins were visualized by immunoblot analysis. A high molecular weight smear compatible with ubiquitinated BMPR-II was seen in the HeLa-K5 lane but not the HeLa lane (Fig. 3f), confirming BMPR-II as a ubiquitination target for K5.
K5 Ubiquitinates BMPR-II on a Membrane-proximal Lysine
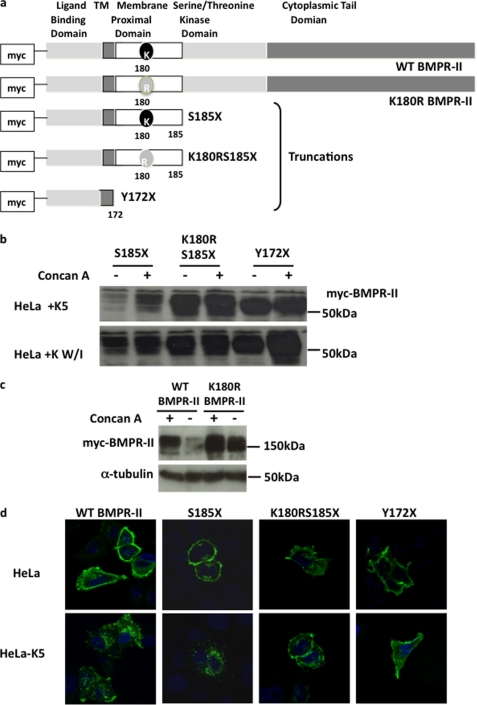
K5 targets MHC Class I by preferentially ubiquitinating a membrane-proximal lysine (19). We investigated whether this was also the case for BMPR-II. The amino acid sequence of BMPR-II reveals that there is a single lysine residue present in the cytoplasmic membrane-proximal region of BMPR-II at position 180 (Fig. 4a). To determine the critical site for ubiquitination of BMPR-II, we created three truncated BMPR-II constructs: one lacking the entire intracellular domain (Y172X), a second truncated at amino acid 185 but retaining the lysine at 180 (S185X), and a final construct also truncated at amino acid 185 but harboring a point mutation at position 180, substituting the lysine for an arginine (K180R/S185X) (Fig. 4a). When co-expressed with K5, only the truncation with the lysine at position 180 (S185X) was degraded, an effect that was reversed with the addition of concanamycin A. K5 was unable to degrade either of the other truncated receptors, Y172X or K180R/S185X, both of which lack the lysine at position 180. K5W/I was unable to degrade any of the truncated receptors (Fig. 4b). Furthermore, we demonstrate that K5 is unable to degrade a full-length BMPR-II harboring the K180R point mutation (Fig. 4c). This suggests that the tail of BMPR-II is not necessary for the action of K5. Taken together, these observations suggest that K5 targets the lysine in the membrane-proximal region of BMPR-II, a process that is dependent on the RING domain of K5. Confocal microscopy demonstrated that all three truncations trafficked to the cell membrane, but in the presence of K5, only the truncated receptor with the lysine remaining in position 180 could be internalized to the cytoplasm in a similar manner to wild type BMPR-II (Fig. 4d).
FIGURE 4.
K5 targets the membrane-proximal lysine of BMPR-II (Lys-180). a, a series of N-terminally Myc-tagged BMPR-II constructs were made: full-length wild type BMPR-II, full-length BMPR-II in which the lysine at position 180 was mutated to arginine, truncated (S185X) BMPR-II, truncated BMPR-II in which the lysine at 180 was mutated to arginine (K180R/S185X), and BMPR-II in which the entire intracellular domain was missing (Y172X). TM, transmembrane. b, immunoblot for Myc protein in HeLa cells following transient transfection with the Myc-tagged BMPR-II tail truncation constructs and either K5 or the K5 RING mutant, K5W/I, before and after concanamycin A (Concan A) treatment. c, immunoblot for Myc protein in HeLa and HeLa-K5 cells after transfection with the full-length constructs, Myc-BMPR-II or Myc-K180R, with and without pretreatment with concanamycin A. d, confocal images were generated after staining for Myc in HeLa and HeLa-K5 cells after transient transfection with the Myc-tagged BMPR-II tail truncated constructs (magnification ×60 oil immersion objective). In d, all blots and images are representative of 3 independent experiments.
