Abstract
The p53 tumor suppressor protein is regulated by multiple post-translational modifications, including lysine methylation. We previously found that monomethylation of p53 at lysine 382 (p53K382me1) by the protein lysine methyltransferase (PKMT) SET8/PR-Set7 represses p53 transactivation of target genes. However, the molecular mechanism linking p53K382 monomethylation to repression is not known. Here we show in biochemical and crystallographic studies the preferential recognition of p53K382me1 by the triple malignant brain tumor (MBT) repeats of the chromatin compaction factor L3MBTL1. We demonstrate that SET8-mediated methylation of p53 at Lys-382 promotes the interaction between L3MBTL1 and p53 in cells, and the chromatin occupancy of L3MBTL1 at p53 target promoters. In the absence of DNA damage, L3MBTL1 interacts with p53K382me1 and p53-target genes are repressed, whereas depletion of L3MBTL1 results in a p53-dependent increase in p21 and PUMA transcript levels. Activation of p53 by DNA damage is coupled to a decrease in p53K382me1 levels, abrogation of the L3MBTL1-p53 interaction, and disassociation of L3MBTL1 from p53-target promoters. Together, we identify L3MBTL1 as the second known methyl-p53 effector protein, and provide a molecular explanation for the mechanism by which p53K382me1 is transduced to regulate p53 activity.
Keywords: Chromatin, p53, Protein Methylation, Protein-Protein Interactions, Signal Transduction, Lysine Methylation, Effector Domain
Introduction
The reversible and dynamic methylation of proteins on the nitrogen side-chain of lysine residues can greatly increase the signaling potential of the modified factor (1, 2). Lysine residues can accept up to three methyl groups, forming mono-, di-, and trimethylated derivatives, with a unique activity frequently being coupled to the specific methylation state. The chemical addition of methyl moieties to histones is not believed to intrinsically affect chromatin structure. Rather, the principal mechanism by which histone lysine methylation is thought to manifest functionally occurs through regulation of modular protein-protein interactions (3–5). In this regard, the protein(s) that recognize a methylated lysine within a specific sequence context can define the functional outcome associated with that specific lysine methylation event. Thus, mechanistic insight into how lysine methylation influences a biological program requires knowledge of the proteins and domains that recognize and transduce this modification.
In addition to histones, several other proteins such as the tumor suppressor p53 undergo lysine methylation, arguing that this modification may be a common mechanism for modulating protein-protein interactions and key cellular signaling pathways (6–8). p53 plays a pivotal role in the regulation of cellular responses to various forms of genotoxic stresses, and its activity is coordinated by a complex network of post-translational modifications (9, 10). We previously demonstrated that the protein lysine methyltransferase (PKMT)5 PR-Set7/SET8 monomethylates p53 exclusively at lysine 382 (p53K382me1), and that the placement of this modification negatively regulates p53 activity (11). Specifically, SET8 suppresses transcriptional activation of p53 rapid response target genes (11); however, the molecular explanation for how p53K382me1 is sensed and transduced to repress p53 is not clear.
In this study, we sought to understand how monomethylation of p53 at Lys-382 negatively regulates p53 function. Recently, p53 transcriptional activity was shown to be restrained at chromatin via an interaction with Cabin1, suggesting that a general mechanism for modulating p53 function might occur at the level of chromatin regulation (12). L3MBTL1 represses transcription by promoting a more compact, inaccessible chromatin state (13–15). This chromatin compaction activity requires the three MBT domains of L3MBTL1, with the middle MBT domain functioning as a binding module of mono- and dimethylated histone lysines that are mostly enriched at silenced chromatin (e.g. H4K20me1/2 and H1K26me1/2) (13, 16–21). Here we have identified that L3MBTL1, via its MBT motifs, binds to p53K382me1. We provide evidence for a model by which L3MBTL1 acts as a dual methyllysine-sensor, coupling the methylated form of p53 to silenced histone methyl marks to render p53 inert at target genes.
EXPERIMENTAL PROCEDURES
Constructs and Reagents
SET8 and p53 constructs were generated as reported previously (11). L3MBTL1 cDNA was cloned into pCAG-Flag and pGEX6p, and pMSCVFlag Puro. L3MBTL1 mutants were generated by site-directed mutagenesis (Stratagene). Antibodies used in this study are the following: αp53K382me1 (11); horseradish peroxidase-p53 (R&D Systems); p53 (DO1) (Santa Cruz Biotechnology); PR-SET7 (Abcam); GST (E5) (Santa Cruz Biotechnology); Flag (M2) and tubulin (Sigma). Methylated p53 and histone peptides were synthesized at the W. M. Keck Facility at Yale University as previously described (11).
Peptide Pull-down Assays
Peptide pull-down assays were performed as previously described (22). Briefly, 1 μg of biotinylated peptides was incubated with 1 μg of protein in binding buffer (50 mm Tris-HCl, pH 7.5, 150–300 mm NaCl, 0.05% Nonidet P-40, 1 mm phenylmethysulfonyl fluoride, and protease inhibitors) overnight at 4 °C with rotation. After a 1-h incubation with streptavidin beads (Amersham Biosciences) and extensive washing, bound proteins were analyzed by SDS-PAGE and Western blotting.
