Abstract
The retinoblastoma tumor suppressor (RB) is a central cell cycle regulator and tumor suppressor. RB cellular functions are known to be regulated by a diversity of post-translational modifications such as phosphorylation and acetylation, raising the possibility that RB may also be methylated in cells. Here we demonstrate that RB can be methylated by SMYD2 at lysine 860, a highly conserved and novel site of modification. This methylation event occurs in vitro and in cells, and it is regulated during cell cycle progression, cellular differentiation, and in response to DNA damage. Furthermore, we show that RB monomethylation at lysine 860 provides a direct binding site for the methyl-binding domain of the transcriptional repressor L3MBTL1. These results support the idea that a code of post-translational modifications exists for RB and helps guide its functions in mammalian cells.
Keywords: Cell Cycle, Post-translational Modification, Protein Methylation, Retinoblastoma (Rb), Transcription Repressor, L3MBTL1, SMYD2
Introduction
The retinoblastoma tumor suppressor gene RB2 is mutated in a large spectrum of human cancers (1, 2). In tumor cells where RB is not directly mutated, and in normal cells during cell cycle progression, the RB protein is functionally inactivated by phosphorylation by cyclin/CDK complexes (3). RB phosphorylation results in the release of E2F transcription factors, allowing cells to progress into the S phase of the cell cycle (4). In addition to phosphorylation by cyclin/CDK complexes, RB activity is controlled by other post-translational events. For instance, Chk2 phosphorylates RB in response to DNA damage (5). In addition, RB is acetylated (6–9), sumoylated (10), and ubiquitinated (11–13) in response to various cellular signals. The consequences of these modifications involve changes in RB protein levels and in the binding affinity for proteins that interact with RB, such as E2F, chromatin-remodeling enzymes, and other regulators of cell cycle progression and cellular differentiation. For example, RB acetylation is thought to inhibit its phosphorylation and to promote its binding to MDM2, which results in the subsequent degradation of EID-1, an inhibitor of differentiation (8, 9).
Recent evidence that non-histone proteins can be methylated supports the idea that methylation may affect gene expression and cellular functions not only by modifying histone tails (14–17) but also by changing the activity of transcription factors, including the p53 tumor suppressor (18–27). These observations suggest that, similar to the “histone code” (28), combinations of post-translational modifications may define codes that affect the function of key regulators of transcription. Based on these observations and evidence suggesting that RB directly interacts with chromatin-modifying agents, including methyltransferases (29–33), it is not surprising that RB was recently shown to be a target for lysine methylation by SET9 (34). Nevertheless, the extent of RB methylation in cells and its consequences for RB function are still largely unknown. In this study, we report that SMYD2 methylates RB at lysine 860. We show that this modification permits direct binding of RB to the lysine methyl-binding protein L3MBTL1, which may alter the function of RB in cells.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Peptides
MYC-SMYD2, MYC-SMYD2(Y290F) (35), FLAG-SMYD2 (19), GST-SET9 (18) were described previously. Details for the construction of human GST-RB fragments, HA-RB(K860R), HA-RB(K870,873,874R), GST-SMYD2, GST-3xMBT, GST-3xMBT mutants, and FLAG-L3MBTL1 wild-type and mutant vectors are available upon request. Biotinylated RB peptides VCNSDRVLK(me0–3) RSEAGSNPPKPLKKLK and H4K20(aa 1–23) (me0–3) peptides were purchased from the Small Scale Peptide Synthesis facility (Yale University). Rabbit polyclonal antibodies to RBK860me1 were generated by Abmart following immunization with the monomethylated peptide 853-CNSDRVLK(me1) RSEAG-865. Other antibodies used in this study are directed against Streptavidin (Abcam), RB (Ref. 36, 4.1, DSHB, University of Iowa), RB C-15 (Santa Cruz Biotechnology), E2F1 (Santa Cruz Biotechnology), MYC (Sigma Aldrich), β-actin (Sigma Aldrich), p107 (Santa Cruz Biotechnology), γH2AX (Cell Signaling Technology), GST (E5, Santa Cruz Biotechnology), FLAG (M2, Sigma Aldrich), α-Tubulin (Sigma Aldrich), L3MBTL1 (Abcam), and H3K4trime (Active Motif).
Cell Culture and Transfections
293T, T98G, Saos-2, U2OS, C2C12, and NIH3T3 cells were maintained in DMEM medium supplemented with 10% bovine growth serum. Cells were transfected with calcium phosphate or using a Nucleofector (Lonza).
Immunoprecipitation
Whole cell extracts from cells were prepared in 50 mm Tris pH 7.4, 250 mm NaCl, 10% glycerol, 0.5% Triton-X-100, and protease inhibitors. RB was immunoprecipitated with RB or RBK860me1 antibodies. Ectopically expressed HA-RB and mutants were precipitated with anti-HA-agarose beads (Sigma Aldrich) and ectopically expressed FLAG-L3MBTL1 and mutants were immunoprecipitated with anti-M2 FLAG resin (Sigma Aldrich). Immunoprecipitated proteins were resolved on SDS-PAGE gels for immunoblot analysis as described (36).