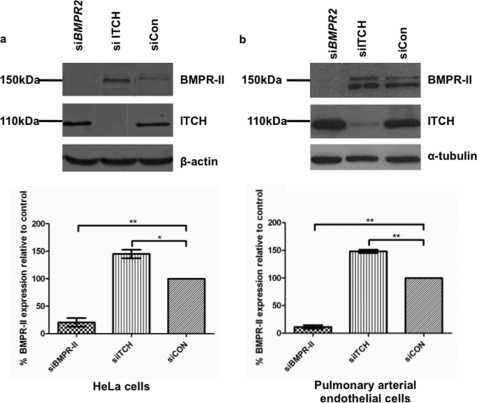
The E3 Ligase, Itch, Contributes to Constitutive Degradation of BMPR-II in Mammalian Cells
To identify the participants in the endogenous, constitutive turnover of BMPR-II, we used siRNAs to knock down known members of the NEDD4-like family of HECT E3 ligases (Nedd4, Nedd4-2, Smurf 1 and 2, Itch (AIP4), WWP1 and -2 and NEDL 1 and 2) in HeLa cells as this ligase family is known to be involved in TGF-β/BMP signaling (33, 34). We observed a consistent increase in BMPR-II protein after knockdown of Itch (AIP4) (Fig. 5a). This observation was replicated in human pulmonary artery endothelial cells (Fig. 5b).
FIGURE 5.
siRNA knockdown of Itch causes an increase in endogenous levels of BMPR-II. a, immunoblots for BMPR-II protein and Itch, demonstrating the effect of BMPR-II or ITCH knockdown in HeLa cells, when compared with control siRNA (siCON). b, similar experiments in pulmonary arterial endothelial cells. Graphs show results of densitometry from n = 3–4 independent experiments (*, p < 0.05 and **, p < 0.01). Error bars indicate S.E.
DISCUSSION
The bone morphogenetic protein type II receptor plays a major role in early development, carcinogenesis, and vascular disease, particularly pulmonary arterial hypertension (PAH). Our studies were initially prompted by the observation that a significant proportion of patients with idiopathic PAH were reported to have latent KSHV infection (8), although this finding is controversial (10, 35). In addition, mutations in BMPR-II are the main cause of familial PAH (4, 5, 36), and reduced expression of BMPR-II protein is found in idiopathic PAH (23) and experimental models of PAH (24). Thus the identification of mechanisms regulating cell surface expression of BMPR-II is of major interest.
K5 is a viral E3 ubiquitin ligase known to be expressed during, and long after, the induction of KSHV lytic replication. K5 targets a number of host cell receptors for ubiquitination and degradation, including the immunoreceptors MHC Class I, Natural Killer-activating ligands MHC Class I-related chains A and B, and activation-induced C-type lectin, ICAM-1, B7.2 (18–20, 37), and the endothelial cell receptors platelet-endothelial cell adhesion molecule and vascular endothelial cadherin (21, 22). Our data are the first to identify that KSHV K5 targets a growth factor signaling receptor pathway. In addition, we confirmed directly that K5 overexpression leads to ubiquitination of BMPR-II. K5 is an E3 ligase with a characteristic cytosolic N-terminal RING-CH domain, responsible for ubiquitination of the target protein. K5 preferentially targets the membrane-proximal lysine for ubiquitination (38, 39). Using a series of truncated BMPR-II constructs, we first confirmed that the cytoplasmic domain of BMPR-II was essential for the action of K5. The most membrane-proximal lysine residue in the cytoplasmic domain of BMPR-II lies at position 180, predicted to be 8 amino acids from the cell membrane. Mutation of this lysine residue to an arginine prevented the degradation of BMPR-II protein and prevented distribution of the mutated BMPR-II to the lysosome. In addition, we confirmed that this lysine residue was essential for BMPR-II degradation in the context of the full-length receptor. Thus we have identified that BMPR-II is susceptible to targeting by E3 ligases at this membrane-proximal lysine. We studied the fate of BMPR-II in the presence of K5 and show that ubiquitinated BMPR-II is targeted to the lysosome for degradation. Degradation of BMPR-II could be rescued by inhibition of the lysosome by concanamycin A, but not by the proteasomal inhibitor, lactacystin. Importantly, concanamycin A treatment of HeLa cells and primary cultured pulmonary vascular cells increased the protein expression of BMPR-II even in the absence of K5. This suggested that the expression of BMPR-II protein is constitutively regulated by the lysosome and that an endogenous mammalian E3 ligase may be involved in this process.