Peptide Synthesis and Affinity Measurements
The human p53 peptides were prepared by solid-phase peptide synthesis with an Applied Biosystem 431A peptide synthesizer. Peptides were assembled on Wang resin using Fmoc synthetic strategy. The coupling reaction was carried out by means of the HBTU-HOBt method. Cleavage of the peptide from the resin was achieved with TFA/water/EDT/TIS (94/2.5/2.5/1.0, v/v) for 3 h at room temperature. After removing the resin by filtration, the filtrate was concentrated by nitrogen gas flushing, and crude peptides were precipitated by diethyl ether. Crude peptides were purified by preparative HPLC on a C18 column with water-acetonitrile system. The purity of the peptides was determined to be over 95% by analytical RP-HPLC. The mass of the peptides was confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Micromass, Beverly, MA).
ITC measurements were performed using a VP-ITC calorimeter (MicroCal, Northhampton, MA). Titrations were carried out in 25 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 2 mm β-mercaptoethanol at 25 °C. The concentration of protein solution was estimated from the absorbance at 280 nm after the protein solutions were dialyzed. The concentration of peptide solution was determined from the base on the weight. The protein and peptide solutions were degassed before each experiment. Experiments were performed by injecting 10 μl of peptide solution (1.0 mm) into a sample cell containing 29–47 μm human L3MBTL1 (190–530). A total 29 injections were performed with a spacing 180 s and reference power of 13 μcal/s. Binding isotherms were plotted and analyzed using Origin software (Microcal Inc). The ITC data were fitted to a one-site binding model.
Protein Purification and X-ray Crystallography
The human L3MBTL1 3xMBT repeats, (residues 190–530), were expressed in Escherichia coli BL21(DE3) pLysS (Stratagene) grown in LB media. Bacteria were harvested by centrifugation after IPTG induction (1 mm) and lysed by sonication. The GST fusion protein was purified on a glutathione-Sepharose 4B column (Amersham Biosciences), cleaved with precision protease and concentrated in Millipore concentrators (Millipore). The protein was further purified by FPLC and concentrated into 20 mm Tris, pH 8.0 buffer, containing 100 mm NaCl, and 5 mm dithiothreitol.
The solution of 0.25 mm L3MBTL1 (residues 190–530) was incubated overnight with p53K382me1 (residues 377–386) peptide in a 1:5 molar ratio prior to crystallization. Crystals of the complex were grown using the hanging drop vapor diffusion method at 18 °C by mixing 2 μl of the protein-peptide solution with 2 μl of precipitant solution containing 0.1 m sodium acetate pH 4.8, 4% PEG 4000, and 0.1 m sodium acetate. Crystals grew in a trigonal space group P3 with three molecules per asymmetric unit. Two of the three molecules, chains A and C were complexes of L3MBTL1 with the p53K382me1 peptide and the third one, chain B, was the ligand-free protein. The complete data sets were collected at 100 K on a “NOIR-1” MBC system detector at beamline 4.2.2 at the Advanced Light Source in Berkeley, CA. The data were processed with D*TREK (23). The molecular replacement solution was generated using the program Phaser (24) and the crystal structure of L3MBTL1 (PDB 2RJE) as a search model. The crystal was detected with a twin fraction of 0.417. The initial models were built with COOT (25) and refined using the program Phenix with twin operator -h,-k,l (26). Statistics are shown in supplemental Table S1. The coordinates have been deposited in the Protein Data Bank under accession number 3OQ5.
Cell Culture and Transfection
U2OS, H1299, 293T, and HCT116 cells were maintained in DMEM medium supplemented with 10% fetal bovine serum or newborn calf serum. Cells were transfected with plasmids or siRNA duplexes by TransIT-LT1, TransIT-293, or DharmaFECT (Dharmacon), respectively, according to the manufacturers' protocols.
Coimmunoprecipitation and Western Blots
Cotransfections for CoIPs were performed at a 1:3 ratio Flag-L3MBTL1: SET8, with 10 μg of total DNA transfected. Ectopically expressed Flag-tagged L3MBTL1 or endogenous L3MBTL1 was immunoprecipitated with anti-Flag M2-agarose from mouse (Sigma) or L3MBTL1 antibody (Abcam) from WCE in cell lysis buffer (50 mm Tris-HCl, pH 7.4, 250 mm NaCl. 0.5% Triton X-100, 10% glycerol, 1 mm DTT, 1 mm PMSF, and protease inhibitors). After incubation at 4 °C overnight, the beads were washed 3× with the same buffer and boiled in 2× sample loading buffer. The immunoprecipitated L3MBTL1 was resolved on SDS-PAGE gel and detected by L3MBTL1 antibody. Coimmunoprecipitated p53 was detected by HRP-αp53 to avoid crossreactivity with IgG heavy chain. p53, p53K382me1, SET8, Flag-L3MBTL1, and tubulin were detected with their respective antibodies.
ChIP Assays
Real-time ChIP assays were performed as previously described (11). ChIP samples were prepared from HCT116 wt or HCT116 p53−/− stably expressing Flag-L3MBTL1 or Flag-L3MBTL1-D355N from U2OS cells. U2OS were treated with NCS (0.5 μg/ml Sigma) for 4 h. Occupancy values were calculated as 0.2% (M2 Flag ChIP for p21) or 1% input. Primer sequences used for ChIP analysis are as follows: p21p53BS, 5′-GTGGCTCTGATTGGCTTTCTG-3′/5′-CTGAAAACAGGCAGCCCAAG-3′; p21 promoter-3, 5′-GCATGTGTGCTTGTGTGAGTGT-3′/5′-GGGAGCAGGCTGTTAAAAGTCA-3′; p21 coding3′, 5′-CCAGTTCATTGCACTTTGATTAGC-3′/5′-GCCTCTACTGCCACCATCTTAAA-3′; PUMA promoter, 5′-GCGAGACTGTGGCCTTGTGT-3′/5′-CGTTCCAGGGTCCACAAAGT-3′; PUMA3′-1, 5′-GCCGAGGTGGGTAGATCTCTT-3′/5′-TCATCGTGTTGCCCAGGAT-3′. Other primer sequences are available upon request.