In Vitro Methyltransferase Assay and Mass Spectrometry
Assays were performed as previously described (37). Briefly, 5 μg of GST-RB were incubated with 5 μg of GST-SMYD2 or GST-SET9, and 0.1 mm S adenosyl-methionine (SAM, Sigma Aldrich) or 55 μCi of [3H]SAM (Perkin Elmer) at 30 °C for 2 h before electrophoresis and autoradiography or mass spectrometry.
Mass Spectrometry Analysis
Samples were first reduced and alkylated with 10 mm Tris(2-carboxyethyl) phosphine hydrochloride (Roche Applied Science) and 55 mm iodoacetamide (Sigma Aldrich), respectively, then digested with endoproteinase Arg-C (Roche Applied Science) according to the manufacturer's specifications. The protein digest was pressure-loaded onto a 100 μm i.d. fused silica capillary (Polymicro Technologies) analytical column with a 5 μm pulled-tip, packed with 10 cm of 5 μm C18 resin (Phenomenex). The analytical column was placed inline with an 1100 quaternary HPLC pump (Agilent Technologies) and the eluted peptides were electrosprayed directly into an LTQ-XL mass spectrometer (Thermo Scientific). The buffer solutions used were 5% acetonitrile/0.1% formic acid (buffer A) and 80% acetonitrile/0.1% formic acid (buffer B). The 120 min elution gradient had the following profile: 10% buffer B at 5 min, to 50% buffer B at 80 min, to 100% buffer B from 90–100 min. A cycle consisted of one full scan mass spectrum (400–2000 m/z) followed by 5 data-dependent electron transfer dissociation (ETD) MS/MS spectra with an isolation width of 2 m/z. Fluoranthene anions were set to a target value of 105 and ETD reaction time was set at 50 ms. Supplemental activation was enabled. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (Thermo Scientific). ETD MS/MS spectra were extracted up to charge state 10+ using RawXtract (version 1.9) (38). ETD MS/MS spectra were searched with the ProLuCID algorithm (39) against the Escherichia coli SGD data base supplemented with the GST-RB (amino acids 829–928) fusion protein sequence and concatenated to a decoy data base in which the sequence for each entry in the original database was reversed (40). Charged reduced precursors were removed from the spectra prior to searching. The ProLuCID search was performed using full enzyme specificity and a static modification of cysteine due to carboxyamidomethylation (57.02146). Differential modifications considered were lysine monomethylation (14.0157), dimethylation (28.0314), and trimethylation (42.0471). ProLuCID search results were assembled and filtered using the DTASelect2.0 algorithm (41) with a false positive rate below one percent.
shRNA-mediated Knockdown
A pGIPZ lentiviral vector containing shRNA against SMYD2 (Open Biosytems) or a control vector were used to infect cells as described (42). For transient knockdown, U2OS were transfected with two rounds of SMYD2 ON-TARGETplus SMARTpool (l-020291–00-0005) or ON-TARGETplus Non-targeting siRNA1 (d-001810–01-05) using DharmaFECT 1 (Thermo Scientific).
RT-PCR and Real-Time PCR
RNA was extracted with TRIzol (Invitrogen) and the RNeasy Mini Kit (Qiagen). RT-PCR and quantitative real-time PCR were performed using the DyNAmo cDNA synthesis kit and the SybrGreenER Mastermix (Invitrogen), respectively. Primer sequences are available upon request.
Cell Cycle and Cell Death and Differentiation Assays
T98G cells were serum starved in DMEM supplemented with 0.1% bovine growth serum and then stimulated with DMEM supplemented with 20% bovine growth serum for 0, 9, and 18 h. For flow cytometry, T98G cells were collected and fixed with 70% ethanol at 4 °C. Cells were then washed with PBS and stained with 10 μg/ml propidium iodide and 100 μg/ml RNase. DNA damage assays were performed in NIH3T3 cells treated with 10 μm etoposide. C2C12 myoblasts were induced to differentiate by growing cells to confluency in medium with 2% horse serum for 0, 1, 2, and 7 days.
Peptide Pull-down Assays
Peptide pull-down assays were performed as previously described (37). Briefly, 1 μg of biotinylated peptide was incubated with 1 μg of protein in binding buffer (50 mm Tris-HCl pH 7.5, 150 mm NaCl, 0.05% Nonidet P-40, and protease inhibitors) overnight at 4 °C with rotation before incubation with streptavidin beads (Amersham Biosciences) and immunoblot analysis.
Isothermal Titration Calorimetry
ITC assays were conducted with a MicroCal VPITC Calorimeter as recently described (43). Data were analyzed with the Origin calorimetry software package assuming a one site binding model. N-values, reflecting the stoichiometry of the RB-3xMBT complex, were between 0.8 and 1.2. Experiments were repeated 2–4 times, and the reported error is the standard deviation of each set of measurements.
