Abstract
p53 is a thermodynamically unstable protein containing a conformationally flexible multiprotein docking site within the DNA-binding domain. A combinatorial peptide chip used to identify the novel kinase consensus site RXSΦ(K/D) led to the discovery of a homologous phosphorylation site in the S10 β-strand of p53 at Ser269. Overlapping peptide libraries confirmed that Ser269 was a phosphoacceptor site in vitro, and immunochemical approaches evaluated whether p53 is phosphorylated in vivo at Ser269. Mutation or phosphorylation of p53 at Ser269 attenuates binding of the p53-specific monoclonal antibody DO-12, identifying an assay for measuring Ser269 phosphorylation of p53 in vivo. The mAb DO-12 epitope of p53 is masked via phosphorylation in a range of human tumor cells with WT p53 status, as defined by increased mAb DO-12 binding to endogenous p53 after phosphatase treatment. Phospho-Ser269-specific monoclonal antibodies were generated and used to demonstrate that p53 phosphorylation is induced at Ser269 after irradiation with kinetics similar to those of p53 protein induction. Phosphomimetic mutation at Ser269 inactivated the transcription activation function and clonogenic suppressor activity of p53. These data suggest that the dynamic equilibrium between native and unfolded states of WT p53 can be modulated by phosphorylation of the conformationally flexible multiprotein binding site in the p53 DNA-binding domain.
Keywords: DNA-binding Protein, E3 Ubiquitin Ligase, Epitope Mapping, Phosphorylation Enzymes, Transcription, Cancer, MDM2, p53, Phosphorylation
Introduction
Exposure to a wide variety of genotoxic and metabolic stresses leads to the activation of the p53 tumor suppressor protein, a sequence-specific DNA-binding protein and stress-activated transcription factor that controls and coordinates the expression of a battery of genes involved in transient growth arrest, DNA and cellular repair, and/or apoptosis (1). The function of the p53 protein is regulated post-translationally by enzymes that catalyze p53 ubiquitination, acetylation, and phosphorylation. In the absence of stress, the specific activity of p53 is suppressed by the action of E3 ubiquitin ligases like MDM2 that promote proteasomal degradation of the protein (2). In response to stress signals like DNA damage, this degradation program is suppressed, and sets of protein kinase pathways, notably ATM, trigger activation of the protein.
The mechanisms of p53 activation by phosphorylation at the most evolutionarily conserved phosphoacceptor sites has been assigned both biochemically and genetically. The phosphoacceptor sites in the p53 transactivation domain are the most highly conserved between vertebrates and invertebrates: phosphorylation of p53 at Ser20 stabilizes the interaction with acetyltransferases like p300 and in turn stimulates DNA-dependent acetylation (3, 4); phosphorylation at Ser15 can stimulate CBP binding and p53 acetylation (5); and phosphorylation at Thr18 can both stabilize p300 binding and reduce MDM2 binding (4, 6). The stabilization of p300 relates to the conversion of intrinsically unstructured activation motifs in p53 to a more helical character with a higher affinity for p300 (7). Mice with mutations at the equivalent Ser20 residue develop spontaneous B-cell lymphomas (8), and Ser15 mutant transgenes develop spontaneous late onset lymphoma (9). The Ser392 phosphoacceptor site in the C-terminal domain of p53 is the second most highly conserved class of phosphoacceptor site but only within vertebrates. Increased phosphorylation of p53 at the Ser392 site occurs in vivo after UV and ionizing radiation (10, 11), and this stimulates the sequence-specific DNA-binding function of p53 (12). Phosphorylation of p53 at Ser392 enhances the stability of the p53 tetramerization domain (13), and phosphomimetic mutation at codon 392 results in enhanced thermostability of the p53 tetramer (14), providing biophysical evidence for conformational changes of this phosphorylation on p53. Genetic studies in mice have shown that mutation of the CK2 site results in enhanced skin or bladder cancer in response to UV damage or carcinogen exposure (15, 16), and mouse embryo fibroblasts from such transgenic mice also have an attenuated p53 transcriptome (17). Further, the enhanced phosphorylation of p53 in the basal/stem cells of UV-irradiated human skin (11) is attributable to the transcriptional activation of ATM by ΔNp63 (18). Although these biochemical and genetic studies provide a paradigm for how phosphorylation can regulate p53 protein function at the most highly conserved phosphorylation sites, the effects of many other covalent modifications on p53, including over 12 other phosphorylation sites and methylation sites, are only just now being defined at the biochemical and genetic level.
There is growing evidence that protein-protein interactions, although driven by globular domains, are regulated by intrinsically disordered motifs or linear peptide docking motifs (19). These linear motifs might acquire structure upon binding to target protein or may themselves induce a specific structure by stabilizing the target protein in a specific conformation. p53 protein is a case in point; it is a thermodynamically unstable protein that has a large set of peptide-docking sites within its structural or unstructured domains that drive key protein-protein interactions that regulate its function, including ubiquitination and phosphorylation (20). The E3 ubiquitin ligase MDM2 is a prime example of this because at least two distinct linear peptide domain interaction sites are required for MDM2 to catalyze p53 ubiquitination. The primary binding site of MDM2 for p53 occurs at a peptide motif (FXXXWXXL) in the intrinsically unstructured N-terminal domain of p53 through an interaction with the N-terminal hydrophobic pocket of MDM2 (21). The second MDM2 interaction site occurs through an interaction between the MDM2 acidic domain and a motif (SXXLXXGXXXF) in the conformationally flexible DNA-binding domain in p53 within the S9-S10 loop/S10 β-sheet (22). This latter site forms the ubiquitination signal for E2-E3 (MDM2)-catalyzed ubiquitin transfer to the p53 tetramer (23). This site is notable in that it is also a site of pronounced conformational flexibility that reveals and “opening” or destabilization of the p53 DNA-binding domain (24). This conformationally-flexible site is also notable in that it forms a docking site for the distinct class of protein kinases that phosphorylate p53 in its transactivation domain (25, 26), indicating that this region forms a multiprotein interaction site.
In order to determine whether p53 is directly phosphorylated at this multiprotein docking site, thus providing another layer of post-translational regulation, we took advantage of a tool involving kinase substrate profiling to define the linear interaction motifs that direct enzyme specificity and substrate utilization. Here we used chip peptide array technology to expand substrate utilization of the calmodulin kinase family members to define potential physiological phosphoacceptor consensus sites using peptides from naturally occurring proteins. Indeed, PepChip technology has been used to successfully characterize the complex changes that occur within the epithelial esophageal kinome during the early transitional stages of carcinogenesis (27) and map the cellular phosphoproteome (28). After delineation of a novel consensus phosphoacceptor site coupled to homology searches for similar motifs in the p53 DNA-binding domain, we identified a novel phosphorylation site in the S10 β-sheet region of p53 at Ser269. This region is notable in being (i) a site of conformational flexibility in mutant gain-of-function p53 (24); (ii) the ubiquitin signal for MDM2-mediated ubiquitination of p53 (23); and (iii) a docking site for a range of protein kinases that phosphorylate the transactivation domain of p53 (25). A range of approaches were used to demonstrate that Ser269 phosphorylation can occur on endogenous WT p53 in cells and that phosphomimetic mutation at codon 269 results in the production of inactive WT p53. An accompanying paper describes the biophysical basis for p53 inactivation by phosphorylation of p53 at Ser269 and involves primarily increases in the thermoinstability of the core DNA-binding domain of p53. These data highlight the existence of a novel kinase pathway that can regulate the dynamic range of conformations in WT p53 and that can produce a mutant-like conformation on WT p53.
MATERIALS AND METHODS
Reagents and Plasmids
N-terminally tagged biotinylated peptides with an SGSG spacer were obtained from Mimotopes (Carlton, Australia). Anti-p53 antibodies were DO-1, DO-12, PAb240, PAb1620, CM-1, Ab-1 (anti-p21, Cell Signaling), and 2A10 (anti-Mdm2). pcDNA 3.1 p53 and pCMV-Mdm2 plasmids were described (23). The p53 gene was cloned into the pExpr vector (IBA Systems), generating a construct with an N-terminal streptavidin tag. Single amino acid mutations were introduced into wild type p53 at Ser269 according to the QuikChange site-directed mutagenesis kit (Stratagene) using pcDNA 3.1 p53, pExpr p53, and pRSET p53 DNA core domain as templates, and the following oligonucleotides were used for mutagenesis (underlined): p53 S269A, 5′-g gga cgg aac gcc ttt gag gtg cg-3′ (forward primer) and 5′-cg cac ctc aaa ggc gtt ccg tcc c-3′ (reverse primer); p53 S269D, 5′-ctg gga cgg aac gac ttt gag gtg cg-3′ (forward primer) and 5′-cg cac ctc aaa gtc gtt ccg tcc cag-3′ (reverse primer).