siRNA-mediated Knockdown of L3MBTL1 or SET8
Knockdown of L3MBTL1 was performed by transfection of U2OS or H1299 cells for 48 h with two sets of Dharmacon on-target plus siRNA duplex targeting human L3MBTL1 (5′-GAUCUUGGUUCCUCUAAUGUU-3′/5′CAUUAGAGGAACCAAGAUCUU-3′) or (5′-GGUCAGUCAUAGUGGAGAAUU-3′/5′-UUCUCCACUAUGACUGACCUU-3′), respectively, by using DharmaFECT according to the manufacturer's protocol. Knockdown of SET8 was performed sequentially with addition of Dharmacon on-target plus siRNA duplex targeting SET8 (5′-AGUCAAAGAUCUAUUCCUAUU-3′/5′-GUACGGAGCGCCAUGAAGUUU-3′) at 0 and 24 h for a total of 48 h. On-target plus siControl siRNA (5′-UGGUUUACAUGUCGACUAA-3′) (Dharmacon) or on-target plus SMARTpool siControl siRNA were used as controls.
Stable Knockdown of L3MBTL1, Growth Curves, and Colony Formation Assays
Stable knockdown of L3MBTL1 was achieved using independent l3mbtl1 short hairpin (sh) RNA constructs obtained from the MISSION TRC-Hs 1.0 (Human) shRNA library (Sigma). For growth curves, cells were counted manually every 24 h for 7 days following initial plating on day one of 1000 cells. Standard error of the mean (S.E.) was calculated at each count. For colony formation assay, 1000 cells were plated for each strain in triplicate. After 2 weeks, the medium was removed, and plates were washed twice with PBS, fixed, and stained with 0.1% crystal violet in methanol, and rinsed with water. The mean colony number per plate was graphed for each strain. The error was calculated as S.E.
Reverse Transcription-PCR and Real-time PCR
Reverse transcription-PCR and real-time PCR were performed as previously described (11). mRNA was prepared using RNeasy plus kit (Qiagen) and reverse transcribed using First Strand Synthesis kit (Invitrogen). Quantitative real-time PCR was performed on the ABI PRISM 7700 Sequence Detection System using Taqman Gene Expression Assay primer/probe sets (Applied Biosystems). Gene expressions were calculated following normalization to GAPDH levels by the comparative cycle threshold method (Ct).
RESULTS
The 3xMBT Repeat of L3MBTL1 Recognizes p53K382me1 in Vitro
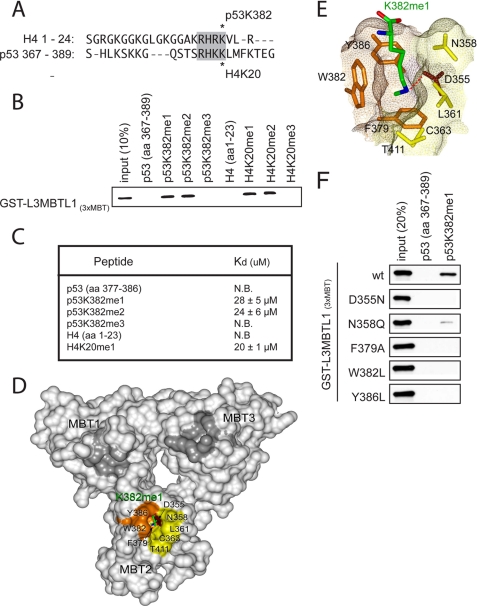
L3MBTL1 binds to H4K20me1 through its second MBT repeat (17, 18, 20, 21). Considering the sequence similarity between H4K20 and p53K382 (Fig. 1A), we tested if L3MBTL1, through the triple MBT domain, recognizes p53K382me1. In vitro peptide pull-down assays were performed with recombinant GST-tagged 3xMBT (L3MBTL13xMBT) repeats and biotinylated p53 peptides (amino acids 367–389) in which Lys-382 was unmodified or modified by mono-, di-, or trimethylation: p53K382me1, p53K382me2, or p53K382me3 respectively. As shown in Fig. 1, B and C, L3MBTL13xMBT bound preferentially to p53K382me1 and p532K382me2 peptides relative to unmodified or trimethylated peptides, similar to its known methyl state binding preference for H4K20. Finally, the binding affinity of L3MBTL13xMBT for p53K382me1 and p53K382me2 peptides was comparable to its affinity for H4K20me1, as determined by isothermal calorimetry (Fig. 1C) (17, 18, 20, 21). Based on these data, we conclude that the 3xMBT domain of L3MBTL1 binds in vitro to p53K382me1.
FIGURE 1.
