Abstract
The mitochondrial amidoxime reducing component mARC is a newly discovered molybdenum enzyme that is presumed to form the catalytical part of a three-component enzyme system, consisting of mARC, heme/cytochrome b5, and NADH/FAD-dependent cytochrome b5 reductase. mARC proteins share a significant degree of homology to the molybdenum cofactor-binding domain of eukaryotic molybdenum cofactor sulfurase proteins, the latter catalyzing the post-translational activation of aldehyde oxidase and xanthine oxidoreductase. The human genome harbors two mARC genes, referred to as hmARC-1/MOSC-1 and hmARC-2/MOSC-2, which are organized in a tandem arrangement on chromosome 1. Recombinant expression of hmARC-1 and hmARC-2 proteins in Escherichia coli reveals that both proteins are monomeric in their active forms, which is in contrast to all other eukaryotic molybdenum enzymes that act as homo- or heterodimers. Both hmARC-1 and hmARC-2 catalyze the N-reduction of a variety of N-hydroxylated substrates such as N-hydroxy-cytosine, albeit with different specificities. Reconstitution of active molybdenum cofactor onto recombinant hmARC-1 and hmARC-2 proteins in the absence of sulfur indicates that mARC proteins do not belong to the xanthine oxidase family of molybdenum enzymes. Moreover, they also appear to be different from the sulfite oxidase family, because no cysteine residue could be identified as a putative ligand of the molybdenum atom. This suggests that the hmARC proteins and sulfurase represent members of a new family of molybdenum enzymes.
Keywords: Enzymes, Mitochondrial Metabolism, Molybdenum, Protein Domains, Sulfotransferase, MOSC, Moco Sulfurase C-terminal Domain, mARC, Mitochondrial Amidoxime Reducing Component
Introduction
In eukaryotes the trace element molybdenum is essential for a number of enzymes where the molybdenum atom is part of the so-called molybdenum cofactor (Moco)2 in the active site of these enzymes (1). Moco is a pterin-based cofactor with a C6-substituted pyrano ring, a terminal phosphate, and a unique dithiolate group that binds the molybdenum atom. Moco-containing enzymes (Mo-enzymes) catalyze important reactions in the global carbon, sulfur, and nitrogen cycles that are characterized by transfer of an oxygen atom to or from a substrate. In mammals, one Mo-enzyme is sulfite oxidase (SO), which catalyzes the last step in the degradation of sulfur-containing amino acids and sulfatides (2). The active SO protein is a homodimer with each monomer of ∼52 kDa consisting of a N-terminal cytochrome b5 (cyt b5)/heme-binding domain and a C-terminal Moco-binding domain, the latter also harboring the dimerization interface. Both the Moco- and the heme-binding domain of mammalian SO are similar to the respective domains of nitrate reductase (NR), which catalyzes the first and rate-limiting step in nitrate assimilation in autotrophic organisms like plants, algae, and fungi (3). In addition to its N-terminal Moco-binding domain and the cytb5/heme-binding domain, each NR monomer possesses a C-terminal FAD-binding domain. Xanthine oxidoreductase (XOR) is another mammalian Mo-enzyme, and it is active as a homodimer with each ∼145-kDa monomer consisting of several distinct domains: an N-terminal domain responsible for binding of two nonidentical iron-sulfur clusters of the [2Fe-2S]-type, followed by a FAD-binding domain and a pair of C-terminal domains that bind the molybdenum center at their interface (4). XOR is a key enzyme in the purine degradation pathway where it catalyzes the oxidation of hypoxanthine to xanthine and of xanthine to uric acid. Another mammalian Mo-enzyme, aldehyde oxidase (AO), is thought to have arisen from a progenitor XOR via gene duplication and neo-functionalization (5, 6). In contrast to XOR, AO does not act on hypoxanthine and xanthine but instead oxidizes a variety of aromatic and nonaromatic heterocycles and aldehydes, which are converted to the respective carboxylic acids.