Kinase Assays
PepChip kinase slides were obtained from Mimotopes Pty. Ltd. Protein kinases were mixed with 10 μm ATP and 300 μCi/ml [γ-33P]ATP (3000 Ci/mmol) in 50 mm Hepes, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol, 0.8 mm EDTA, 5% glycerol, and 0.01% Brij-35, in the presence or absence of 75 μm Box V peptide (30). The reaction mixtures were spotted onto the PepChip slide and covered before they were transferred into a humidified chamber and incubated at 30 °C for 3 h. The PepChip slides were washed once with phosphate-buffered saline containing 1% Triton X-100 and twice with 2 m NaCl containing 1% Triton X-100. [γ-33P]ATP incorporation was visualized using a PhosphorImager (Storm 840, Amersham Biosciences). Peptide sequence information pertaining to the individual spots was obtained by overlaying the image with the reference grid provided by Pepscan Systems B.V. (Lelystad, Netherlands). Peptide kinase reactions were carried out as described previously (30), using 150 ng of purified recombinant protein kinase and 1 μg of peptide substrate.
Peptide ELISA
Biotinylated unphosphorylated and phosphorylated peptides were captured onto ELISA wells coated with streptavidin and blocked with 3% BSA in PBS-Tween as described previously (31).
Cell Culture, Transfection, and Analysis
All tissue culture medium was obtained from Invitrogen and was supplemented with 10% fetal bovine serum. H1299 cells were cultured in RPMI 1640 medium; A375, HEK-293, and MCF-7 cells were cultured in DMEM, and HCT-116 p21 null cells were grown in McCoy's medium. Cells were harvested and lysed using urea lysis buffer as described previously (30) unless otherwise stated. For luciferase assays, H1299 cells were transfected the following day with 30 ng of pCMV Renilla and 70 ng of either p21-luc or Bax-luc and plasmids encoding p53 wild type, p53S269A, or p53S269D. Cells were lysed with lysis buffer according to the dual luciferase assay kit (Promega), and luciferase activity was quantified using a luminometer (Fluoroskan Ascent FL). For cellular fractionation, cells were treated with 10 J cm2 UVC and grown for a further 6 h at 37 °C prior to fractionation using the S-PEK subcellular fractionation kit (Calbiochem). For immunoprecipitation, cell lysates (100 ng) were precleared with protein G beads (Sigma) for 1 h before incubation with 1 μg of DO-1, PAb1620, or PAb240 at 4 °C. For immunoprecipitation of endogenous phospho-Ser269 p53, mouse monoclonal antibodies were preadsorbed to protein G beads overnight before incubation with precleared lysates and washed and eluted as above. For clonogenic survival assays, H1299 cells were transfected with pcDNA p53 constructs, and colonies were selected using Geneticin as described previously (32).
Phosphatase Treatment of Nitrocellulose Membranes
Nitrocellulose membranes were incubated in 50 mm Tris, pH 7.5, 5 mm DTT, 0.1 mm EDTA, 2 mm MnCl2, and 80 units of λ-protein phosphatase (Sigma) for 1 h at 30 °C (33). The degree of epitope unmasking was determined by quantifying the band intensity using Scion Image software.
RESULTS
In Vitro Kinase Screens Identify a Novel Phosphoacceptor Site at Ser269 in the DNA-binding Domain of p53
Because protein-protein interactions are driven in part by intrinsically disordered linear peptide motifs, we previously screened a peptide library with MDM2 protein to acquire novel MDM2 consensus peptide binding motifs. These peptide motifs were scanned for homology to sites in the tumor suppressor protein p53, thus identifying a second MDM2 binding site in the p53 DNA-binding domain (22, 35). Similarly, in this report, we screened selected kinase superfamily members using a kinase-peptide array that contains 192 naturally occurring phosphoacceptor sites to define a broader consensus site for protein kinases. These consensus sites in turn could be scanned for homology to motifs in p53 that might reveal novel p53 phosphorylation sites.
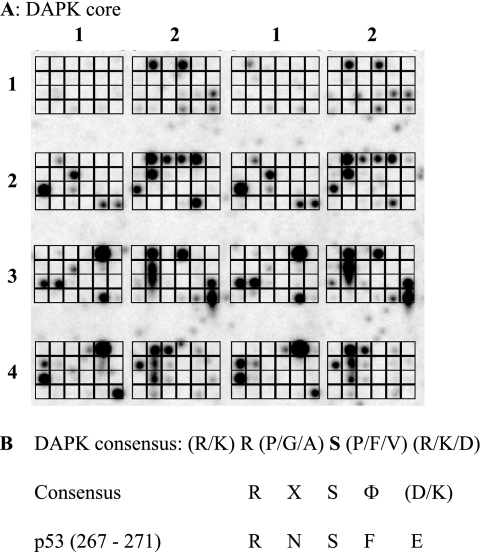
When kinases are screened in this assay, a range of peptide substrates were identified (Fig. 1A shows a representative screen using DAPK-1 (i.e. DAPK core kinase domain)). DAPK targeted 68 of the 192 peptides on the chip (Fig. 1A), and these peptides covered a wide range of cellular proteins (supplemental Table I). Although the library of peptides on the chip does not comprehensively represent the diversity of the human proteome and known substrates of DAPK, such as p21, p53, and myosin light chain, are not represented, there were several peptide hits that were in keeping with the function of DAPK in regulating the cytoskeleton, such as vimetin. The majority of peptides targeted by DAPK core contained a serine residue as the phosphoacceptor (52 of 68 peptides), whereas only a small proportion contained a threonine as the phosphoacceptor residue (9 of the 68 phosphorylated peptides). Several peptides contained more than one potential phosphoacceptor site (e.g. peptide 1, 4, or 8 (LRRSSSVGY, PGGSTPVSS, or KTTASTRKV)), and in such cases, the central residue was taken as the phosphoacceptor. Analysis of the relative abundance of the various amino acid residues within the peptides targeted by DAPK (Table 1) showed a strong selection for peptides with leucine and lysine residues at the −4-position and a strong preference for basic residues, such as lysine and arginine, at the −3- and −2-positions. This observation is in keeping with previous studies that show that DAPK is a basophilic kinase and prefers substrates containing a high proportion of basic residues, such as lysine and arginine, upstream of the phosphoacceptor site (30, 36, 37). The most prominent residues at the −1-position were more non-polar/hydrophobic residues, such as proline, glycine, and alanine. Downstream of the phosphoacceptor serine, there was a high prevalence of basic residues, such as arginine and lysine, as well as proline residues at the +1- and +2-positions; however, there was also an increased abundance of valine and phenylalanine residues. This is also in keeping with previous findings that showed that DAPK has a strong preference for hydrophobic leucine, valine, and phenylalanine residues downstream of the phosphorylated serine (36, 37). There was also a positive preference for alanine residues at the +3-position and acidic residues, such as glutamic and aspartic acid, at +2 and +3. The relative abundance of the various amino acids was tallied to generate a consensus sequence, and the simplest derived consensus for DAPK substrates was determined to be (K/L)(R/K)R(P/G/A)S(P/V/F)(R/K/D)(A/E/P)(K/S/P) (Table 1). The minimal consensus site of RXSF(D/K) obtained (Fig. 1B) was then screened for homology to p53 and identified a site in the conformationally flexible motif, 267RNSFE271.
FIGURE 1.
Pepchip kinase array screen identifies a novel phosphoacceptor site in the DNA-binding domain of p53. A, in vitro kinase consensus site definition using peptide arrays. A Pepchip array (total of 192 peptides) was incubated with 30 ng of DAPK core and [γ-33P]ATP for 3 h at 30 °C, and phosphorylated peptides were detected by storage phosphor screen. The array is arranged in a grid from 1 to 4 in the y axis and 1 and 2 in the x axis (in duplicate). B, consensus sites for protein kinases. Phosphorylated peptides from A (supplemental Table I) were analyzed as indicated in Table 1 to develop the consensus site summarized (RXSF(D/K)), which matches the Ser269 site in the p53 tumor suppressor protein.