L3MBTL1(3xMBT) recognizes p53 monomethylated at lysine 382 (p53K382me1) in vitro. A, sequence alignment of the H4 N terminus and p53 C-terminal tail. Homology surrounding the SET8/53BP1 binding site is highlighted. Asterisks indicate methylation sites. B, L3MBTL1(3xMBT) binds to p53K382me1 and p53K382me2 peptides. Shown are the results from Western analysis of peptide pull-down assays with the indicated biotinylated peptides and GST-L3MBTL1(3xMBT). C, dissociation constants (Kd) of p53 or histone H4 peptides for L3MBTL1(3xMBT) were determined by ITC. Kd of H4K20me1 is shown as control. D, L3MBTL1(3xMBT) repeats are shown as a solid surface with the residues comprising the p53K382me1-binding pocket of MBT2 labeled and colored orange, brown, and yellow for aromatic, acidic, and hydrophobic/polar residues, respectively. Lysine 382me1 of the p53K382me1 peptide is shown as a stick model with C, O, and N atoms colored green, red, and blue, respectively. The unoccupied semi-aromatic pockets MBT1 and MBT3 are colored gray. E, close-up view of the K382me1 binding cage. A dotted red line depicts the hydrogen bond between the methylammonium proton and the carboxyl group of Asp-355. F, point mutations in the MBT2 repeat of L3MBTL1 abolish the p53K382me1 binding activity of L3MBTL1(3xMBT) in vitro. Western analysis of the indicated GST-L3MBTL1(3xMBT) mutants in peptide pull-down assays are shown.
Structural Basis of L3MBTL1 Recognition of p53K382me1
To gain insight into the p53K382me1 - L3MBTL1 interaction at the molecular level we solved the crystal structure of L3MBTL13xMBT bound to the p53K382me1 peptide. The crystal structure of the 3xMBT repeats in complex with a peptide corresponding to residues 377–387 of p53 monomethylated at Lys-382 was determined at a 2.5 Å resolution. The protein structure comprises three globular domains arranged into a three-leaved propeller-like fold (Fig. 1D and supplemental Table S1). Each MBT repeat is composed of two subunits. The N-terminal subunit contains short helices and long loops, whereas the C-terminal subunit consists of four anti-parallel β-strands folded into a barrel-like shape. The N-terminal subunit of each repeat packs against the C-terminal subunit of the neighboring repeat resulting in an arrangement in which the N and C termini of 3xMBT are nearby. The overall structure of the protein is in agreement with the previously reported structures of L3MBTL13xMBT (20, 21, 27).
Of the three molecules comprising an asymmetric unit, two were complexes of 3xMBT with the p53K382me1 peptide (chains A and C) and one was the ligand-free protein (chain B). The structure of p53K382me1-bound 3xMBT superimposes well with the structure of the unbound protein (root mean square deviation of 0.3/0.4 Å between chain B and chains A/C, over Cα atoms), indicating that binding to the p53K382me1 peptide does not induce significant conformational changes. In the complex (chains A and C), the p53K382me1 peptide was bound by the second MBT domain only (MBT2), despite the fact that all three repeats have conserved amino acid sequences and structures, which was also observed for the interaction of L3MBTL13xMBT with mono- and dimethylated histone peptides (20, 21). Although the electron density was not seen for most of the peptide residues, it was clear for K382me1. The fully extended side chain of K382me1 inserted almost perpendicularly to the protein surface into a deep cavity created by aromatic Phe-379, Trp-382, and Tyr-386 residues, hydrophobic/polar Asn-358, Leu-361, Cys-363, and Thr-411 residues, and an acidic Asp-355 (Fig. 1E). The carboxylate of Asp-355 forms a hydrogen bond with one of protons of the methylammonium group as well as a salt bridge with this positively charged moiety. A close comparison of the bound and unbound states (chains C and B) reveals that the binding pocket is pre-formed, with each protein residue occupying essentially the same position in both states (supplemental Fig. S1). This binding mode is similar to the mechanism by which L3MBTL13xMBT, Tudor domain and engineered plant homeodomain (PHD) fingers, recognize mono- and dimethylated lysine residues of histones (16, 20, 28), suggesting conservation for readout of the low-methylation state.
To test the importance of the MBT2 binding pocket in recognition of p53K382me1, a mutational analysis of the MBT pockets was performed. As shown in Fig. 1F, mutation of any of the individual residues implicated in the structure analysis to be important for MBT2 binding to p53K382me1, abrogated p53K382me1-recognition. In contrast, analogous mutations within the MBT1 and MBT3 repeats did not affect binding to p53K382me1 (data not shown). Taken together, we conclude that the second MBT repeat of L3MBTL1 binds to p53K382me1 with a molecular mode of action similar to L3MBTL1 recognition of H4K20me1.
SET8-mediated p53K382 Methylation Augments L3MBTL1 Binding to p53 in Cells
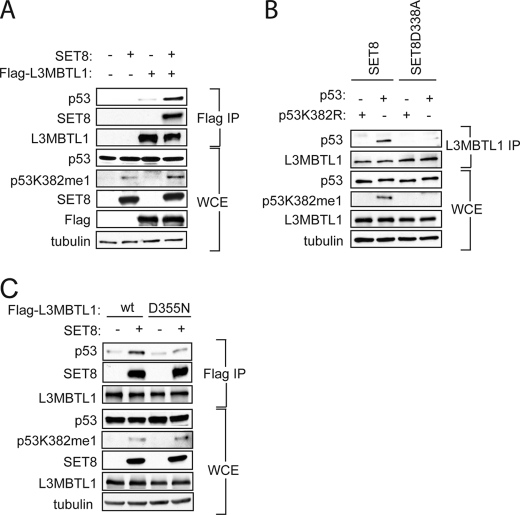
Next, the importance for p53 methylation in mediating p53-L3MBTL1 interaction in cells was investigated. We overexpressed Flag-tagged L3MBTL1 with or without SET8 in 293T cells for coimmunoprecipitation (co-IP) assays. As shown in Fig. 2A, far more endogenous p53 was co-IPed in cells co-transfected with SET8 and L3MBTL1 relative to L3MBTL1 alone (compare lanes 3 and 4), suggesting that monomethylation of p53 at Lys-382 augments the ability of L3MBTL1 to bind to p53 in cells.