Cell Fractionation
2 × 107 cells were incubated in Buffer A (10 mm HEPES pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 0.1% Triton X-100, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 0.1 mm PMSF) for 10 min on ice. Nuclei were collected by centrifugation at 1,300 × g at 4 °C for 5 min and the supernatant (S1) was further clarified by centrifugation at 16,000 g at 4 °C for 10 min to yield a cytoplasmic fraction (S2). The nuclei were washed once in Buffer A and then lysed in Buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 0.1 mm PMSF) for 15 min on ice followed by centrifugation at 1,700 g at 4 °C for 5 min. The supernatant (S3) consisting of nucleoplasm was removed and the pellet (P3) was resuspended in Laemmli buffer and boiled for 20 min to yield the chromatin fraction (Chr).
Statistical Significance
Student's t-tests were conducted to determine statistical significance. * denotes p value of <0.05.
RESULTS
SMYD2 Methylates RB in Vitro
The SMYD2 methyltransferase was originally identified as an enzyme whose activity could suppress cell proliferation (35) and directly regulate p53 function (19). We tested whether SMYD2 could also methylate the RB protein, a major regulator of cell cycle progression at the G1/S phase of the cell cycle. We found that recombinant SMYD2 could methylate a C-terminal fragment of RB and had minimal or no activity against the N-terminal and large pocket fragments of the protein (Fig. 1, A and B, left, and data not shown). We next identified lysine 860 (Lys-860 or K860) as the residue of RB that was methylated in vitro by SMYD2 (Fig. 1B, right). In contrast, SET9 methylated RB within the large RB pocket and not in the C-terminal fragment containing Lys-860 (Fig. 1C), suggesting a certain level of specificity for the methylation of RB by SMYD2 at Lys-860. The conservation of RB K860 and surrounding sequences in multiple species and the fact that this region does not seem to be important in maintaining the structure of RB (44) suggest that this amino acid may have an important functional role (Fig. 1D).
FIGURE 1.
Methylation of human RB by SMYD2 at lysine 860 in vitro. A, schematic representation showing RB N-terminal region (N), its pocket (A, I, B), and its C-terminal domain. Known acetylated (Ac) lysines (positions 873 and 874) and the lysine methylated by SMYD2 (position 860) are shown. B, RB is methylated by SMYD2 at lysine 860 in vitro. Autoradiograms (3H) of histone methyltransferase (HMTase) assays with recombinant RB fragments (GST-RB) and SMYD2. Single amino acid substitutions (K-R) are indicated. Coomassie Blue staining shows the amount of RB protein loaded. C, C-term RB methylation is specific to SMYD2. Autoradiogram of an HMTase assay with GST-RB fragments and SET9. D, RB lysine 860 is highly conserved. Conservation of the sequences surrounding RB lysine 860 (in red) in several species.
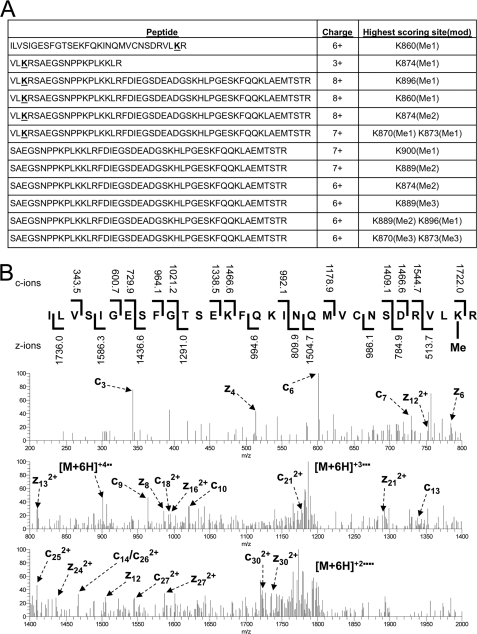
To determine if RB K860 was mono-, di-, or trimethylated by SMYD2, an RB C-terminal fragment methylated in vitro by SMYD2 was subjected to mass spectrometry analysis. While several methylated peptides were identified using this sensitive detection method, this analysis identified peptides containing monomethylated RB K860 (Fig. 2), indicating that SMYD2 monomethylates RB at Lys-860 in vitro, similar to its activity on p53 (19).
FIGURE 2.
Mass spectrometry analysis of RB methylation by SMYD2 in vitro. A, RB is monomethylated by SMYD2. Modified peptides identified, charge state, and highest scoring site of modification (degree of methylation) in the RB/SMYD2 mass spectrometry analysis are shown. Lysine 860 is bolded and underlined. B, electron transfer dissociation (ETD) MS/MS spectrum matching charge state 6+ for the peptide ILVSIGESFGTSEKFQKINQMVCNSDRVLK(Me)R and corresponding fragmentation map. Matched c- and z-ions are indicated on the fragmentation map along with their m/z values. Matched c- and z-ions as well as charge-reduced precursors are labeled on the MS/MS spectrum. Lysine monomethylation can be localized to Lys-860 and not Lys-844 or Lys-847 based on site-determining ions.