TABLE 1.
Quantifying the abundance and positioning of the amino acid residues within the DAPK peptide targets (data derived from supplemental Table I)
The derived consensus sequence is as follows: (K/L)(R/K)R(P/G/A)S(P/F/V)(R/K/D)(A/E/P)(K/S/P).
| −4 | −3 | −2 | −1 | Phosphoacceptor site | +1 | +2 | +3 | +4 | |
|---|---|---|---|---|---|---|---|---|---|
| Ser | 6 | 5 | 2 | 52 | 5 | 6 | 2 | 8 | |
| Thr | 3 | 4 | 1 | 9 | 4 | 2 | 4 | 6 | |
| Tyr | 1 | 7 | 1 | ||||||
| Lys | 24 | 14 | 4 | 3 | 5 | 9 | 6 | 9 | |
| Leu | 23 | 4 | 5 | 5 | 4 | 4 | 2 | 4 | |
| Arg | 2 | 22 | 20 | 2 | 2 | 13 | 3 | 3 | |
| Pro | 9 | 4 | 4 | 17 | 11 | 4 | 7 | 7 | |
| Gly | 5 | 7 | 9 | 4 | 2 | 5 | 3 | ||
| Ala | 1 | 8 | 2 | 4 | 12 | 3 | |||
| Val | 1 | 1 | 2 | 7 | 3 | 4 | 3 | ||
| Ile | 2 | 2 | 3 | 2 | 3 | 4 | |||
| Asp | 1 | 3 | 4 | 5 | 7 | 3 | 1 | ||
| Glu | 2 | 3 | 2 | 4 | 5 | 8 | 5 | ||
| Asn | 2 | 1 | 2 | 1 | |||||
| Gln | 2 | 2 | 5 | 2 | 2 | 4 | 1 | 4 | |
| Phe | 1 | 4 | 7 | 1 | 2 | 4 | |||
| Trp | 1 | 3 | 1 | 1 | 1 | ||||
| Met | 2 | 2 | 1 | 2 | |||||
| His | 1 | 2 |
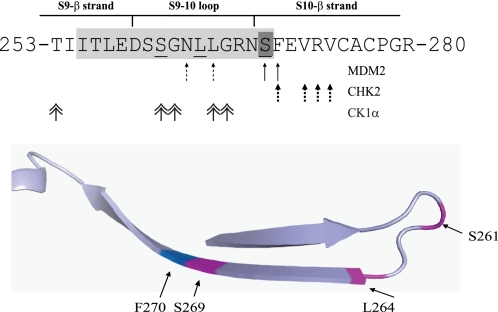
This motif in p53 is notable in that it is one site of three conformational flexible epitopes that are “cryptic” on WT p53 but “exposed” on mutant p53 in human cancers (24). The biophysical basis for this equilibrium is now thought to be a function of the intrinsic thermodynamic instability of the core DNA-binding domain of p53 that is further destabilized by mutation (38). This allostery operating in p53 is now thought to be consistent with the ensemble model of allostery (39), where proteins exist in a range of conformational states under regulatable equilibrium. This motif in p53 also forms the second MDM2 binding site, whose mutation stimulates p53 ubiquitination in cells (22) and which forms the ubiquitination signal in the allosteric MDM2-mediated ubiquitination of p53 (23, 40). This region also forms docking sites for a range of protein kinases that catalyze phosphorylation of p53 in its transactivation domain (reviewed in Figs. 2 and 11). The specific contacts for these many binding proteins like CHK2, MDM2, and CK1 are distinct, as highlighted in Fig. 2. This is consistent with recent data showing that small linear and intrinsically unstructured peptide motifs, such as those in the C terminus of p53, can interact with a target protein through distinct and specific amino acid side chain contacts (41). Thus, this Ser269 site on p53 can be solvent-exposed, is in dynamic equilibrium, and is a potential phosphoacceptor site that could have pleotropic effects on p53 folding and function.
FIGURE 2.
The multiprotein docking sites within the p53 DNA-binding domain. Protein-protein contacts in the conformationally flexible ubiquitination signal from the DNA-binding domain of p53. The key residues involved in MDM2, Chk2, and CK1a binding (arrows) and the contact sites for the DO-12 monoclonal antibody (green) are highlighted. The bottom panel indicates the positioning of key residues in the region known to influence p53 stability.
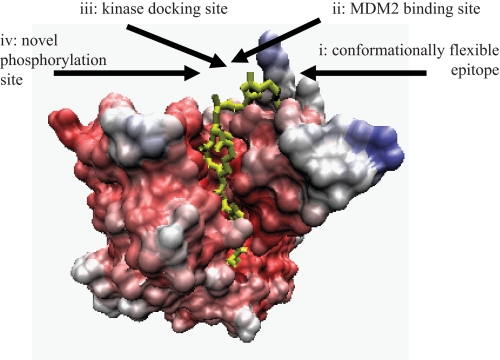
FIGURE 11.
Features of the regulatory motif in the p53 DNA-binding domain. The multiprotein binding site in the p53 core DNA-binding domain (highlighted in yellow) has a conformationally flexible motif (24) (i), MDM2-docking site or ubiquitination signal (22, 40) (ii), kinase docking site (25, 26, 29) (iii), and novel phosphorylation site at Ser269 (this study) (iv).
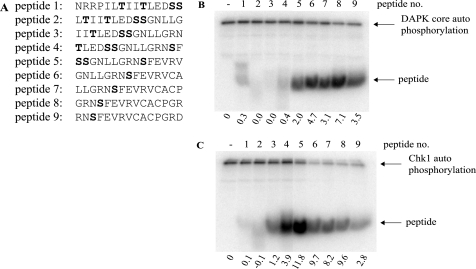
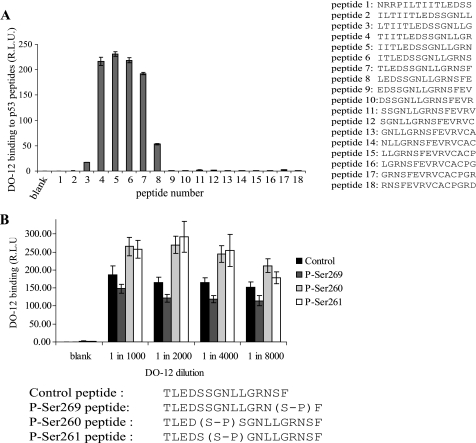
In addition to the potential phosphoacceptor site at Ser269 defined from the homology screen above, the conformationally flexible motif of p53 contains two further, putative phosphoacceptor sites at Ser260 and Ser261 (Fig. 2). Peptides derived from this region of p53 containing Ser269 or Ser260/Ser261 phosphoacceptor sites (Fig. 3A) were used in kinase assays to determine whether these serine residues are bona fide phosphoacceptor sites. Only peptides containing the Ser269 site can function as substrates for DAPK (Fig. 3B, lanes 5–9 versus lanes 1–4), consistent with the kinase-peptide scan in Fig. 1, where a bulky hydrophobic residue flanking the phosphoacceptor site plays a role in phosphorylation. By contrast, another kinase we evaluated (Chk1) is a relatively poor kinase toward peptides containing the Ser269 phosphoacceptor site, whereas it demonstrates better specificity for the peptide with double phosphoacceptor sites at Ser260/261 (Fig. 3C, lanes 2–5 versus lanes 6–9). Together, these data indicate that the flexible linker of p53 has a phosphoacceptor site at Ser269 in vitro that shares homology with members of the calcium-calmodulin kinase superfamily that can target the RXSF(D/E) motif.
FIGURE 3.
The multiprotein docking peptide from the ubiquitin signal in the p53 DNA-binding domain contains an in vitro phosphoacceptor site. A, overlapping peptides corresponding to the conformationally flexible ubiquitination signal (with Ser260/261 phosphoacceptor sites in peptides 1–5 or Ser269 phosphoacceptor site in peptides 5–9) were incubated in kinase reactions containing [γ-32P]ATP and either DAPK core (B) or Chk1 (C) for 30 min at 30 °C. Reaction products were resolved via 20% SDS-PAGE and dried, and phosphopeptides were visualized by a PhosphorImager (highlighted by arrows). The intensity of peptide phosphorylation was quantified and normalized to kinase autophosphorylation and is expressed below each panel.