According to the structure of the molybdenum center, eukaryotic Mo-enzymes fall into two families: the SO family, represented by SO and NR, and the xanthine oxidase (XO) family, represented by XOR and AO. In the SO family, the molybdenum is coordinated by the two dithiolene sulfurs of the pterin, one apical and one equatorial oxo ligand, and a protein-derived cysteinyl sulfur. In the XO family, the molybdenum is ligated to the two dithiolene sulfurs, one apical oxo ligand, one equatorial hydroxyl group, and an equatorial sulfido group (7). The molybdenum-sulfur bond is essential for activity and is inserted in a post-translational reaction by a specific enzyme, the so-called Moco sulfurase (8–10). The N-terminal domain of this enzyme is responsible for the mobilization of sulfur from l-cysteine (11), whereas the C-terminal domain binds Moco and is presumed to function as a scaffold for sulfuration of the cofactor as required by the enzymes of the XO family (12).
A hitherto unknown Mo-enzyme has recently been identified in mammals (13) that shares significant sequence similarity to the C-terminal domain of Moco sulfurases and therefore has been referred to as MOSC (Moco sulfurase C-terminal domain) protein (14). Purification of the native MOSC-2 isoform from the outer membrane of pig liver mitochondria has shown that it plays a key role in the activation of N-hydroxylated prodrugs by reducing inactive amidoxime prodrugs to the respective active amidine drug (13). Because of this activity and also the fact that it acts in concert with heme/cyt b5 and NADH-dependent FAD/cytochrome b5 reductase (cyt b5R), the enzyme has been renamed mARC (mitochondrial amidoxime reducing component). Interestingly, the cofactor composition of this N-reductive system consisting of mARC, cyt b5, and cyt b5R is identical to that of eukaryotic NR proteins (Moco, heme, and FAD). Moreover, in the N-reductive system, the electrons pass from the electron donor NADH via FAD-containing cyt b5R and heme-containing cyt b5 to the mARC protein, which harbors the molybdenum-active site where the respective substrate is reduced. Thus, not only the cofactor composition but also the order of cofactors and the direction of electron flow are identical to NR. However, although many N-hydroxylated compounds have been found to serve as substrates for native and recombinant mARC proteins (15–17), the physiological substrates, and thus the physiological function, of mARC proteins are as yet unknown. The present study aimed to provide the first detailed biochemical information about the heterologously expressed human mARC isoforms hmARC-1 and hmARC-2 with particular focus on the characterization of their molybdenum center.
EXPERIMENTAL PROCEDURES
Construction of Expression Vectors
Full-length cDNAs of hmARC-1 (1011 base pairs corresponding to NCBI reference sequence NM_022746, MOSC-1) and hmARC-2 (1005 base pairs corresponding to NCBI reference sequence NM_017898, MOSC-2) were obtained as described earlier (15–17). For cloning of N-terminally truncated hmARC-1, the full-length open reading frame was truncated at its 5′ end by PCR using the primers hmARC-1_Start_N-del (5′-ATA TAT GGA TCC ATG CAG CAG GTG GGC ACA GTG GCG-3′) and hmARC-1_Stop (5′-AAA TTT AAG CTT TTA CTG GCC CAG CAG GTA CAC AGG-3′). For cloning of N-terminally truncated hmARC-2, the full-length open reading frame was truncated at its 5′ end using the primers hmARC-2_Start_N-del (5′-ATA TAT GGA TCC ATG CAG CAG GTG GGC ACC GTG GCG AAG-3′) and hmARC-2_Stop (5′-ATA ATT AAG CTT CTA CAC CAT CCG ATA CAC AGG GTC-3′). By use of these primers, a BamHI restriction site and a new in-frame ATG start codon were introduced at the 5′ end, and a HindIII restriction site was introduced at the 3′ end of each cDNA. The newly generated open reading frames of 861 base pairs for hmARC-1 and 858 base pairs for hmARC-2 encode truncated versions of these proteins, where the predicted N-terminal mitochondrial targeting sequences were removed. We note that in hmARC-1 two reproducible polymorphisms were identified that resulted in substitution of lysine 187 by methionine and alanine 165 by threonine when compared with the reference sequence. For generating the cysteine-to-serine variants of hmARC-1 (C69S, C79S, C119S, C149S, C161S, C246S, C270S, C273S, and C300S) and hmARC-2 (C210S) by PCR mutagenesis, the primers carrying the respective desired mutation were used (supplemental Table S1).