FIGURE 2.
L3MBTL1 binds to p53K382me1 in vivo. A, SET8 augments the interaction between p53 and L3MBTL1 in vivo. Western analysis with the indicated antibodies of the Flag IPs or whole cell extract (WCE) from 293T cells expressing the indicated proteins is shown. Tubulin levels in the WCE are shown as loading control. B, monomethylation on p53K382 by SET8 is important for the association of L3MBTL1 and p53 in vivo. Western analysis with the indicated antibodies of endogenous L3MBTL1 IPs or WCE from H1299 cells expressing wild-type p53 or p53K382R mutant in the presence of SET8 or SET8(D338A) as indicated. Tubulin is shown as a loading control. C, intact MBT2 domain is required for interaction between L3MBTL1 and p53 in vivo. Western analysis with the indicated antibodies of the Flag IPs or WCE from 293T cells expressing wild-type F-L3MBTL1 or F-L3MBTL1(D355N) mutant ± SET8. Tubulin levels in the WCE are shown as loading control.
To specifically address the role of p53K382 methylation in the p53-L3MBTL1 interaction, we performed co-IPs in the p53-null strain H1299 reconstituted for p53 with either wild-type (wt) p53 or a p53K382R mutant that cannot be methylated by SET8. SET8 or a SET8 catalytic mutant SET8 (D338A) (29, 30) was cotransfected with the wt or mutant p53 into H1299 cells and endogenous L3MBTL1 was immunoprecipitated. As shown in Fig. 2B, p53 was clearly present in the L3MBTL1 IP in cells cotransfected with wt p53 and wt SET8, whereas in all other transfection combinations, p53 was not detected in the L3MBTL1 IP. These results argue that SET8 monomethylation of p53 at lysine 382 is important for in vivo interaction of p53 with L3MBTL1.
To further investigate the importance of p53K382 methylation and its recognition by MBT2 of L3MBTL1 in the L3MBTL1-p53 interaction, the ability of L3MBTL1 or a L3MBTL1(D355N) mutant, shown to disrupt binding to p53K382me1 in vitro (Fig. 1), to co-IP endogenous p53 in a SET8-dependent fashion was determined. The increase in interaction between L3MBTL1 and p53 induced by SET8 was diminished in the L3MBTL1(D355N) mutant IP (Fig. 2C). We note that SET8 does facilitate binding between L3MBTL1 and p53 in a methylation-independent fashion, potentially via formation of a trimeric complex (data not shown). Together, we conclude that recognition of p53K382me1 by the second MBT repeat of L3MBTL1 is important for L3MBTL1-p53 interactions in cells.
L3MBTL1 Represses p53 Target Genes at Basal Conditions
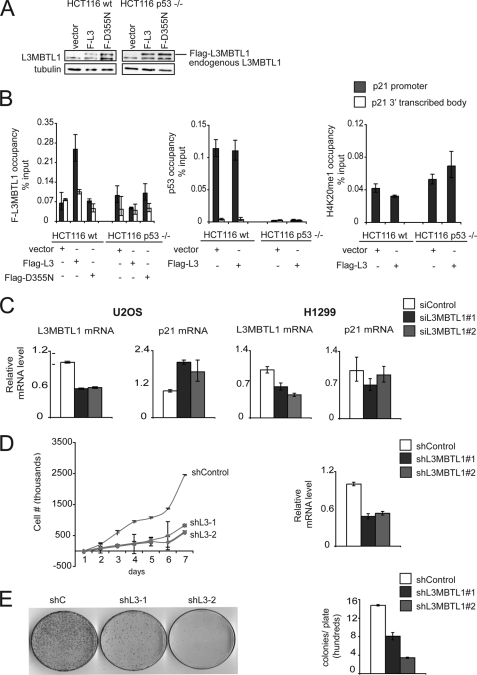
We previously demonstrated that induction of p53 target genes, such as p21, is negatively regulated by SET8-mediated monomethylation at p53K382 under both basal conditions and in response to DNA damage (11). L3MBTL1 has been implicated in chromatin compaction and promotion of a repressive chromatin state (13, 15). We therefore postulated that the repressive effect of p53K382me1 at these promoters might in part be due to binding to and stabilizing L3MBTL1 at p53 target genes. To test this hypothesis, we determined in chromatin immunoprecipitation (ChIP) assays the occupancy of stably expressing Flag-tagged L3MBTL1 (introduced by retroviral transduction (Fig. 3A)), endogenous p53 and H4K20me1 at the p21 and PUMA genes in wild-type HCT116 cells and p53−/− HCT116 cells (31). As shown in Fig. 3B and supplemental Fig. S2, both p53 and L3MBTL1 occupy the p21 and PUMA promoter under basal conditions, but are not detected in the p21- and PUMA-transcribed body. Moreover, in p53−/− cells, L3MBTL1 is not detected at the two promoters (Fig. 3B, supplemental Fig. S2). H4K20me1, which is generated by SET8 (29, 30) and can also be bound by L3MBTL1 (13), is detected at low levels at promoters and does not show a difference in signal between p53+/+ and p53−/− cells (Fig. 3B, supplemental Fig. S2). Together, these data suggest that p53 can play a role in stabilizing L3MBTL1 at its target promoters.
FIGURE 3.