SMYD2 Methylates RB in Cells
To test if SMYD2 methylates RB at Lys-860 in cells, we developed polyclonal antibodies that specifically recognize monomethylated Lys-860 in the RB protein (RBK860me1). The specificity of these antibodies was verified in vitro against non-methylated, mono-, di-, and tri-methylated RB K860 and histone H4K20 peptides (Fig. 3A). Using these antibodies in immunoblot experiments, we were able to detect RB methylation in 293T cells that had been transfected with tagged HA-RB and MYC-SMYD2 (Fig. 3B). Furthermore, this methylation was dependent on the methyltransferase catalytic domain of SMYD2 because RB methylation was not detected in similar conditions where MYC-SMYD2 was substituted by MYC-SMYD2(Y290F), a catalytically inactive derivative of SMYD2 (Fig. 3B). Methylation was specific to lysine 860 as MYC-SMYD2 failed to methylate HA-RB(K860R), a form of RB where lysine 860 is mutated to arginine, arguing for a preferential activity of SMYD2 at Lys-860 (Fig. 3B). As an additional control, we expressed a form of RB with lysine to arginine substitutions in residues close to lysine 860 (HA-RB(K870,873, 874R)), two of which can be acetylated in cells (6–9). We found that these mutations did not affect the capacity of MYC-SMYD2 to methylate RB (Fig. 3B). Thus, RB is monomethylated at lysine 860 by SMYD2 in cells when both proteins are overexpressed. Similarly, we found that endogenous RB molecules were methylated by MYC-SMYD2 (Fig. 3C). To test whether endogenous RB methylation at lysine 860 was dependent on endogenous SMYD2 expression, we used shRNA constructs to knockdown SMYD2 in U2OS cells and found that a decrease of ∼80% in SMYD2 mRNA levels resulted in a visible decrease in the amount of methylated RB that could be immunoprecipitated with the antibody against RBK860me1 (Fig. 3D). Similar experiments using stable knockdown in 293T cells and transient knockdown in U2OS cells produced similar results (supplemental Fig. S1). Together, these experiments indicate that endogenous RB is monomethylated by SMYD2 at Lys-860 in cells.
FIGURE 3.
Methylation of human RB by SMYD2 at lysine 860 in vivo. A, RBK860me1 antibody recognizes monomethyl Lys-860 RB. The specificity of polyclonal antibodies recognizing monomethylated Lys-860 in the RB protein (RBK860me1) was assessed by dot blot analysis with biotinylated peptides. A peptide from histone H4 (amino acids 1–20) serves as a negative control. Streptavidin was used as a positive control and a loading control. B, overexpressed SMYD2 monomethylates overexpressed RB at lysine 860 in 293T cells. Whole cell extracts from 293T cells transfected with the indicated plasmids were immunoprecipitated with HA resin and analyzed by immunoblot with RB, RBK860me1, and E2F antibodies. Controls include lysine to arginine substitutions in RB at Lys-860 and Lys-870, -873, -874 as well as a mutant form of SMYD2 (Y290F) with decreased methyltransferase activity. Inputs for HA-RB and MYC-SMYD2 are shown. C, overexpressed SMYD2 monomethylates endogenous RB at lysine 860. Whole cell extracts of 293T cells were immunoprecipitated with antibodies against RB or control IgG antibodies followed by immunoblot analysis with RB and RBK860me1 antibodies. Inputs for RB and MYC-SMYD2 are shown. D, methylation of endogenous RB at K860 in U2OS cells is decreased upon knockdown of SMYD2. shRNA molecules against SMYD2 (shSMYD2) were stably expressed from a lentiviral vector. Cells infected with an empty lentivirus serve as a control. Whole cell extracts were immunoprecipitated with antibodies against RBK860me1 or control IgG antibodies followed by immunoblot analysis with RB. Inputs for RB and actin are shown. The efficiency of the knockdown was measured by quantitative RT-PCR. One representative quantification of the knockdown is indicated.
RB Methylation at Lys-860 Is Increased in Response to Anti-proliferative Signals
Because RB normally inhibits the G1/S transition of the cell cycle (2), we hypothesized that RB methylation at Lys-860 may be regulated during G1 arrest. To test this idea, we performed a cell cycle re-entry experiment. Cell cycle re-entry from quiescence upon serum stimulation was confirmed by increased RB phosphorylation, as judged by a shift in migration, increased levels of the E2F target p107, and cell cycle profiles (Fig. 4A). Interestingly, the amount of RB immunoprecipitated with the antibody to RBK860me1 was higher in quiescent cells compared with cells re-entering the cell cycle while RB levels remained constant (Fig. 4A), indicating that RB methylation at Lys-860 is regulated during cell cycle re-entry.
FIGURE 4.