A cDNA with the full-length open reading frame of cyt b5 (453 base pairs, NCBI reference sequence NM_030579; also referred to as CYP5B), which is presumed to be located at the outer mitochondrial membrane, was amplified by PCR using reverse transcribed HepG2 mRNA as template and the primers human_cytb5_for (5′-TAT TAT GGA TCC ATG TCC GGT TCA ATG GCG ACT GCG-3′) and human_cytb5_rev (5′-ATA ATA CCC GGG TCA GGA GGA TTT GCT TTC CGA TGT G-3′). To remove the coding sequence for the putative membrane domain at the C-terminal end, a second PCR was performed using the primer combination human_cytb5_for and human_cytb5_C-del (5′-ATA ATA CCC GGG TTA TGC CCA GCA ACT TTT GCA TGT ATC-3′), which resulted in a cDNA of 375 base pairs and which introduced a BamHI restriction site at the 5′ end and a XmaI site and a new TAA stop codon at the 3′ end.
A full-length open reading frame of cyt b5R isoform 2 (837 base pairs, NCBI reference sequence NM_007326) was amplified by PCR using reverse transcribed HepG2 mRNA as template and the primers human_cytb5Red_Iso2_for (5′-TAT TAT GGA TCC ATG AAG CTG TTC CAG CGC TCC-3′) and human_cytb5Red_rev (5′-ATA ATA CCC GGG TCA GAA GAC GAA GCA GCG CTC CGT G-3′). The used primers introduced a BamHI restriction site at the 5′ end and a XmaI site at the 3′ end. Via their respective restriction sites, all of the cDNAs were subcloned into the plasmid pQE80 (Qiagen), which allows expression of N-terminally His6-tagged fusion proteins in Escherichia coli. The correctness of all cDNA sequences was confirmed by DNA sequencing at GATC (Konstanz, Germany).
Expression and Purification of Recombinant Proteins Used in This Work
Standard expression of the hmARC-1 and hmARC-2 proteins and their cysteine variants was performed in freshly transformed E. coli TP1000 cells (18), whereas expression of cyt b5 and cyt b5R was performed in E. coli DL41 cells. The cells were grown aerobically in LB medium in the presence of 100 μg/ml ampicillin at 22 °C to an A600 of 0.1 before induction. TP1000 cells were induced with 15 μm isopropyl-β-d-thiogalactopyranoside and additionally supplemented with 1 mm sodium molybdate to initiate expression of hmARC-1 and hmARC-2. DL41 cells were induced with 50 μm isopropyl-β-d-thiogalactopyranoside to start expression of cyt b5 and cyt b5R. The cells expressing cyt b5 were supplemented with 1 mm aminolevolinic acid to support heme synthesis. After induction the cells were grown for an additional 20 h at 22 °C. Expression of hmARC-1 and hmARC-2 in E. coli RK5206 and RK5204 strains (19) was performed basically as described for expression in TP1000 cells, but in the absence of sodium molybdate. The cells were harvested by centrifugation and stored at −70 °C until use. Cell lysis was achieved by two passages through a French pressure cell followed by sonication for 3 min. After centrifugation at 41,000 × g for 45 min at 4 °C, His6-tagged proteins were purified on a nickel-nitrilotriacetic acid superflow matrix (Qiagen) under native conditions at 4 °C according to the manufacturer's instructions and eluted in elution buffer (50 mm sodium phosphate, pH 8.0, containing 300 mm sodium chloride, 250 mm imidazole, 10% glycerol). Expression and purification of His6-tagged Arabidopsis thaliana AAO1 in the yeast Pichia pastoris was performed as described in Ref. 20, whereas expression and purification of His6-tagged Chlamydomonas reinhardtii Moco carrier protein Moco carrier protein (MCP) was performed as described in Ref. 21.