KSHV encodes a panel of immune modulatory proteins (39). Down-regulation of immunoreceptors by K5 presumably contributes to reduced immune surveillance of KSHV, and a reduction in endothelial junctional molecules may allow the virus to pass easily between neighboring cells. Why then should KSHV target BMPR-II? One possible explanation is that the degradation of BMPR-II in the infected endothelial cell contributes to apoptosis resistance, helping to facilitate KSHV persistence within the host. In support of this, mice with conditional ablation of endothelial BMPR-II demonstrate increased pulmonary endothelial proliferation. A proportion of these mice develop PAH (40). In addition, infected endothelial cells resistant to apoptosis could facilitate KSHV lytic infection and increase titers.
A further possibility is that BMPR-II is involved in regulation of the host immune response. The identification of BMPR-II as an additional substrate for Itch adds support to this hypothesis (41). Although TGF-β signaling has an established role in the immune response (42), the role of BMPs is largely unexplored, although BMP4 and BMP2 are involved in thymic homeostasis by regulating T cell lineage commitment and differentiation (43). Ablation of BMPR-II in the pulmonary endothelium or transgenic overexpression of a dominant negative BMPR-II in vascular smooth muscle predisposes mice to pulmonary hypertension accompanied by cellular inflammation (40, 45). Autoimmunity is common in PAH patients, and immune mechanisms for PAH pathobiology are rapidly achieving prominence (46).
A previous study has demonstrated that BMPR-II in association with Dullard (a phosphatase) is degraded via the proteasomal pathway (47). In that study, an E3 ligase was implicated but never determined. Here we show that in the presence of K5, BMPR-II is degraded by the lysosomal pathway and not by the proteasome. The disparity in results may be explained because we studied endogenous BMPR-II degradation, whereas Satow et al. (47) used an overexpression system to study BMPR-II, which may preferentially activate the proteasome to degrade excess BMPR-II.
Importantly, in our studies, concanamycin A treatment of HeLa cells and primary cultured pulmonary vascular cells increased the abundance of BMPR-II in the absence of K5. Moreover, inactive forms of K5 enhanced cell surface expression of BMPR-II. These data suggested that the expression of BMPR-II protein is constitutively regulated by the lysosome and that an endogenous mammalian E3 ligase may be involved in this process. Several members of the NEDD4 family of HECT E3 ligases are known to play a role in the regulation of the BMP and TGF-β pathways. Smurf 1 and 2 belong to the NEDD4 family and regulate Smad protein degradation (48), and through an interaction with the inhibitory Smads, the TGF-β receptor complex (44). Through siRNA screening, we identified Itch as an endogenous mammalian HECT E3 ligase that is involved in BMPR-II degradation. Knockdown of Itch consistently led to increased expression of BMPR-II in HeLa cells and pulmonary artery endothelial cells. It remains to be confirmed that Itch actually ubiquitinates BMPR-II in the same way as K5 and whether other ligases are involved in BMPR-II degradation. Itch has been implicated in a number of important cellular processes, including the regulation of autoimmunity, Th2 cell differentiation, and activation of TGF-β receptor signaling via Smad 2 (41).
In summary, we show for the first time that the viral E3 ligase, K5, regulates the BMP signaling pathway via ubiquitination and lysosomal degradation of BMPR-II. Moreover, we identify a constitutive lysosomal pathway regulating cell surface expression of BMPR-II and implicate the likely involvement of the mammalian E3 ligase, Itch, in this process.
This work was supported by a Wellcome Trust Research Training fellowship (to H. J. D.). Additional support was provided by a British Heart Foundation Programme Grant (to N. W. M.) and a Wellcome Trust grant (to P. J. L.). L. M. B. was supported by Cancer Research UK Project Grant C7934/A9468 (to D. J. B. and G. B. N.).
- BMP
- bone morphogenetic protein
- BMPR
- BMP receptor
- PAH
- pulmonary arterial hypertension
- KSHV
- Kaposi sarcoma-associated herpesvirus
- HUVEC
- human umbilical vein endothelial cell
- ICAM
- intercellular adhesion molecule
- r
- recombinant
- HECT
- homologous to E6-AP carboxyl terminus.