L3MBTL1 represses p53 target genes under normal conditions. A, Western blot with L3MBTL1 antibodies of WCE from the indicated cell lines transduced with control, Flag-L3MBTL1, or Flag-L3MBTL1-D355N retrovirus. B, p53 stabilizes L3MBTL1 at target genes under basal conditions. Chromatin immunoprecipitation (ChIP) assays to determine occupancy at the promoter and 3′ transcribed body region of the p21 gene in wt and p53−/− HCT116 cells stably expressing Flag-L3MBTL1 or Flag-L3MBTL1-(D355N) as indicated. ChIPs: Flag (left), p53 (middle), H4K20me1 (right). Occupancy values (% input) were determined by real-time PCR. C, p21 expression increases upon L3MBTL1 depletion in the presence of endogenous p53 and in the absence of DNA damage. U2OS or the p53-negative cell line H1299 treated with control or two independent L3MBTL1 siRNAs and L3MBTL1 and p21 mRNA levels were determined by real-time PCR. D, L3MBTL1 depletion attenuates cellular proliferation rates. Left panel, 7-day growth curve time course for U2OS cells stably expressing control or two independent L3MBTL1 shRNAs. Error bars indicate S.E. from three experiments. Right panel, real-time PCR of L3MBTL1 mRNA levels of cells used in the growth curve experiment. E, L3MBTL1 depletion attenuates colony growth. Left panel, colony formation panel of cells as in D. Representative plates are shown for control and the two L3MBTL1 stable knockdown lines. Right panel, quantitation of colonies counted for each cell line. Error bars indicate S.E. from three independent experiments.
Next, the role of L3MBTL1 in regulating p21 mRNA expression was determined. Endogenous L3MBTL1 was depleted by RNA interference employing two independent L3MBTL1-targeting siRNAs. As shown in Fig. 3C, in the absence of DNA damage (a condition that stimulates p21 expression), depletion of L3MBTL1 induced an ∼2-fold increase in p21 transcription in the p53 positive U2OS cells. In contrast, no up-regulation of p21 transcript was observed upon L3MBTL1 knock-down in p53 negative H1299 cells. Together, these results suggest that: 1) L3MBTL1 normally plays a role in maintaining p21 repressed, 2) this repression is p53 dependent, and 3) the molecular mechanism for this repression is in part due to p53 stabilization of L3MBTL1 at chromatin of p53 target genes.
Stable depletion of L3MBTL1 in U2OS cells with two independent shRNAs resulted in a markedly slower cell proliferation rate relative to cells expressing the control shRNA (Fig. 3D). Moreover, colony formation assays indicated that fewer and smaller colonies develop in L3MBTL1-depleted cells relative to control cells (Fig. 3E). These data suggest that L3MBTL1 plays a role in promoting proliferation of transformed cells, consistent with its ability to repress p53 target gene expression.
Regulation of p21 Expression by L3MBTL1 Requires SET8-mediated p53K382me1
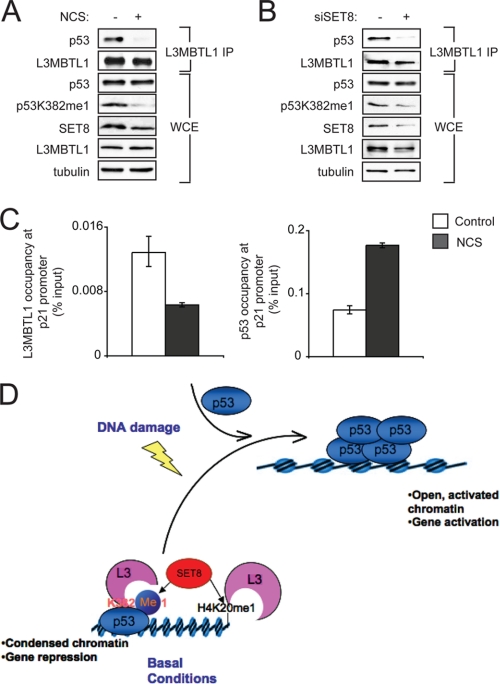
The level of total p53 protein increases with genotoxic stress, whereas SET8 and p53K382me1 levels decrease (11); p53 occupancy on the p21 and other target promoters also increases with genotoxic stress as part of the p53-dependent induction of a DNA damage gene expression program. Based on these observations, we predicted that L3MBTL1 would disassociate from p53 upon DNA damage. To test this hypothesis, cells were treated with the radiomimetic NCS, to induce DNA double strand breaks, or vehicle control, and the interaction between endogenous L3MBTL1 and p53 was determined in co-IP assays. At basal conditions (no DNA damage), p53 was present in the L3MBTL1 IP (Fig. 4A). However, upon NCS treatment, which elicits a decrease in p53K382me1 levels (Fig. 4A), p53 was not detected in the L3MBTL1 IP. Depletion of SET8 by siRNA to emulate DNA damage-induced reduction of SET8 also abrogated the endogenous interaction between L3MBTL1 and p53 (Fig. 4B). Further, L3MBTL1 occupancy at the p21 promoter decreases in response to NCS treatment, in contrast to p53 binding at the p21 promoter, which as expected increases concomitant with DNA damage (Fig. 4C) (11). Taken together, we conclude that the interaction between p53K382me1 and L3MBTL1 at the promoters of p53 target genes is one mechanism by which SET8-mediated methylation of p53 acts to repress p53 transactivation under basal conditions. Upon DNA damage, SET8 levels are down-regulated, p53K382me1 and H4K20me1 levels are reduced, L3MBTL1 dissociates from the promoter, and transactivation is initiated by p53.
FIGURE 4.