Determination of Protein Concentrations
Concentrations of total soluble protein were determined either by use of Roti Quant solution (Roth, Karlsruhe, Germany) according to Ref. 22 or by use of the BCA protein assay kit (Pierce) according to the manufacturer's instructions.
SDS-PAGE
SDS-PAGE was carried out according to Ref. 23, using a 5% stacking gel and 10–15% separating gels. Staining of electrophoresed proteins was performed in the presence of Coomassie Brilliant Blue R250 (Serva, Heidelberg, Germany). Molecular mass standards and protein samples were pretreated with β-mercaptoethanol for 5 min at 95 °C prior to loading onto the gel.
Molecular Mass Determination
After purification of the recombinant proteins under native conditions, size exclusion chromatography was performed with an Äkta Basic system (Amersham Biosciences) using the analytical Superdex 200 column (Amersham Biosciences). The column was equilibrated in 50 mm Hepes-HCl with 250 mm sodium chloride, pH 6.5, and 200 μg of purified hmARC-1, hmARC-2, cyt b5, or cyt b5R was separated at a flow rate of 0.3 ml/min. The molecular mass was determined using a calibration curve obtained from the retention times of standard proteins (aldolase, 161 kDa; albumin, 67 kDa; ovalbumin, 43 kDa; chymotrypsinogen A, 25 kDa; and ribonuclease A, 13.7 kDa). Each time v0 was determined with blue dextran 2000 (2000 kDa).
nit-1 NADH Nitrate Reductase Reconstitution Assay
Extracts from the Moco-deficient Neurospora crassa nit-1 mutant were prepared as described by in Ref. 24 and stored in aliquots at −70 °C. All of the reconstitutions were performed in nit-1 buffer (50 mm sodium phosphate, 200 mm sodium chloride, and 5 mm EDTA, pH 7.2) in the presence of 4 mm reduced glutathione and 10 mm sodium molybdate where appropriate. The reconstitution assay was performed in a 40-μl reaction volume containing 20 μl of gel-filtrated nit-1 extract. Complementation was carried out anaerobically overnight at 4 °C. After the addition of 20 mm NADPH and incubation for 10 min, reconstituted NADPH-nitrate reductase activity was determined as described (24).
Chemical Detection of Molybdenum Cofactor and Molybdopterin
Moco and its metal-free precursor molybdopterin (MPT) were detected and quantified by converting them to the stable oxidation product FormA-dephospho according to Ref. 25. Oxidation, dephosphorylation, diethyl-(2-hydroxy-propy)amino ethyl chromatography, and HPLC analysis were performed as described in detail (26). FormA-dephospho was quantified by comparison with a standard isolated from xanthine oxidase for which the absorptivity was ϵ380 = 13,200 m−1 cm−1 (25).
UV-visible Spectroscopy
UV-visible absorption spectra of purified hmARC-1, hmARC-2, cyt b5, and cyt b5R were recorded using a Ultrospec 2100 pro spectrophotometer (Amersham Biosciences). Wavescans were performed in the range between 200 and 800 nm with the relevant absorption ranges between 300 and 550 nm (hmARC-1 and hmARC-2) and between 300 and 600 nm (cyt b5 and cyt b5R) being presented in the respective figures.