Regulation of RB methylation at lysine 860 in vivo. A, methylation of RB accumulates in G0. Quiescent T98G cells were stimulated with 20% serum and RB methylation was measured by immunoprecipitation of whole cell extracts with RBK860me1 antibody and immunoblot analysis with RB antibody. Note that RB becomes hyperphosphorylated as cells re-enter the cell cycle, and that the levels of the RB/E2F target p107 increase as cells progress into S phase. Actin serves as a loading control. Propidium iodide FACS profiles (bottom) show cell cycle re-entry. B, methylated RB accumulates as C2C12 myoblasts differentiate. Whole cell extracts from differentiating C2C12 myoblasts were immunoprecipitated with RBK860me1 antibody followed by immunoblot for RB. Input levels of RB are shown along with actin as a loading control (top). A replicate of this experiment is shown with RB loading equalized from day 0 to day 2 (middle). Photographs at days 0 and 2 show the differentiation into myotubes (bottom). C, RB methylation accumulates with DNA damage. Whole cell extracts from NIH3T3 cells treated with etoposide for the indicated time periods were immunoprecipitated with RBK860me1 followed by immunoblot analysis. Immunoblotting input with the phospho-H2AX epitope (γH2AX) serves as a positive control for the accumulation of DNA damage. Input levels of RB are shown along with actin as a loading control.
We next tested whether RB methylation at Lys-860 is affected when cells exit the cell cycle during differentiation. To this end, we grew mouse C2C12 muscle cells under differentiation conditions. Differentiation was monitored by visual inspection of the culture and the appearance of fused myotubes and hypophosphorylation of RB (Fig. 4B). Under these conditions, RB levels and acetylated RB levels have been shown to increase transiently as cells enter the differentiation program (8). Similarly, we found a concomitant increase in total RB levels and methylated RB levels as C2C12 cells differentiated (Fig. 4B). This increase was not observed when RB levels were normalized (Fig. 4B), indicating that there is an increase in both the total pool of RB and methylated RB in C2C12 cells as they exit the cell cycle and undergo the first stages of muscle differentiation.
Finally, we tested whether RB methylation by SMYD2 was affected by DNA damage, as was shown for p53 (19). We treated NIH 3T3 cells with the DNA damaging agent etoposide and verified the induction of DNA damage by immunoblot analysis with antibodies to the phosphorylated form of histone H2AX (γH2AX) (Fig. 4C). We found that the amount of RB that could be immunoprecipitated with the antibody to RBK860me1 increased after etoposide treatment while RB levels remained constant, indicating that RB methylation at Lys-860 is induced by DNA damage (Fig. 4C). Thus, RB methylation at Lys-860 is dynamic and regulated during several cellular processes, suggesting that it may play a functional role in mammalian cells.
RB Methylation at Lys-860 Regulates RB Binding to the Transcriptional Repressor L3MBTL1
The observation that the amount of methylated RB increases under cytostatic conditions, and the fact that co-expression of SMYD2 and RB in RB mutant Saos-2 cells causes further repression of E2F target genes when compared with RB alone (supplemental Fig. S2), raised the question of how methylation of RB might affect its function. One possibility was that RB methylation may change its binding to specific cellular partners. Whereas E2F1 binds to the C terminus of RB (6), Lys-860 contributes a weak interaction with E2F1 (44), and we found no reproducible differences in the RB/E2F1 interaction with or without SMYD2, in cells and in vitro (Fig. 3C and supplemental Fig. S3 and data not shown). This observation did not exclude that the binding to other partners may be affected by the methylation event.
L3MBTL1 is a histone methyl-binding protein that can condense chromatin and repress gene expression, including the expression of RB targets (45, 46). The MBT domain of L3MBTL1 has been found to bind methyl groups, with a preference for mono- and dimethylated lysines (45, 47, 48). Whereas there is no published evidence that RB and L3MBTL1 directly interact, we hypothesized that RB methylation may serve as a signal to increase a potential interaction between RB and L3MBTL1. We found that a recombinant 3xMBT domain from L3MBTL1 bound more efficiently to RB peptides mono- and dimethylated at Lys-860 than it did to non- and trimethylated peptides (Fig. 5A). In addition, mutations in amino acids of L3MBTL1 predicted to alter its ability to recognize methylated peptides (46, 49, 50) decreased its ability to bind to monomethylated RB at Lys-860 (Fig. 5B). Isothermal titration calorimetry assays further confirmed the specificity of L3MBTL1 for mono- and dimethylated RB K860 (Fig. 5, C and D), with an affinity slightly higher than what was described with mono- and dimethylated histone peptides (46).
FIGURE 5.
RB methylation at Lys-860 increases its interaction with the MBT domains of L3MBTL1 in vitro. A, 3xMBT from L3MBTL1 preferentially binds mono- and dimethyl K860 RB fragments. Pull-down assays of biotinylated RB and H4 peptides containing varying degrees of methylation with GST-tagged 3xMBT from L3MBTL1. B, mutations in key residues of the methyl-binding pocket of 3xMBT L3MBTL1 reduce binding to methylated RB fragments. Pull-down assays of biotinylated RBK860me0 and RBK860me1 peptides with 3xMBT L3MBTL1 mutants. Input levels of the RB fragment are shown to the left of each blot. C and D, calorimetry assays confirm and quantify binding of methylated RB to 3xMBT L3MBTL1. Isothermal titration calorimetry assays were performed to measure the affinity of the interaction between RB and the 3xMBT domain of L3MBTL1. Only the interaction with the monomethylated RB peptide is shown in C. All the data are quantified in D; no Kd could be determined for K860me0 and K860me3 because of the low affinity of the interaction.