The endogenous interaction between L3MBTL1 and p53 is disrupted upon DNA damage. A, Western analysis with the indicated antibodies of L3MBTL1 IPs or WCE prepared from 293T cells, ± 0.5 μg/ml the radiomimetic NCS. Note that p53 levels do not increase upon DNA damage in 293T cells. B, SET8 depletion abrogates the interaction between endogenous L3MBTL1 and p53. Western analysis with the indicated antibodies of the L3MBTL1 IPs or WCE prepared from 293T cells ± SET8 siRNA (11). C, L3MBTL1 occupancy at the p21 promoter decreases upon DNA damage. ChIP assays of endogenous L3MBTL1 at the p21 promoter in U2OS cells ± treatment with 0.5 μg/ml NCS. Occupancy values (% input) were determined by real-time PCR. D, model for SET8-mediated repression of p53 through promotion of p53-L3MBTL1 binding. In the absence of DNA damage, SET8 generates a population of p53 monomethylated at Lys-382. p53K382me1 binds to and stabilizes chromatin compaction factor L3MBTL1 at the promoter regions of p53 target genes, which results in a repressive chromatin state. Upon DNA damage, Lys-382 monomethylation levels decrease by as yet unknown mechanism (possibly via demethylation, higher methylation of p53K382, or simply is diluted out by the newly stabilized p53 protein) and this leads to L3MBTL1 dissociation from p53-target genes and a chromatin environment favorable for active gene transcription.
DISCUSSION
In summary, we have demonstrated by biochemical, structural, and cellular approaches that L3MBTL1 functions as an effector of p53K382 monomethylation. Signaling via lysine methylation of histone proteins is a principal chromatin-regulatory mechanism that influences fundamental cellular programs(1). Like histones, p53 is subject to a wide array of regulatory post-translational modifications that influence its biological properties (6, 8, 10). Several lysines within the unstructured tail of p53 are methylated and the state of methylation at a specific lysine residue regulates various aspects of p53 function, including gene activation, gene repression, protein localization, and protein stability (11, 32–37). Dimethylation of p53 at Lys-370 and Lys-382 represent two examples of how a p53 methylation event is sensed at the molecular level; in both of these cases, the modification acts as a template for an interaction with the DNA damage response mediator 53BP1. In the context of binding to p53K370me2, 53BP1 is a co-activator of p53 transcription in response to DNA damage (36), whereas 53BP1 recognition of p53K382me2 stabilizes p53 at DNA double strand breaks and might function in communicating exposure of cells to genotoxic stress to p53 (37).
In mammals, L3MBTL1 has been linked to transcription repression. For example, L3MBTL1 interacts with the ETS transcription factor TEL to repress TEL-responsive promoters (15). In addition, L3MBTL1 is present in a complex with RB (retinoblastoma) protein, and functions to negatively regulate expression of a number of E2F target genes by compacting chromatin, an activity that requires MBT domain binding to repressive mono- and dimethylation marks on histones (13). The observations that L3MBTL1 functions in part via H4K20me1 recognition and the similarity in sequence surrounding H4K20 and p53K382 led us to test binding of L3MBTL1 to p53K382me1. We propose that under basal conditions, in the absence of DNA damage, L3MBTL1 is stabilized at chromatin of p53-target genes via synergistic binding to p53K382me1 and H4K20me1 (or similar repressive mark such as H1K26me1) (see model, Fig. 4D). The stabilization and subsequent action of L3MBTL1 at these p53 target genes results in chromatin compaction and thereby, L3MBTL1, by regulating chromatin structure, indirectly renders target-DNA-bound p53 inert. In response to DNA damage, p53K382me1 levels decrease (Fig. 4 and Ref. 11), resulting in destabilization of L3MBTL1 from p53-target genes and therefore a more favorable chromatin state, which in turn promotes transcription (Fig. 4D). Cabin1 binding to promoter-bound p53 was recently proposed to negatively regulate a subpopulation of p53 target genes by a mechanism similar to the one proposed in our model (Fig. 4D and Ref. 12). These observations suggest a broader paradigm in which sequence-specific transcription factors are bound to target DNA in the absence of active transcription, but rendered quiescent by accessory factors. Such a mechanism may be important in the context of rapid activation of target genes and/or maintaining certain DNA sequences accessible.
In addition to p53, the tumor suppressor protein RB is also mono methylated by a protein lysine methyltransferase (Smyd2), and this methylation event promotes interaction with L3MBTL1.6 Furthermore, the interaction between L3MBTL1 and methylated RB may play a role in repression of RB target genes.6 Thus, L3MBTL1 might act broadly to inhibit different gene expression programs through its ability to recognize methylated non-histone nuclear proteins. Here we have demonstrated that L3MBTL1 binds to p53 in a modular manner that is dependent on SET8-mediated monomethylation at p53K382 and propose that this interaction inhibits p53 activity by regulating chromatin dynamics.
Acknowledgments
We thank D. Reinberg and K. Glass for helpful discussion and P. Bar-On for technical assistance.
This work was supported, in whole or in part, by Grants GM079641 (to O. G.) and CA113472 (to T. G. K.) from the National Institutes of Health and the Intramural Research Program of the National Institutes of Health (to E. A.).
The atomic coordinates and structure factors (code 3OQ5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
J. Sage, personal communication.
- PKMT
- protein lysine methyltransferase
- MBT
- malignant brain tumor
- L3MBTL1
- lethal 3 malignant brain tumor-like 1
- ITC
- isothermal calorimetry
- WCE
- whole cell extract
- p53K382
- p53 lysine 382
- H4K20
- histone H4 lysine 20
- H1K26
- histone H1 lysine 26
- RB
- retinoblastoma
- me1
- monomethylation
- me2
- dimethylation
- me3
- trimethylation
- NCS
- neocarzinostatin.