In Gel AO Activity Staining
After electrophoresis on native polyacrylamide gels, enzymatic activity of AAO1 was determined by in-gel activity staining as described (20) in 100 mm phosphate buffer, pH 7.5, containing 1 mm indole-3-carboxaldehyde as substrate, 0.1 mm phenazine methosulfate as electron carrier and 1 mm 3[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium-bromide as indicator.
Sulfuration and Desulfuration of hmARC-1, hmARC-2, and AAO1
For sulfuration experiments, recombinant proteins (hmARC-1, hmARC-2, and AAO1), 0.5 m solutions of sodium sulfide and sodium dithionite, and a 0.2 m solution of DTT were made anaerobic by degassing under vacuum and purging with nitrogen. Sulfide, dithionite, and DTT were then added through a septum to the anaerobic protein samples to final concentrations of 32 mm for sodium sulfide, 16 mm for sodium dithionite, and 40 mm for DTT. The reaction mixture was incubated for 18 h anaerobically at 15 °C and rebuffered with 0.1 m potassium phosphate, pH 6.0, for the mARC proteins and with PBS for AAO1 on Sephadex G-50 NICKTM columns (Amersham Biosciences) for subsequent analyses. For desulfuration experiments, hmARC-1, hmARC-2, and AAO1 were treated with 50 mm potassium cyanide and incubated aerobically for 30 min at 22 °C. Prior to further analysis, hmARC proteins were rebuffered with 0.1 m potassium phosphate, pH 6.0, and AAO1 was rebuffered with PBS on Sephadex G-50 NICKTM columns (Amersham Biosciences).
Inductively Plasma-coupled Mass Spectrometry
For analysis of the molybdenum concentrations, a temperature- and pressure-controlled microwave digestion was performed (CEM Mars 5). For sample digestion 1 ml of sample, 5 ml of concentrated HNO3, and 1 ml of H2O2 were incubated overnight in closed Teflon vessels. The digests were heated in two steps to 180 °C (130 and 180 °C). The temperature of 180 °C was maintained for 20 min. Subsequently, the digests were decanted and transferred into 15- or 25-ml flasks, respectively, and filled with ultra pure water. All of the samples were diluted 1:10 prior to analysis. For molybdenum analysis, a Micromass Platform inductively plasma-coupled mass spectrometry was used. The hexapole ion optic cell was rinsed with hydrogen as a collision gas and with helium as a deceleration gas (both 4.1 ml/min). The hexapole bias was set to −2.0 V. The rate of plasma flow was 15.4 liters/min with an intermediate gas of 1.45 liters/min and a nebulizer gas flow rate of 0.94 liters/min. The radio-frequency power was 1.3 kW. The sample was introduced by using a Meinhard nebulizer (pumped) at a flow rate of 1 ml/min. The instrument was calibrated for the molybdenum masses 92, 95, 96, and 98 with plasma standard solutions in 1% HNO3 by a seven-point calibration up to 200 μg/liter. The detection limits are 0.17, 0.18, 0.23, and 0.25 μg/liter, respectively. As internal references, 103-rhodium and 105-rhenium were used (10 μg/liter).
Quantification of Protein-bound Heme and FAD via Extinction Coefficients
For the determination of heme binding to cyt b5, the absorption at 413 nm was monitored, and the heme-to-protein ratio was calculated using an extinction coefficient of 117 mm−1 cm−1 (27). Binding of FAD to cyt b5R was determined at 450 nm, and the FAD-to-protein ratio was calculated using an extinction coefficient of 11.3 mm−1 cm−1 (28).
Determination of NADH Cytochrome b5 Reductase Activity
Activity of NADH-dependent cyt b5R was determined using the ferricyanide reduction assay according to (29) with minor modifications. One unit is defined as the amount of cyt b5R required to reduce 1 μmol of ferricyanide/min.
Determination of the Cytochrome b5 Heme Content
The heme content of purified cyt b5 proteins was estimated by recording the sodium dithionite-reduced spectrum minus the oxidized spectrum of the protein (30).