In Vivo Evidence for p53 Phosphorylation at Ser269 Defined by DO-12 mAb Epitope Masking
We next evaluated whether in vivo evidence could be acquired for Ser269 phosphorylation. If so, this would make biochemical and cellular analysis of Ser269 modification more physiologically relevant. Two approaches were taken to examine Ser269 phosphorylation in cells: (i) epitope masking using a monoclonal antibody whose binding site overlaps with Ser269 and (ii) phosphospecific antibody generation to phospho-Ser269. A notable feature of the conformationally flexible motif containing Ser269 is that it overlaps with the DO-12 monoclonal antibody epitope (Fig. 2) (24); this epitope (amino acids 255–270) is normally constrained in wild type native p53 or when wild type p53 is bound to DNA, but the monoclonal antibody can bind to and immunoprecipitate mutant p53 due to conformational changes that expose the epitope. This change in epitope exposure due to p53 mutations is thought to be due to the changes in the dynamic range in p53 conformational states, according to the ensemble model of allostery.
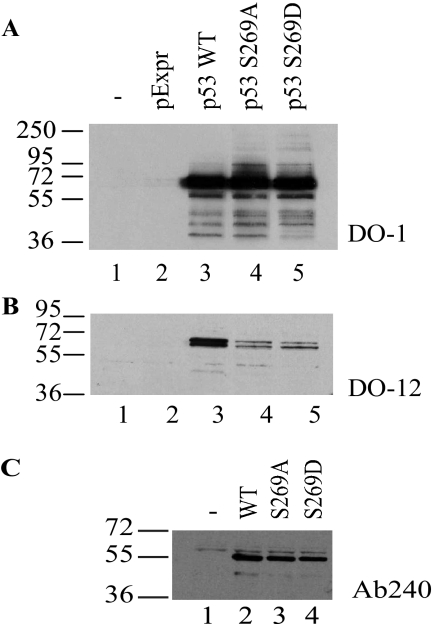
The Ser269 residue is located at the end of the DO-12 epitope (amino acids 255–270) (Fig. 2). Key phosphorylation sites on p53 are often found within the epitopes of several monoclonal antibodies (such as DO-1 (calcium-calmodulin kinase superfamily site at Ser20) (31), PAb421 (PKC site at Ser371/376) (42), ICA-9 (CK2 site, Ser392), or MDM2 (2A10, ATM site at Ser395)) (43), and monoclonal antibody binding to these epitopes is masked by phosphorylation. As such we evaluated whether mutation or substitution of p53 at Ser269 attenuated DO-12 monoclonal antibody binding. If so, this would provide an indirect assay for determining whether phosphorylation occurs at Ser269 on endogenous p53 in vivo. DO-12 mAb binding to peptides derived from this region of p53 confirmed that Ser269 and Phe270 comprise a portion of the DO-12 epitope (Fig. 4A), raising the possibility that Ser269 phosphorylation could affect mAb-DO-12 binding. Phosphopeptides were generated within the DO-12 epitope that contain phosphate at Ser269, Ser261, or Ser260. When DO-12 binding to these substituted peptides was measured, only Ser269 phosphorylation attenuated the binding of the monoclonal antibody (Figs. 4B and 5). By contrast, Ser261 or Ser260 phosphopeptides actually exhibited elevated binding stability toward DO-12 (Fig. 4B). Mutagenesis of codon 269 in full-length p53 to alanine or the phosphomimetic aspartate resulted in masking of the DO-12 epitope (Fig. 5, A and B), indicating that the integrity of Ser269 is required for DO-12 to bind stably. As a control, binding of another conformationally sensitive p53 monoclonal antibody, PAb240 (epitope 209–214) (44), was examined, and PAb240 binding was not inhibited by mutation of codon 269 (Fig. 5C), indicating that mutation and phosphorylation at Ser269 specifically masks the DO-12 epitope.
FIGURE 4.
Attenuation of DO-12 epitope binding by Ser269 phosphorylation. A, DO-12 binding to the p53 DNA-binding domain. The region bound by the DO-12 monoclonal antibody was mapped by ELISA using overlapping peptides derived from the DNA-binding domain of p53 (A, right). B, serine 269 phosphorylation attenuates DO-12 binding. The effect of phosphorylation on the binding affinity of DO-12 to peptides (amino acids 255–270 of human p53) derived from the DNA-binding domain of p53 was defined by ELISA and is shown as a function of DO-12 antibody titration. The peptide sequences and modifications at serine 260, serine 261, or serine 269 are shown below. R.L.U., relative light units. Error bars, S.D.
FIGURE 5.
Mutagenesis of full-length p53 at codon 269 attenuates DO-12 binding. H1299 cells were transfected with pExpr expression vectors encoding p53, p53S269A, or p53S269D mutants, and the proteins were examined for reactivity toward DO-1 (A; total p53), DO-12 (B; epitope 255–270), or PAb240 (C; epitope 209–214) by immunoblotting.
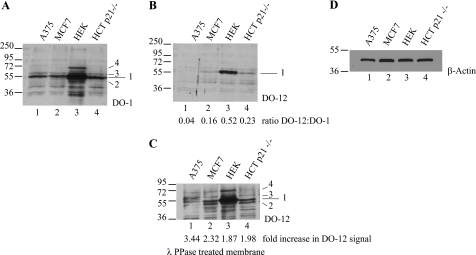
A range of cell lines containing wild type p53 protein (including malignant melanoma (A375), mammary adenocarcinoma (MCF-7), colorectal carcinoma (HCT-116), and human embryonic kidney cells (HEK-293)) were lysed in denaturing urea buffer to preserve endogenous p53 phosphorylation sites, and total p53 protein levels were measured (Fig. 6A, lanes 1–4). Immunoblotting for total p53 protein levels demonstrated that comparable levels of p53 protein were detected in A375, MCF-7, and HCT-116 cells (Fig. 6A, lanes 1, 2, and 4), with significantly greater levels of p53 protein detected in HEK-293 cells (Fig. 6A, lane 3). By contrast, although a significant pool of DO-12-reactive p53 was detected in HEK-293 cells, only faint bands corresponding to p53 protein were detected in MCF-7, A375, and HCT-116 cells (Fig. 6B, lanes 1–4). Because phosphorylation or mutation at Ser269 can attenuate DO-12 antibody binding, we hypothesized that reductions in mAb-DO-12 binding to p53 in these cell lines might be due to phosphorylation masking the epitope. To examine this, nitrocellulose membranes were incubated with λ-protein phosphatase, as described previously (33), to remove any potential phosphate groups from the p53 protein prior to immunoblotting with DO-12. After phosphatase treatment, a significant increase in the intensity of the DO-12-reactive p53 protein pool was observed in HEK-293, MCF-7, and HCT-116 cell extracts (Fig. 6C). The reactivity of the p53 monoclonal antibody DO-1 did not change under these conditions (data not shown). These data suggest that DO-12-sensitive phosphorylation of p53 can be detected in a substantial pool of the total p53 protein (see ratios of DO-12/DO-1 below the lanes in Fig. 6B).
FIGURE 6.
In vivo phosphorylation of endogenous p53 at serine 269; detection by phosphatase unmasking of antibody epitopes. A–C, phosphatase activity exposes the DO-12 epitope on endogenous p53. Lysates from A375, MCF-7, HEK-293, and HCT-116 p21−/− cells were resolved by electrophoresis, and immunoblots were probed with antibody for total p53 (A; DO-1), DO-12-reactive p53 (B), or DO-12-reactive p53 on blots pretreated with λ-phosphatase prior to incubation with DO-12 (C). The various p53-cross-reactive bands are indicated (1–4). Band intensity was quantified by Scion Image software, and the ratio of DO-12 to DO-1 signal and the degree of DO-12 unmasking are indicated below B and C, respectively. Equal sample loading was determined by immunoblotting with β-actin (D).