To determine if methylated RB associates with L3MBTL1 in cells, we first determined if we could detect methylated RB in chromatin where L3MBTL1 has been shown to function as a repressor (45). Indeed, the analysis of different cellular fractions from 293T cells transfected with HA-RB and MYC-SMYD2 revealed monomethyl RB at Lys-860 in the cytoplasmic (S2), nuclear (S3), and most importantly, chromatin (Chr) fractions (Fig. 6A). Next we investigated the binding of methylated RB to L3MBTL1 in cells through co-immunoprecipitation experiments. We found that FLAG-L3MBTL1 co-immunoprecipitated more HA-RB in the presence of MYC-SMYD2 (Fig. 6B). Furthermore, similar to the observations with recombinant peptides (Fig. 5), the interaction between RB and L3MBTL1 was dependent on an intact 3xMBT domain (Fig. 6C). Finally, this interaction was decreased in cells expressing MYC-SMYD2 and HA-RB(K860R) compared with cells expressing wild-type HA-RB, indicating that methylation at lysine 860 was required for the optimal binding of L3MBTL1 to RB (Fig. 6D).
FIGURE 6.
RB methylation at Lys-860 increases its interaction with the MBT domains of L3MBTL1 in vivo. A, monomethyl Lys-860 RB can be localized to chromatin. 293T cells were fractionated by detergent lysis to generate a cytoplasmic S2 fraction, a nuclear S3 fraction, and a chromatin (Chr) fraction. Fractions were then analyzed by immunoblot with antibodies against RB and RBK860me1. Tubulin and H3K4tri-me were used to assess the purity of the fractionation. B, overexpression of SMYD2 increases binding of RB to L3MBTL1. 293T cells were transfected with the indicated expression plasmids. Whole cell extracts were immunoprecipitated with FLAG resin and analyzed by immunoblot with antibodies against RB, RBK860me1, and L3MBTL1. Inputs for FLAG-L3MBTL1, MYC-SMYD2, and RB are shown. C, D355N mutation in the methyl-binding pocket of L3MBTL1 reduces the affinity of L3MBTL1 for RB. 293T cells were transfected with the indicated expression plasmids. IP was performed as in B and analyzed by immunoblot with antibodies against RB, RBK860me1, and L3MBTL1. Inputs for FLAG-L3MBTL1, FLAG-L3MBTL1(D355N), RB, and MYC-SMYD2 are shown. D, RB(K860R) mutation reduces the affinity of RB for L3MBTL1. 293T cells were transfected with the indicated expression plasmids. Immunoprecipitation was performed as in B and C and analyzed by immunoblot with antibodies against HA, RBK860me1, and L3MBTL1. Inputs for FLAG-L3MBTL1, HA-RB, HA-860, and MYC-SMYD2 are shown. Tubulin serves as a loading control.
DISCUSSION
Regulation of RB activity in cells is thought to be largely, although not entirely (51), due to post-translational modifications. Here we describe a previously unidentified post-translational modification of RB, methylation of Lys-860 by SMYD2. We also show that RB K860 methylation facilitates a direct interaction between RB and the methyl-binding protein L3MBTL1. The identification of this modification provides further evidence for the existence of an “RB code” whereby specific patterns of modifications affect one another and dictate the myriad of RB cellular functions.
Our data indicate that SMYD2 methylates RB at Lys-860 in vitro and in cells but does not exclude the possibility that SMYD2 may methylate RB at different sites under specific conditions. It is also possible that SMYD2, or another methyltransferase, may generate K860me2 and/or K860me3 residues. Furthermore, other methyltransferase activities could control the functions of RB in distinct cellular contexts, an idea supported by our finding that SET9 can also methylate RB in vitro, albeit at a different site than SMYD2. Future experiments will continue to examine the spectrum of RB methylation in relation to cellular context and other post-translational modifications.
Early studies of SMYD2 described its ability to inhibit cell cycle progression (35). The identification of RB as a SMYD2 substrate, and the accumulation of methylated RB molecules during conditions where RB represses cell cycle genes, may provide a mechanism for this effect, perhaps in conjunction with the effects of SMYD2 on p53 (19). Nevertheless, additional experiments will be required to test this model. In particular, the genetic interactions between SMYD2, RB, and p53 during embryonic development at a time when many cells exit the cell cycle remain to be tested. In addition, a clear role for SMYD2 in tumorigenesis has yet to be identified.