REFERENCES
- 1.Shi Y., Whetstine J. R. (2007) Mol. Cell. 25, 1–14 [DOI] [PubMed] [Google Scholar]
- 2.Sims R. J., 3rd, Reinberg D. (2006) Genes Dev. 20, 2779–2786 [DOI] [PubMed] [Google Scholar]
- 3.Bannister A. J., Kouzarides T. (2004) Methods Enzymol. 376, 269–288 [DOI] [PubMed] [Google Scholar]
- 4.Daniel J. A., Pray-Grant M. G., Grant P. A. (2005) Cell Cycle. 4, 919–926 [DOI] [PubMed] [Google Scholar]
- 5.Baker L. A., Allis C. D., Wang G. G. (2008) Mutat. Res. 647, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J., Berger S. L. (2008) Curr. Opin. Genet. Dev. 18, 152–158 [DOI] [PubMed] [Google Scholar]
- 7.Zhang K., Dent S. Y. (2005) J. Cell. Biochem. 96, 1137–1148 [DOI] [PubMed] [Google Scholar]
- 8.Morgunkova A., Barlev N. A. (2006) Cell Cycle. 5, 1308–1312 [DOI] [PubMed] [Google Scholar]
- 9.Gottifredi V., Shieh S. Y., Prives C. (2000) Cold Spring Harb. Symp. Quant. Biol. 65, 483–488 [DOI] [PubMed] [Google Scholar]
- 10.Appella E., Anderson C. W. (2001) Eur. J. Biochem. 268, 2764–2772 [DOI] [PubMed] [Google Scholar]
- 11.Shi X., Kachirskaia I., Yamaguchi H., West L. E., Wen H., Wang E. W., Dutta S., Appella E., Gozani O. (2007) Mol. Cell. 27, 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang H., Choi S. Y., Cho E. J., Youn H. D. (2009) Nat. Struct. Mol. Biol. 16, 910–915 [DOI] [PubMed] [Google Scholar]
- 13.Trojer P., Li G., Sims R. J., 3rd, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S. D., Wang Y. H., Reinberg D. (2007) Cell 129, 915–928 [DOI] [PubMed] [Google Scholar]
- 14.Trojer P., Reinberg D. (2008) Cell Cycle 7, 578–585 [DOI] [PubMed] [Google Scholar]
- 15.Boccuni P., MacGrogan D., Scandura J. M., Nimer S. D. (2003) J. Biol. Chem. 278, 15412–15420 [DOI] [PubMed] [Google Scholar]
- 16.Guo Y., Nady N., Qi C., Allali-Hassani A., Zhu H., Pan P., Adams-Cioaba M. A., Amaya M. F., Dong A., Vedadi M., Schapira M., Read R. J., Arrowsmith C. H., Min J. (2009) Nucleic Acids Res. 37, 2204–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm C., de, Ayala, Alonso A. G., Rybin V., Steuerwald U., Ly-Hartig N., Fischle W., Müller J., Müller C. W. (2007) EMBO Rep. 8, 1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalakonda N., Fischle W., Boccuni P., Gurvich N., Hoya-Arias R., Zhao X., Miyata Y., Macgrogan D., Zhang J., Sims J. K., Rice J. C., Nimer S. D. (2008) Oncogene 27, 4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Fischle W., Wang W., Duncan E. M., Liang L., Murakami-Ishibe S., Allis C. D., Patel D. J. (2007) Mol. Cell. 28, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min J., Allali-Hassani A., Nady N., Qi C., Ouyang H., Liu Y., MacKenzie F., Vedadi M., Arrowsmith C. H. (2007) Nat. Struct. Mol. Biol. 14, 1229–1230 [DOI] [PubMed] [Google Scholar]
- 22.Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pflugrath J. W., (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 24.McCoy A. J., Storoni L. C., Read R. J. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 1220–1228 [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 27.Wang W. K., Tereshko V., Boccuni P., MacGrogan D., Nimer S. D., Patel D. J. (2003) Structure 11, 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang J., Feng Q., Ketel C. S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J. A., Zhang Y. (2002) Curr. Biol. 12, 1086–1099 [DOI] [PubMed] [Google Scholar]
- 30.Nishioka K., Rice J. C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Lis J. T., Allis C. D., Reinberg D. (2002) Mol. Cell. 9, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 31.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 32.Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., Reinberg D. (2004) Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 33.Ivanov G. S., Ivanova T., Kurash J., Ivanov A., Chuikov S., Gizatullin F., Herrera-Medina E. M., Rauscher F., 3rd, Reinberg D., Barlev N. A. (2007) Mol. Cell. Biol. 27, 6756–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J., Dorsey J., Chuikov S., Pérez-Burgos L., Zhang X., Jenuwein T., Reinberg D., Berger S. L. (2010) J. Biol. Chem. 285, 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., Dorsey J. A., Kubicek S., Opravil S., Jenuwein T., Berger S. L. (2006) Nature 444, 629–632 [DOI] [PubMed] [Google Scholar]
- 36.Huang J., Sengupta R., Espejo A. B., Lee M. G., Dorsey J. A., Richter M., Opravil S., Shiekhattar R., Bedford M. T., Jenuwein T., Berger S. L. (2007) Nature 449, 105–108 [DOI] [PubMed] [Google Scholar]
- 37.Kachirskaia I., Shi X., Yamaguchi H., Tanoue K., Wen H., Wang E. W., Appella E., Gozani O. (2008) J. Biol. Chem. 283, 34660–34666 [DOI] [PMC free article] [PubMed] [Google Scholar]