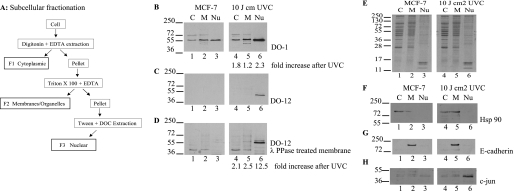
To further analyze in which cellular compartment p53 phosphorylation occurs at serine 269, as defined by DO-12 epitope masking, lysates were isolated from MCF7 cells into cytosol, membrane, and nuclear compartments (Fig. 7A). MCF7 cells were used because this cell line has the most evidence of epitope masking of endogenous p53 protein and is a standard cell line used to study the DNA damage-induced p53 pulse (45). The disadvantage of this methodology compared with the urea denaturing method (Fig. 6) is that it uses non-denaturing buffers that could liberate phosphatases and result in dephosphorylation of p53 protein in the crude lysate. Nevertheless, differentially extracted lysates defined as F1–F3 (Fig. 7A) were acquired from MCF7 cells and immunoblotted for total p53 (Fig. 7B, lanes 1–3), for DO-12-reactive p53 (Fig. 7C), and for DO-12-reactive p53 after phosphatase treatment (Fig. 7D, lanes 1–3). The total p53 protein was distributed into all three compartments shown (Fig. 7B, lanes 1–3) with little evidence of DO-12-reactive p53 protein (Fig. 7C, lanes 1–3). This is consistent with the data acquired using denaturing urea lysis (Fig. 6) and suggests that the buffers used do not permit dephosphorylation of the DO-12 epitope. DO-12-reactive p53 protein could not be detected after phosphatase treatment of blots presumably because of the large dilutions used in the chemical fractionations (Fig. 7A) relative to whole cell denaturing lysis buffer (Fig. 6). As such, we also evaluated whether DNA damage changes DO-12 epitope masking of p53 and in which compartment this occurred. In response to irradiation at doses that activate p53 (data not shown), there is an increase in p53 protein in the nuclear fraction (Fig. 7B, lane 6 versus lanes 1–5), consistent with previous reports that endogenous p53 protein is largely stored in cytosolic fractions and is transported into the nucleus after DNA damage by a dynein-dependent pathway (46). Under conditions where p53 protein is stabilized by irradiation, we could detect DO-12-reactive p53 protein in the nucleus (Fig. 7C, lane 6 versus lanes 1–5), and this was increased by phosphatase treatment of the immunoblots (Fig. 7D, lane 6 versus lanes 1–5). The loading controls for the subcellular fractionation are highlighted in Fig. 7, E–H. These data suggest that p53 can be phosphorylated in the basal state in the DO-12 epitope but that DNA damage induces a mixed pool of DO-12-reactive (i.e. non-phosphorylated) and DO-12-non-reactive p53 protein (i.e. phosphorylated).
FIGURE 7.
Serine 269-phosphorylated p53 has a selective subcellular localization. A, schematic representation of the fractionation methodology. B–D, effects of irradiation on p53 phosphorylation at the DO-12 epitope in MCF7 cells. MCF-7 cells were untreated or irradiated with UVC and grown for a further 6 h before fractionation into cytosolic (C), membrane/organelle (M), and nuclear fractions (Nu). Fractions were resolved by electrophoresis and immunoblotting with DO-1 (B) and DO-12 before (C) and after phosphatase treatment of the membrane (D) to define p53 localization. Band intensity was quantified by Scion Image software, and the -fold increase in antibody signal following UVC irradiation is indicated below B and D, respectively. Equivalent sample loading is shown by Coomassie staining of the various fractions (E), and fractionation of the distinct subcellular fractions was demonstrated by probing for known cytoplasmic (Hsp90), membrane (E-cadherin), and nuclear (c-Jun) marker proteins (F, G, and H, respectively).
In Vivo Evidence for p53 Phosphorylation at Ser269 Defined by the Use of Novel Phosphospecific Ser269 mAbs
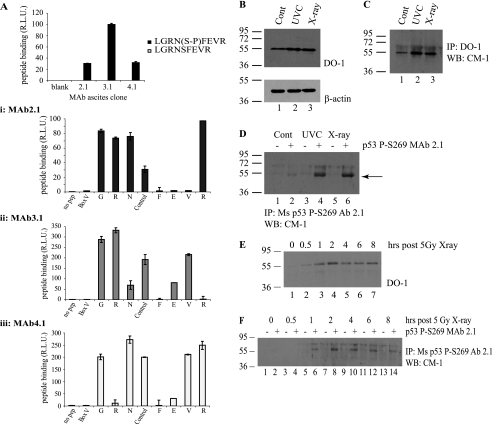
Although phosphatase-sensitive epitope masking of p53 has been a standard method of defining phosphorylation of p53 at some sites, phosphospecific antibodies are the more generally useful tool now used to measure steady-state levels of a post-translational modification. We have previously generated phosphospecific monoclonal antibodies as tools for investigating p53 phosphorylation in vivo (47), including Ser392 and Ser315 (48, 49). A panel of phosphospecific monoclonal antibodies was generated to phospho-Ser269-containing peptides that demonstrated specificity for the peptide from the MDM2 ubiquitination signal (LGRNpSFEVR, where pS represents phosphoserine) (Fig. 8A). An alanine scan phosphopeptide screen indicated that the phospho-Ser269-specific monoclonal antibodies displayed a requirement for key amino acids surrounding the phosphoserine residue (Fig. 8A, i–iii). We used these monoclonal antibodies to determine whether similar evidence could be acquired for radiation-induced Ser269 phosphorylation of p53 protein that correlated with DO-12 epitope masking.
FIGURE 8.
In vivo phosphorylation of p53 at serine 269; detection using phosphospecific antibodies. A, phosphospecific antibody generation to serine 269. Monoclonal antibodies (mAb 2.1, 3.1, and 4.1) were generated toward phospho-Ser269 peptides, and peptide ELISA was used to demonstrate the phosphospecificity of the antibodies. A, i–iii, serine 269 mAb (i, mAb 2.1; ii, mAb 3.1; iii, mAb 4.1) epitopes were mapped via ELISA using an alanine scan mutation of GRNpSFEVR phosphopeptide. B–D, Ser269-phosphospecific mAb 2.1 can immunoprecipitate p53 from MCF7 lysates following DNA damage. MCF7 cells were exposed to 5 grays of x-ray or 10 J cm2 UV irradiation, and induction of p53 protein levels was analyzed 6 h later by immunoblotting with DO-1 (B, showing 10 μg of cellular lysate probed with DO-1 (top) and β-actin (bottom) as a loading control). Total p53 protein and Ser269-phosphorylated p53 were isolated from these lysates by immunoprecipitation with DO-1 (C) or mAb 2.1 (D), respectively, and an increase in Ser269 phosphorylation can be detected (D, lanes 4 and 6). Protein G beads were used as a control for both DO-1 and mAb 2.1 immunoprecipitation and are shown in D in lanes marked with minus signs. This increase in phosphorylation with phosphospecific antibody after irradiation is consistent with the pool of DO-12 non-reactive p53 protein in the nucleus of cells after UV radiation (Fig. 7, C versus D). E–F, time scale of p53 Ser269 phosphorylation following DNA damage. MCF-7 cells were irradiated with 5 grays of x-ray radiation and harvested at the indicated time points. Increases in total p53 (E) or phospho-Ser269 p53 (F) protein levels were determined by immunoblotting cellular lysates with DO-1 (E) or immunoprecipitating phospho-Ser269 p53 from cellular lysates using protein G beads without (−) or with mAb 2.1 (+), followed by immunoblotting with polyclonal CM-1 (F). R.L.U., relative light units; IP, immunoprecipitation; IB, immunoblot.
When cells were exposed to UV or x-ray radiation, the standard induction of p53 protein could be confirmed (Fig. 8B). Immunoprecipitation of p53 with a monoclonal antibody and direct blotting with a polyclonal antibody could confirm that the immunoprecipitation assay could be used to measure increases in p53 protein levels (Fig. 8C). When the monoclonal antibody 2.1 was used under the same conditions, an increase in Ser269-phosphorylated p53 protein could be detected whether using x-ray or UV irradiation (Fig. 8D). A time course measuring increases in p53 protein levels (Fig. 8E) or phospho-Ser269 p53 protein levels (Fig. 8F) indicated that the kinetics of induction of p53 phosphorylation at Ser269 was similar to that of total p53 protein. Thus, using either a mAb epitope masking assay or direct phospho-Ser269-specific mAb binding assay, we present evidence that phosphorylation at Ser269 can occur in vivo and that it increases in response to DNA damage.