L3MBTL1 is a mono- and dimethyl histone-binding protein that has the ability to condense chromatin and repress transcription (52). A possible biochemical link between L3MBTL1 and RB has been suggested by the fact that both proteins have been found in the same large complexes in flies and human cells (45, 53). In addition, L3MBTL1 has the ability to bind to and repress the transcription of some E2F target genes (45). However, these studies did not determine if RB and L3MBTL1 directly interacted. Our studies indicate that RB can bind directly to the 3xMBT domain of L3MBTL1. Moreover, this direct interaction is facilitated by RB methylation at Lys-860. These associations between RB and L3MBTL1 suggest a model whereby RB methylation by SMYD2 may serve to recruit L3MBTL1 to the promoters of specific RB/E2F target genes to repress their transcription. Similar observations were recently made with p53, which indicate that the L3MBTL1/p53 interaction serves to control the repression of p53 target genes in the absence of cellular stress (55). Because L3MBTL1 has been shown to homodimerize (54), future studies will determine if L3MBTL1 bound to methylated RB (and p53) also has the potential to bind methylated histones on the same monomer or as a dimer. This would provide a direct physical link between the “RB code,” the “histone code,” and structurally repressed chromatin. Because L3MBTL1 is the homolog of the Drosophila tumor suppressor protein dL(3)MBT (52), future experiments should also investigate the tumor suppressor function of L3MBTL1 in mammals.
Acknowledgments
We thank Dr. Jim Schilling and Ken Lau (Stanford University) for help with the initial mass spectrometry analysis. We would like to also give our thanks to Drs. Philipp Tucker, Shelly Berger, and Danny Reinberg for reagents and to Drs. Steve Artandi, Joe Lipsick, and Anne Brunet for useful discussions and comments on the manuscript. We thank Chenwei Lin for help with the microarray analysis.
The work was supported, in whole or in part, by National Institutes of Health Grants P41 RR011823 (to J. R. Y.), NCI T32 CA009523 (to A. A.), CIRM RB1-01385 (to J. S. and O. G.), and NIA F30 AG034741 (to L. A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Experimental Methods.
- RB
- retinoblastoma protein
- SMYD2
- SET-, and MYND-containing protein 2
- MBT
- malignant brain tumor
- CDK
- cyclin-dependent kinase
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1.Knudsen E. S., Knudsen K. E. (2008) Nat. Rev. Cancer 8, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkhart D. L., Sage J. (2008) Nat. Rev. Cancer 8, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherr C. J., McCormick F. (2002) Cancer Cell 2, 103–112 [DOI] [PubMed] [Google Scholar]
- 4.Weinberg R. A. (1995) Cell 81, 323–330 [DOI] [PubMed] [Google Scholar]
- 5.Inoue Y., Kitagawa M., Taya Y. (2007) EMBO J. 26, 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markham D., Munro S., Soloway J., O'Connor D. P., La Thangue N. B. (2006) EMBO Rep. 7, 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leduc C., Claverie P., Eymin B., Col E., Khochbin S., Brambilla E., Gazzeri S. (2006) Oncogene 25, 4147–4154 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen D. X., Baglia L. A., Huang S. M., Baker C. M., McCance D. J. (2004) EMBO J. 23, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan H. M., Krstic-Demonacos M., Smith L., Demonacos C., La Thangue N. B. (2001) Nat. Cell Biol. 3, 667–674 [DOI] [PubMed] [Google Scholar]
- 10.Ledl A., Schmidt D., Müller S. (2005) Oncogene 24, 3810–3818 [DOI] [PubMed] [Google Scholar]
- 11.Ying H., Xiao Z. X. (2006) Cell Cycle 5, 506–508 [DOI] [PubMed] [Google Scholar]
- 12.Munakata T., Liang Y., Kim S., McGivern D. R., Huibregtse J., Nomoto A., Lemon S. M. (2007) PLoS Pathog. 3, 1335–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalejta R. F., Shenk T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3263–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang K., Lin W., Latham J. A., Riefler G. M., Schumacher J. M., Chan C., Tatchell K., Hawke D. H., Kobayashi R., Dent S. Y. (2005) Cell 122, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Reinberg D. (2001) Genes Dev. 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 16.Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 17.Shukla A., Chaurasia P., Bhaumik S. R. (2009) Cell Mol. Life Sci. 66, 1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., Reinberg D. (2004) Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 19.Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., Dorsey J. A., Kubicek S., Opravil S., Jenuwein T., Berger S. L. (2006) Nature 444, 629–632 [DOI] [PubMed] [Google Scholar]