REFERENCES
- 1.Miyazono K., Maeda S., Imamura T. (2005) Cytokine Growth Factor Rev. 16, 251–263 [DOI] [PubMed] [Google Scholar]
- 2.Beppu H., Kawabata M., Hamamoto T., Chytil A., Minowa O., Noda T., Miyazono K. (2000) Dev. Biol. 221, 249–258 [DOI] [PubMed] [Google Scholar]
- 3.Liu D., Wang J., Kinzel B., Müeller M., Mao X., Valdez R., Liu Y., Li E. (2007) Blood 110, 1502–1510 [DOI] [PubMed] [Google Scholar]
- 4.International, PPH Consortium Lane K. B., Machado R. D., Pauciulo M. W., Thomson J. R., Philips J. A., 3rd, Loyd J. E., Nichols W. C., Trembath R. C. (2000) Nat. Genet. 26, 81–84 [DOI] [PubMed] [Google Scholar]
- 5.Thomson J. R., Machado R. D., Pauciulo M. W., Morgan N. V., Humbert M., Elliott G. C., Ward K., Yacoub M., Mikhail G., Rogers P., Newman J., Wheeler L., Higenbottam T., Gibbs J. S., Egan J., Crozier A., Peacock A., Allcock R., Corris P., Loyd J. E., Trembath R. C., Nichols W. C. (2000) J. Med. Genet. 37, 741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long L., MacLean M. R., Jeffery T. K., Morecroft I., Yang X., Rudarakanchana N., Southwood M., James V., Trembath R. C., Morrell N. W. (2006) Circ. Res. 98, 818–827 [DOI] [PubMed] [Google Scholar]
- 7.West J., Fagan K., Steudel W., Fouty B., Lane K., Harral J., Hoedt-Miller M., Tada Y., Ozimek J., Tuder R., Rodman D. M. (2004) Circ. Res. 94, 1109–1114 [DOI] [PubMed] [Google Scholar]
- 8.Cool C. D., Rai P. R., Yeager M. E., Hernandez-Saavedra D., Serls A. E., Bull T. M., Geraci M. W., Brown K. K., Routes J. M., Tuder R. M., Voelkel N. F. (2003) N. Engl. J. Med. 349, 1113–1122 [DOI] [PubMed] [Google Scholar]
- 9.Katano H., Ito K., Shibuya K., Saji T., Sato Y., Sata T. (2005) J. Infect. Dis. 191, 743–745 [DOI] [PubMed] [Google Scholar]
- 10.Henke-Gendo C., Mengel M., Hoeper M. M., Alkharsah K., Schulz T. F. (2005) Am. J. Respir. Crit. Care Med. 172, 1581–1585 [DOI] [PubMed] [Google Scholar]
- 11.Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L., Cogliano V. (2009) Lancet Oncol. 10, 321–32219350698 [Google Scholar]
- 12.Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., d'Agay M. F., Clauvel J. P., Raphael M., Degos L., et al. (1995) Blood 86, 1276–1280 [PubMed] [Google Scholar]
- 13.Bull T. M., Cool C. D., Serls A. E., Rai P. R., Parr J., Neid J. M., Geraci M. W., Campbell T. B., Voelkel N. F., Badesch D. B. (2003) Eur. Respir. J. 22, 403–407 [DOI] [PubMed] [Google Scholar]
- 14.Tuder R. M., Groves B., Badesch D. B., Voelkel N. F. (1994) Am. J. Pathol. 144, 275–285 [PMC free article] [PubMed] [Google Scholar]
- 15.Regezi J. A., MacPhail L. A., Daniels T. E., Greenspan J. S., Greenspan D., Dodd C. L., Lozada-Nur F., Heinic G. S., Chinn H., Silverman S., Jr., et al. (1993) J. Oral Pathol. Med. 22, 292–297 [DOI] [PubMed] [Google Scholar]
- 16.Krishnan H. H., Naranatt P. P., Smith M. S., Zeng L., Bloomer C., Chandran B. (2004) J. Virol. 78, 3601–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coscoy L., Sanchez D. J., Ganem D. (2001) J. Cell Biol. 155, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coscoy L., Ganem D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8051–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coscoy L., Ganem D. (2001) J. Clin. Invest. 107, 1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishido S., Choi J. K., Lee B. S., Wang C., DeMaria M., Johnson R. P., Cohen G. B., Jung J. U. (2000) Immunity 13, 365–374 [DOI] [PubMed] [Google Scholar]
- 21.Mansouri M., Douglas J., Rose P. P., Gouveia K., Thomas G., Means R. E., Moses A. V., Früh K. (2006) Blood 108, 1932–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansouri M., Rose P. P., Moses A. V., Früh K. (2008) J. Virol. 82, 9615–9628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atkinson C., Stewart S., Upton P. D., Machado R., Thomson J. R., Trembath R. C., Morrell N. W. (2002) Circulation 105, 1672–1678 [DOI] [PubMed] [Google Scholar]