An Inactivating Function for Ser269 Phosphorylation of p53
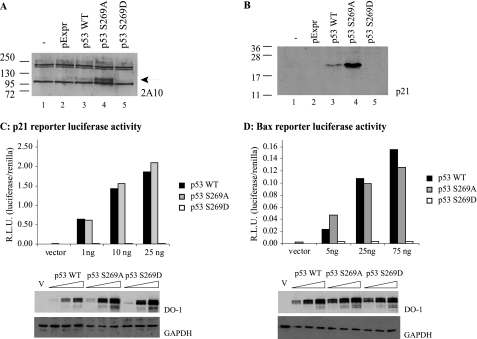
In order to propose a function for Ser269 phosphorylation of p53, mutagenesis was performed to produce a phosphomimetic p53 mutant. However, one of the difficulties with studying p53 protein-protein interactions in the core DNA-binding domain by mutation is that changes in the majority of codons “inactivate” p53, making such an analyses difficult to interpret. Examination of codon 269 mutations in human cancers shows the predominant changes, including Serine mutation to Thr, Ile, Gly, Leu, Asn, Arg, and Cys (see the International Agency for Research on Cancer Web site). Because these mutations are presumably inactivating, we examined whether we could first identify “neutral” mutations at codon 269 that do not inactivate p53. The p53 mutant p53S269A did not lose activity; indeed, the specific activity was elevated relative to wild type p53 as defined by MDM2 protein induction (Fig. 9A, lane 4 versus lane 3) or p21 protein induction (Fig. 9B, lane 4 versus lane 3). The enhanced function of p53S269A was not observed using transient transfection of bax or p21 luc reporters; nevertheless, the S269A mutant was as active as WT p53 (Fig. 9, C and D), indicating that a mutation does not a priori “inactivate” p53 protein function.
FIGURE 9.
The effects of codon 269 mutation on p53 activity in cells; phosphomimetic substitution of serine 269 inactivates p53. The effect of serine 269 mutation on expression of the p53 downstream target genes, Mdm2 and p21, was examined by immunoblotting 20 μg of lysates derived from H1299 cells transfected with pcDNA vector control, wild type p53, p53S269A, or p53S269D mutants using the anti-MDM2 monoclonal antibody 2A10 (A) or the p21 monoclonal antibody Ab-1 (B). C and D, p53 function from reporters containing p53-responsive binding sites. H1299 cells were transfected with the p21 or Bax reporter plasmids alone or with (1–75 ng of) pExpr p53, S269A, or S269D expression vectors. Total DNA was normalized with pExpr vector control plasmid DNA. Twenty-four hours later, cells were lysed, and luciferase activity from p21 (C) or Bax (D) reporters was determined using dual luciferase reporter assays. Data were normalized by expressing luciferase activity (luciferase/Renilla) in relative light units (R.L.U.). The lower panels depict p53 protein levels after transfection by immunoblotting. GAPDH immunoblotting was used as a loading control.
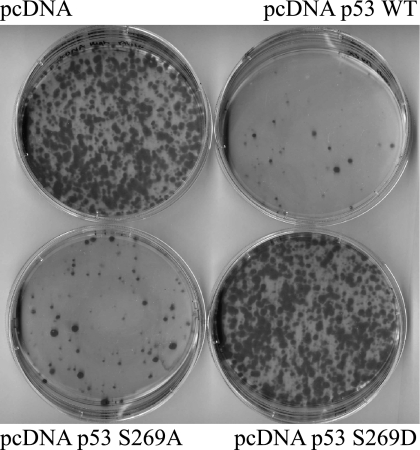
As such, we also examined whether a phosphomimetic mutation at codon 269 would likewise stimulate, inactivate, or demonstrate no change in p53 activity. The mutant p53S269D was unable to induce MDM2 or p21 proteins (Fig. 9, A and B), suggesting that adding a negative charge at codon 269 can inactivate wild type p53. These data suggest that Ser269 phosphorylation will most likely produce a functionally mutant wild type p53 protein and therefore represent an inactivating phosphorylation. The transcriptional activities of p53S269D mutant toward luciferase cassettes containing p21 and Bax promoter elements were also tested. Transfection of the phosphomimetic p53S269D mutant was unable to induce transcription from p21 and Bax promoter elements (Fig. 9, C and D). A clonogenic assay was also used to evaluate whether the phosphomimetic mutation altered WT p53 function. Compared with the wild type or p53S269A, which exhibit growth-suppressing activities (Fig. 10, B and C versus A), the S269D mutation inactivated the growth-suppressive function of p53 (Fig. 10, D versus B). These cellular findings (Figs. 9 and 10) indicate that phosphomimetic mutation of p53 at Ser269 inhibits WT p53 function and together suggest that phosphorylation of p53 in vivo at serine 269 produces a pool of transcriptionally attenuated p53 protein.
FIGURE 10.
Phosphomimetic mutation of Ser269 inhibits p53 growth-suppressive activity. H1299 cells were transfected with 1 μg of pcDNA plasmid containing wild type, S269A, or S269D. Twenty-four hours later, cells were trypsinized and diluted and then plated in medium containing 0.5 mg/ml Geneticin and grown for a further 2 weeks. Colonies were fixed and stained with Giemsa dye.
DISCUSSION
There is growing evidence that a majority of protein-protein interactions in higher eukaryotes are regulated by intrinsically disordered motifs (50). p53 protein has large regions containing intrinsically unstructured motifs, and the core DNA-binding domain is thermodynamically unstable. These regions regulate protein-protein interactions that control p53 activity. One recently identified conformationally flexible motif harbors both the MDM2 ubiquitination signal and protein kinase docking sites in the p53 DNA-binding domain (Fig. 11). This multifunctional protein interaction site thus has a potential to play a fundamental role in p53 activation and its inhibition. In order to determine whether p53 is phosphorylated within this multiprotein docking site in the MDM2 ubiquitination signal, we took advantage of a tool involving kinase substrate profiling to define the linear interaction motifs that direct enzyme specificity and substrate utilization. Here we used chip peptide array technology to expand substrate utilization of the calmodulin kinase family members, including DAPK and Chk2, to define potential physiological phosphoacceptor consensus sites using peptides from naturally occurring proteins. PepChip technology has been used to successfully characterize the complex changes that occur within the epithelial esophageal kinome during the early transitional stages of carcinogenesis (27) and map the cellular phosphoproteome (28). After delineation of a novel consensus phosphoacceptor site coupled to homology searches for similar motifs in the p53 DNA-binding domain, we identified a novel phosphorylation site in the S10 β-sheet region of p53 at Ser269. This region (Fig. 11) is notable in being (i) a site of conformational flexibility in mutant gain-of-function p53 (24); (ii) the ubiquitin signal for MDM2-mediated ubiquitination of p53 (23); and (iii) a docking site for a range of protein kinases that phosphorylate the transactivation domain of p53 (25). A range of approaches were used to demonstrate that Ser269 phosphorylation can occur on endogenous WT p53 in cells and that phosphomimetic mutation results in the production of inactive WT p53. An accompanying paper (51) describes the biophysical basis for p53 inactivation by phosphorylation of p53 at Ser269 and involves primarily increases in the thermoinstability of the core DNA-binding domain of p53. These data highlight the existence of a novel kinase pathway that can regulate the dynamic range of conformations in WT p53 and that can produce a mutant-like conformation on WT p53.