- 20.Ivanov G. S., Ivanova T., Kurash J., Ivanov A., Chuikov S., Gizatullin F., Herrera-Medina E. M., Rauscher F., 3rd, Reinberg D., Barlev N. A. (2007) Mol. Cell. Biol. 27, 6756–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estève P. O., Chin H. G., Benner J., Feehery G. R., Samaranayake M., Horwitz G. A., Jacobsen S. E., Pradhan S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X. D., Huang B., Li M., Lamb A., Kelleher N. L., Chen L. F. (2009) EMBO J. 28, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ea C. K., Baltimore D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18972–18977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mowen K. A., Tang J., Zhu W., Schurter B. T., Shuai K., Herschman H. R., David M. (2001) Cell 104, 731–741 [DOI] [PubMed] [Google Scholar]
- 25.Huq M. D., Tsai N. P., Khan S. A., Wei L. N. (2007) Mol. Cell Proteomics 6, 677–688 [DOI] [PubMed] [Google Scholar]
- 26.Subramanian K., Jia D., Kapoor-Vazirani P., Powell D. R., Collins R. E., Sharma D., Peng J., Cheng X., Vertino P. M. (2008) Mol. Cell 30, 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J., Dorsey J., Chuikov S., Pérez-Burgos L., Zhang X., Jenuwein T., Reinberg D., Berger S. L. (2010) J. Biol. Chem. 285, 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Fischle W., Cheung W., Jacobs S., Khorasanizadeh S., Allis C. D. (2004) Novartis Found. Symp. 259, 3–17; discussion 17-21, 163–169 [PubMed] [Google Scholar]
- 29.Ferreira R., Naguibneva I., Pritchard L. L., Ait-Si-Ali S., Harel-Bellan A. (2001) Oncogene 20, 3128–3133 [DOI] [PubMed] [Google Scholar]
- 30.Yoshimoto T., Boehm M., Olive M., Crook M. F., San H., Langenickel T., Nabel E. G. (2006) Exp. Cell Res. 312, 2040–2053 [DOI] [PubMed] [Google Scholar]
- 31.Vandel L., Nicolas E., Vaute O., Ferreira R., Ait-Si-Ali S., Trouche D. (2001) Mol. Cell. Biol. 21, 6484–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steele-Perkins G., Fang W., Yang X. H., Van Gele M., Carling T., Gu J., Buyse I. M., Fletcher J. A., Liu J., Bronson R., Chadwick R. B., de la Chapelle A., Zhang X., Speleman F., Huang S. (2001) Genes Dev. 15, 2250–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen S. J., Schneider R., Bauer U. M., Bannister A. J., Morrison A., O'Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R. E., Kouzarides T. (2001) Nature 412, 561–565 [DOI] [PubMed] [Google Scholar]
- 34.Munro S., Khaire N., Inche A., Carr S., La Thangue N. B. (2010) Oncogene 29, 2357–2367 [DOI] [PubMed] [Google Scholar]
- 35.Brown M. A., Sims R. J., 3rd, Gottlieb P. D., Tucker P. W. (2006) Mol. Cancer 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho V. M., Schaffer B. E., Karnezis A. N., Park K. S., Sage J. (2009) Oncogene 28, 1393–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald W. H., Tabb D. L., Sadygov R. G., MacCoss M. J., Venable J., Graumann J., Johnson J. R., Cociorva D., Yates J. R., 3rd. (2004) Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 39.Carvalho P. C., Xu T., Han X., Cociorva D., Barbosa V. C., Yates J. R., 3rd (2009) Bioinformatics 25, 2734–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 41.Tabb D. L., McDonald W. H., Yates J. R., 3rd. (2002) J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sage J., Miller A. L., Pérez-Mancera P. A., Wysocki J. M., Jacks T. (2003) Nature 424, 223–228 [DOI] [PubMed] [Google Scholar]
- 43.Burke J. R., Deshong A. J., Pelton J. G., Rubin S. M. (2010) J. Biol. Chem. 285, 16286–16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubin S. M., Gall A. L., Zheng N., Pavletich N. P. (2005) Cell 123, 1093–1106 [DOI] [PubMed] [Google Scholar]
- 45.Trojer P., Li G., Sims R. J., 3rd, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S. D., Wang Y. H., Reinberg D. (2007) Cell 129, 915–928 [DOI] [PubMed] [Google Scholar]
- 46.Min J., Allali-Hassani A., Nady N., Qi C., Ouyang H., Liu Y., MacKenzie F., Vedadi M., Arrowsmith C. H. (2007) Nat. Struct. Mol. Biol. 14, 1229–1230 [DOI] [PubMed] [Google Scholar]
- 47.Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalakonda N., Fischle W., Boccuni P., Gurvich N., Hoya-Arias R., Zhao X., Miyata Y., Macgrogan D., Zhang J., Sims J. K., Rice J. C., Nimer S. D. (2008) Oncogene 27, 4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Fischle W., Wang W., Duncan E. M., Liang L., Murakami-Ishibe S., Allis C. D., Patel D. J. (2007) Mol. Cell 28, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eryilmaz J., Pan P., Amaya M. F., Allali-Hassani A., Dong A., Adams-Cioaba M. A., Mackenzie F., Vedadi M., Min J. (2009) PLoS ONE 4, e7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burkhart D. L., Ngai L. K., Roake C. M., Viatour P., Thangavel C., Ho V. M., Knudsen E. S., Sage J. (2010) Mol. Cell. Biol. 30, 1729–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonasio R., Lecona E., Reinberg D. (2010) Semin. Cell Dev. Biol. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis P. W., Beall E. L., Fleischer T. C., Georlette D., Link A. J., Botchan M. R. (2004) Genes Dev. 18, 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boccuni P., MacGrogan D., Scandura J. M., Nimer S. D. (2003) J. Biol. Chem. 278, 15412–15420 [DOI] [PubMed] [Google Scholar]
- 55.West L. E., Roy S., Lachmi-Weiner K., Hayashi R., Shi X., Appella E., Kutatelodze T. G., Gozani O. (2010) J. Biol. Chem. 285, 37725–37732 [DOI] [PMC free article] [PubMed] [Google Scholar]