- 24.Long L., Crosby A., Yang X., Southwood M., Upton P. D., Kim D. K., Morrell N. W. (2009) Circulation 119, 566–576 [DOI] [PubMed] [Google Scholar]
- 25.Beppu H., Mwizerwa O. N., Beppu Y., Dattwyler M. P., Lauwers G. Y., Bloch K. D., Goldstein A. M. (2008) Oncogene 27, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 26.Kim I. Y., Lee D. H., Lee D. K., Ahn H. J., Kim M. M., Kim S. J., Morton R. A. (2004) Oncogene 23, 7651–7659 [DOI] [PubMed] [Google Scholar]
- 27.Pouliot F., Blais A., Labrie C. (2003) Cancer Res. 63, 277–281 [PubMed] [Google Scholar]
- 28.Miller G., Heston L., Grogan E., Gradoville L., Rigsby M., Sun R., Shedd D., Kushnaryov V. M., Grossberg S., Chang Y. (1997) J. Virol. 71, 314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hewitt E. W., Duncan L., Mufti D., Baker J., Stevenson P. G., Lehner P. J. (2002) EMBO J. 21, 2418–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehner P. J., Hoer S., Dodd R., Duncan L. M. (2005) Immunol. Rev. 207, 112–125 [DOI] [PubMed] [Google Scholar]
- 31.Dodd R. B., Allen M. D., Brown S. E., Sanderson C. M., Duncan L. M., Lehner P. J., Bycroft M., Read R. J. (2004) J. Biol. Chem. 279, 53840–53847 [DOI] [PubMed] [Google Scholar]
- 32.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 33.Lin X., Liang M., Feng X. H. (2000) J. Biol. Chem. 275, 36818–36822 [DOI] [PubMed] [Google Scholar]
- 34.Kuratomi G., Komuro A., Goto K., Shinozaki M., Miyazawa K., Miyazono K., Imamura T. (2005) Biochem. J. 386, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laney A. S., De Marco T., Peters J. S., Malloy M., Teehankee C., Moore P. S., Chang Y. (2005) Chest 127, 762–767 [DOI] [PubMed] [Google Scholar]
- 36.Deng Z., Morse J. H., Slager S. L., Cuervo N., Moore K. J., Venetos G., Kalachikov S., Cayanis E., Fischer S. G., Barst R. J., Hodge S. E., Knowles J. A. (2000) Am. J. Hum. Genet. 67, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas M., Boname J. M., Field S., Nejentsev S., Salio M., Cerundolo V., Wills M., Lehner P. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boname J. M., Thomas M., Stagg H. R., Xu P., Peng J., Lehner P. J. (2010) Traffic 11, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aresté C., Blackbourn D. J. (2009) Trends Microbiol. 17, 119–129 [DOI] [PubMed] [Google Scholar]
- 40.Hong K. H., Lee Y. J., Lee E., Park S. O., Han C., Beppu H., Li E., Raizada M. K., Bloch K. D., Oh S. P. (2008) Circulation 118, 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melino G., Gallagher E., Aqeilan R. I., Knight R., Peschiaroli A., Rossi M., Scialpi F., Malatesta M., Zocchi L., Browne G., Ciechanover A., Bernassola F. (2008) Cell Death Differ. 15, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 42.Li M. O., Flavell R. A. (2008) Cell 134, 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hager-Theodorides A. L., Outram S. V., Shah D. K., Sacedon R., Shrimpton R. E., Vicente A., Varas A., Crompton T. (2002) J. Immunol. 169, 5496–5504 [DOI] [PubMed] [Google Scholar]
- 44.Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 45.Tada Y., Majka S., Carr M., Harral J., Crona D., Kuriyama T., West J. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 292, L1556–L1563 [DOI] [PubMed] [Google Scholar]
- 46.Hassoun P. M., Mouthon L., Barberà J. A., Eddahibi S., Flores S. C., Grimminger F., Jones P. L., Maitland M. L., Michelakis E. D., Morrell N. W., Newman J. H., Rabinovitch M., Schermuly R., Stenmark K. R., Voelkel N. F., Yuan J. X., Humbert M. (2009) J. Am. Coll. Cardiol. 54, S10–19 [DOI] [PubMed] [Google Scholar]
- 47.Satow R., Kurisaki A., Chan T. C., Hamazaki T. S., Asashima M. (2006) Dev. Cell 11, 763–774 [DOI] [PubMed] [Google Scholar]
- 48.Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]