Most phosphorylation sites characterized to date play a role in p53 activation. These include (i) phosphorylation in the transactivation domain (the BOX-I domain) at Thr18 and Ser20, which stabilizes the activator p300 (3, 4) and/or destabilizes the inhibitory protein MDM2 (31, 52); (ii) phosphorylation at the C-terminal CK2 site, which stimulates specific DNA binding (42); (iii) PKC phosphorylation in the C terminus that can create a 14-3-3 phosphopeptide binding motif and stimulate the specific DNA binding function of p53 (53); and (iv) CDK phosphorylation at Ser315 after DNA damage that can stimulate the specific DNA-binding function of p53 (49). There are also a small growing number of phosphorylation events that are linked to p53 inactivation. These include (i) phosphorylation by the CK2 component of the COP9 signalosome that triggers p53 degradation (54); (ii) phosphorylation at Ser315 by a GSK3-dependent pathway that can stimulate p53 nuclear export (55); and (iii) phosphorylation by Aurora kinase at either Ser215 or Ser315, which was reported to inactivate p53 by an undefined mechanism (56, 57). Thus, despite the fact that there are a growing number of inactivating phosphorylation sites on p53, there are few mechanistic data on how this regulates protein-protein interactions that lead to p53 inactivation. In this report, in analyzing a novel phosphorylation site on p53 using aspartate, phosphomimetic mutants, our data indicate that Ser269 phosphorylation would be inactivating with respect to p53-dependent transcription and clonogenic suppression.
We do not know as of yet the physiological kinase that targets the Ser269 site on p53 and whether DAPK or CHK1 could actually target Ser269 in vivo. Previous work on protein kinases have shown dual functions: (i) the CK1 enzyme can either activate p53 (via virus or TGF-β pathway) (26, 58) or suppress p53 functions (59, 60), depending upon the input signal, and (ii) the same phosphorylation site on p53 (Ser315) can be activating after DNA damage (CDK2) (49, 61) or inactivating in cycling cells (by GSK3 or by Aurora) (55), suggesting that context is important for kinase signaling. Thus, by this logic, if DAPK is a Ser269 kinase in vivo, then it would be consistent with its previously defined dual function; DAPK can be activating for p53 with an ARF-oncogene signal input (62), but it could be inactivating for p53 given an EGF input signal where DAPK inactivates TSC2, resulting in mTOR stimulation (63). However, we do not know that DAPK is the real in vivo kinase for Ser269. DAPK is part of the calcium-calmodulin superfamily, which can target the XSFX motif (25) (Table 1). Thus, we do not know that DAPK and CHK1 are, necessarily, the Ser269 kinases in cells; this is the work we are currently investigating to see whether the calcium-calmodulin superfamily or other families represent the in vivo kinase for the Ser269 site.
The interesting feature of Ser269 phosphorylation is that, although basal phosphorylation can be detected in a range of tumor cell lines with a wild type p53 status, DNA damages, such as that induced by UV or x-rays, induce Ser269 phosphorylation with kinetics that parallel p53 protein induction by DNA damage. These data would suggest that Ser269-phosphorylated p53 would be in a transcriptionally inactive state. Reasons for inducing this modification of p53 after DNA damage might include the following: (i) cells need to make “inactive” mutant-like WT p53 protein in order to begin to attenuate WT p53 functions and begin the process of regrowth after repairing damaged DNA, and/or (ii) creating this inactive and unfolded conformation to phospho-Ser269 p53 might begin the process of p53 ubiquitination and degradation that occurs in undamaged as well as damaged cells. Although the mechanistic stages that play a role in p53 ubiquitination and degradation in cells are undefined, it is known that unfolded mutant p53 is more prone to MDM2-dependent ubiquitination in vivo (64), and a trimeric complex of MDM2-Hsp90-CHIP can unfold the WT p53 tetramer in vitro (65). However, the ubiquitin ligase CHIP is also implicated in ubiquitination and degradation of mutant p53 (34, 65). Such unfolding of WT p53 might play a role in how it is ubiquitinated and/or degraded in vivo by many distinct ubiquitin ligases. Thus, there are widespread implications for the existence of cellular kinase pathways that can promote WT p53 inactivation by phosphorylation of this conformationally flexible motif in the p53 DNA-binding domain. In an accompanying paper (51), we report on the molecular basis for how p53 can be inactivated by Ser269 phosphorylation through the characterization of aspartate phosphomimetic mutant and through computational modeling. Our data suggest that Ser269 phosphorylation destabilizes the normal folding of the p53 DNA-binding domain and provides a mechanism for cells to create a mutant-like conformation to the WT p53 tetramer. Understanding the functions of phospho-Ser269 WT p53 in cells, defining how the altered conformation of Ser269-phospho-p53 alters in the interactome of p53, and defining the kinase/phosphatase pathways that target this site on p53 will begin to define how this phosphorylation site regulates the activation and inactivation of the p53 response.
Footnotes
This work was supported by CRUK Programme Grants C483 (Grant Agency of the Czech Republic) and 6354 (Ministry of Education, Youth, and Sports of the Czech Republic) (to T. R. H.). Development of monoclonal antibodies was supported by Moravian-Biotechnology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table I.
REFERENCES
- 1.Horn H. F., Vousden K. H. (2004) Nature 427, 110–111 [DOI] [PubMed] [Google Scholar]
- 2.Brooks C. L., Gu W. (2006) Mol. Cell. 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dornan D., Shimizu H., Burch L., Smith A. J., Hupp T. R. (2003) Mol. Cell. Biol. 23, 8846–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dornan D., Hupp T. R. (2001) EMBO Rep. 2, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert P. F., Kashanchi F., Radonovich M. F., Shiekhattar R., Brady J. N. (1998) J. Biol. Chem. 273, 33048–33053 [DOI] [PubMed] [Google Scholar]
- 6.Brown C. J., Srinivasan D., Jun L. H., Coomber D., Verma C. S., Lane D. P. (2008) Cell Cycle 7, 608–610 [DOI] [PubMed] [Google Scholar]
- 7.Teufel D. P., Bycroft M., Fersht A. R. (2009) Oncogene 28, 2112–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacPherson D., Kim J., Kim T., Rhee B. K., Van Oostrom C. T., DiTullio R. A., Venere M., Halazonetis T. D., Bronson R., De Vries A., Fleming M., Jacks T. (2004) EMBO J. 23, 3689–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armata H. L., Garlick D. S., Sluss H. K. (2007) Cancer Res. 67, 11696–11703 [DOI] [PubMed] [Google Scholar]
- 10.Wallace M., Coates P. J., Wright E. G., Ball K. L. (2001) Oncogene 20, 3597–3608 [DOI] [PubMed] [Google Scholar]
- 11.Finlan L. E., Nenutil R., Ibbotson S. H., Vojtesek B., Hupp T. R. (2006) Cell Cycle 5, 2489–2494 [DOI] [PubMed] [Google Scholar]
- 12.Hupp T. R., Lane D. P. (1995) J. Biol. Chem. 270, 18165–18174 [DOI] [PubMed] [Google Scholar]
- 13.Sakaguchi K., Sakamoto H., Lewis M. S., Anderson C. W., Erickson J. W., Appella E., Xie D. (1997) Biochemistry 36, 10117–10124 [DOI] [PubMed] [Google Scholar]
- 14.Nichols N. M., Matthews K. S. (2002) Biochemistry 41, 170–178 [DOI] [PubMed] [Google Scholar]
- 15.Bruins W., Zwart E., Attardi L. D., Iwakuma T., Hoogervorst E. M., Beems R. B., Miranda B., van Oostrom C. T., van den Berg J., van den Aardweg G. J., Lozano G., van Steeg H., Jacks T., de Vries A. (2004) Mol. Cell. Biol. 24, 8884–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruins W., Jonker M. J., Bruning O., Pennings J. L., Schaap M. M., Hoogervorst E. M., van Steeg H., Breit T. M., de Vries A. (2007) Carcinogenesis 28, 1814–1823 [DOI] [PubMed] [Google Scholar]
- 17.Bruins W., Bruning O., Jonker M. J., Zwart E., van der Hoeven T. V., Pennings J. L., Rauwerda H., de Vries A., Breit T. M. (2008) Mol. Cell. Biol. 28, 1974–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig A. L., Holcakova J., Finlan L. E., Nekulova M., Hrstka R., Gueven N., DiRenzo J., Smith G., Hupp T. R., Vojtesek B. (2010) Mol. Cancer 9, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neduva V., Linding R., Su-Angrand I., Stark A., de Masi F., Gibson T. J., Lewis J., Serrano L., Russell R. B. (2005) PLoS Biol. 3, e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hupp T. R., Walkinshaw M. (2007) Nat. Struct. Mol. Biol. 14, 885–887 [DOI] [PubMed] [Google Scholar]
- 21.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 22.Shimizu H., Burch L. R., Smith A. J., Dornan D., Wallace M., Ball K. L., Hupp T. R. (2002) J. Biol. Chem. 277, 28446–28458 [DOI] [PubMed] [Google Scholar]
- 23.Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Mol. Cell. 23, 251–263 [DOI] [PubMed] [Google Scholar]
- 24.Vojtesek B., Dolezalova H., Lauerova L., Svitakova M., Havlis P., Kovarik J., Midgley C. A., Lane D. P. (1995) Oncogene 10, 389–393 [PubMed] [Google Scholar]
- 25.Craig A. L., Chrystal J. A., Fraser J. A., Sphyris N., Lin Y., Harrison B. J., Scott M. T., Dornreiter I., Hupp T. R. (2007) Mol. Cell. Biol. 27, 3542–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLaine N. J., Oster B., Bundgaard B., Fraser J. A., Buckner C., Lazo P. A., Meek D. W., Höllsberg P., Hupp T. R. (2008) J. Biol. Chem. 283, 28563–28573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Baal J. W., Diks S. H., Wanders R. J., Rygiel A. M., Milano F., Joore J., Bergman J. J., Peppelenbosch M. P., Krishnadath K. K. (2006) Cancer Res. 66, 11605–11612 [DOI] [PubMed] [Google Scholar]
- 28.Diks S. H., Parikh K., van der Sijde M., Joore J., Ritsema T., Peppelenbosch M. P. (2007) PLoS ONE 2, e777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craig A., Scott M., Burch L., Smith G., Ball K., Hupp T. (2003) EMBO Rep. 4, 787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser J. A., Hupp T. R. (2007) Biochemistry 46, 2655–2673 [DOI] [PubMed] [Google Scholar]
- 31.Craig A. L., Burch L., Vojtesek B., Mikutowska J., Thompson A., Hupp T. R. (1999) Biochem. J. 342, 133–141 [PMC free article] [PubMed] [Google Scholar]
- 32.Pohler E., Craig A. L., Cotton J., Lawrie L., Dillon J. F., Ross P., Kernohan N., Hupp T. R. (2004) Mol. Cell. Proteomics 3, 534–547 [DOI] [PubMed] [Google Scholar]
- 33.Maya R., Oren M. (2000) Oncogene 19, 3213–3215 [DOI] [PubMed] [Google Scholar]
- 34.Lukashchuk N., Vousden K. H. (2007) Mol. Cell. Biol. 27, 8284–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burch L., Shimizu H., Smith A., Patterson C., Hupp T. R. (2004) J. Mol. Biol. 337, 129–145 [DOI] [PubMed] [Google Scholar]
- 36.Burch L. R., Scott M., Pohler E., Meek D., Hupp T. (2004) J. Mol. Biol. 337, 115–128 [DOI] [PubMed] [Google Scholar]
- 37.Velentza A. V., Schumacher A. M., Weiss C., Egli M., Watterson D. M. (2001) J. Biol. Chem. 276, 38956–38965 [DOI] [PubMed] [Google Scholar]
- 38.Joerger A. C., Fersht A. R. (2007) Oncogene 26, 2226–2242 [DOI] [PubMed] [Google Scholar]
- 39.Boehr D. D., Nussinov R., Wright P. E. (2009) Nat. Chem. Biol. 5, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G. W., Rudiger S., Veprintsev D., Freund S., Fernandez-Fernandez M. R., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson J., Hupp T. (2010) Cell Cycle 9:10, 1878–1881 [DOI] [PubMed] [Google Scholar]
- 42.Pospísilová S., Brázda V., Kucharíková K., Luciani M. G., Hupp T. R., Skládal P., Palecek E., Vojtesek B. (2004) Biochem. J. 378, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maya R., Balass M., Kim S. T., Shkedy D., Leal J. F., Shifman O., Moas M., Buschmann T., Ronai Z., Shiloh Y., Kastan M. B., Katzir E., Oren M. (2001) Genes Dev. 15, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephen C. W., Helminen P., Lane D. P. (1995) J. Mol. Biol. 248, 58–78 [DOI] [PubMed] [Google Scholar]
- 45.Batchelor E., Mock C. S., Bhan I., Loewer A., Lahav G. (2008) Mol. Cell. 30, 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giannakakou P., Sackett D. L., Ward Y., Webster K. R., Blagosklonny M. V., Fojo T. (2000) Nat. Cell. Biol. 2, 709–717 [DOI] [PubMed] [Google Scholar]
- 47.Craig A. L., Bray S. E., Finlan L. E., Kernohan N. M., Hupp T. R. (2003) Methods Mol. Biol. 234, 171–202 [DOI] [PubMed] [Google Scholar]
- 48.Blaydes J. P., Hupp T. R. (1998) Oncogene 17, 1045–1052 [DOI] [PubMed] [Google Scholar]
- 49.Blaydes J. P., Luciani M. G., Pospisilova S., Ball H. M., Vojtesek B., Hupp T. R. (2001) J. Biol. Chem. 276, 4699–4708 [DOI] [PubMed] [Google Scholar]
- 50.Neduva V., Russell R. B. (2006) Curr. Opin. Biotechnol. 17, 465–471 [DOI] [PubMed] [Google Scholar]
- 51.Fraser J., Blackwell E., Madhu K., Bramham J., Verma C., Walkinshaw M., Hupp T. R. (2010) J. Biol. Chem. 285, 37773–37786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schon O., Friedler A., Bycroft M., Freund S. M., Fersht A. R. (2002) J. Mol. Biol. 323, 491–501 [DOI] [PubMed] [Google Scholar]
- 53.Waterman M. J., Stavridi E. S., Waterman J. L., Halazonetis T. D. (1998) Nat. Genet. 19, 175–178 [DOI] [PubMed] [Google Scholar]
- 54.Bech-Otschir D., Kraft R., Huang X., Henklein P., Kapelari B., Pollmann C., Dubiel W. (2001) EMBO J. 20, 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qu L., Huang S., Baltzis D., Rivas-Estilla A. M., Pluquet O., Hatzoglou M., Koumenis C., Taya Y., Yoshimura A., Koromilas A. E. (2004) Genes Dev. 18, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Q., Kaneko S., Yang L., Feldman R. I., Nicosia S. V., Chen J., Cheng J. Q. (2004) J. Biol. Chem. 279, 52175–52182 [DOI] [PubMed] [Google Scholar]
- 57.Katayama H., Sasai K., Kawai H., Yuan Z. M., Bondaruk J., Suzuki F., Fujii S., Arlinghaus R. B., Czerniak B. A., Sen S. (2004) Nat. Genet. 36, 55–62 [DOI] [PubMed] [Google Scholar]
- 58.Cordenonsi M., Montagner M., Adorno M., Zacchigna L., Martello G., Mamidi A., Soligo S., Dupont S., Piccolo S. (2007) Science 315, 840–843 [DOI] [PubMed] [Google Scholar]
- 59.Huart A. S., MacLaine N. J., Meek D. W., Hupp T. R. (2009) J. Biol. Chem. 284, 32384–32394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inuzuka H., Tseng A., Gao D., Zhai B., Zhang Q., Shaik S., Wan L., Ang X. L., Mock C., Yin H., Stommel J. M., Gygi S., Lahav G., Asara J., Xiao Z. X., Kaelin W. G., Jr., Harper J. W., Wei W. (2010) Cancer Cell. 18, 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Prives C. (1995) Nature 376, 88–91 [DOI] [PubMed] [Google Scholar]
- 62.Raveh T., Droguett G., Horwitz M. S., DePinho R. A., Kimchi A. (2001) Nat. Cell. Biol. 3, 1–7 [DOI] [PubMed] [Google Scholar]
- 63.Stevens C., Lin Y., Harrison B., Burch L., Ridgway R. A., Sansom O., Hupp T. (2009) J. Biol. Chem. 284, 334–344 [DOI] [PubMed] [Google Scholar]
- 64.Shimizu H., Saliba D., Wallace M., Finlan L., Langridge-Smith P. R., Hupp T. R. (2006) Biochem. J. 397, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller P., Hrstka R., Coomber D., Lane D. P., Vojtesek B. (2008) Oncogene 27, 3371–3383 [DOI] [PubMed] [Google Scholar]