Determination of the N-Reductive Activity of hmARC-1 and hmARC-2
N-Reductive activities of hmARC-1 and hmARC-2 with benzamidoxime, N-hydroxy-l-arginine, or N-hydroxy-sulfonamides as substrates were determined as described previously with minor modifications (16, 17). In the case of benzamidoxime, either 100 mm potassium phosphate buffer, pH 6.0 (for experiments shown in Figs. 5 and 6), or 20 mm MES buffer, pH 6.0 (for results shown in Table 3), was used for activity measurements. Apparent kinetic parameters Km and Vmax were estimated using nonlinear regression analysis (Sigma Plot 5.0; SPSS Science, Chicago, IL). In the case of N-hydroxy-cytosine, incubation mixtures contained 76.2 pmol of the respective hmARC enzyme, 76.2 pmol of cyt b5, and 7.6 pmol of NADH cyt b5R as well as the indicated concentration of substrate and 1.0 mm NADH in a total volume of 150 μl of 20 mm MES buffer, pH 6.0. Incubations were carried out under aerobic conditions at 37 °C in a shaking water bath. After a preincubation period of 3 min at 37 °C, the reaction was initiated by the addition of NADH and terminated after 15 min by the addition of methanol. Precipitated proteins were sedimented by centrifugation, and the supernatant was analyzed by HPLC for residual substrate and the newly formed product. Minor contaminations of cytosine in the substrate were subtracted for calculation of the enzymatic conversion rates. The HPLC system used for the separation of N-hydroxy-cytosine, and the cytosine reaction product consisted of a Waters 600 controller (Milford, CT) equipped with an autosampler (Waters 717 plus) and a variable wavelength UV detector (Waters 2487 dual absorbance detector). The peak areas were integrated with the EZChrom chromatography data system (EZChrom Elite version 2.8.3; Scientific Software Inc., San Ramon, CA). Separation was carried out isocratically with 95% (v/v) ammonium acetate buffer and 5% (v/v) methanol using a Nucleodur® 100–5 C18 ec, 4.6 × 250-mm column (Macherey-Nagel, Düren, Germany) with a security guard cartridge system C18, 3 × 4 mm (Phenomenex, Torrance) as a precolumn at a flow rate of 1.0 ml/min. The effluent was monitored at 267 nm. The retention times were 5.0 ± 0.1 min (N-hydroxy-cytosine) and 3.9 ± 0.1 min (cytosine). The injected sample volume was 10 μl. Solvents used in the analysis were filtered through a Sartorius membrane filter (0.45 μm; Sartorius AG, Goettingen, Germany) and degassed by sonication.
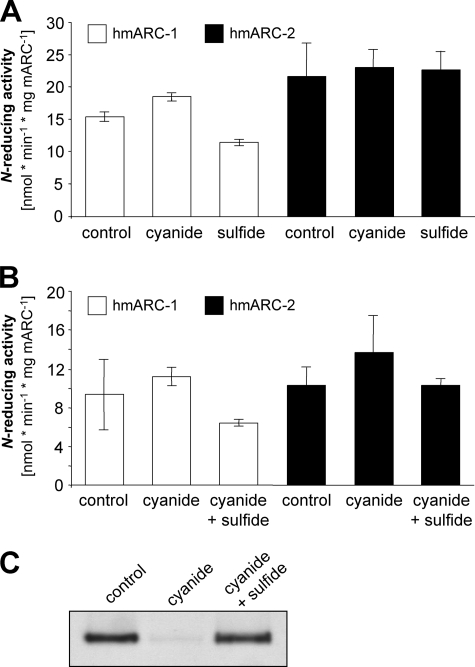
FIGURE 5.
Influence of cyanide and sulfide/dithionite treatment on the N-reductive activity of hmARC-1 and hmARC-2. A, the N-reductive activity determined either after cyanide treatment or after sulfide/dithionite treatment, respectively, of the purified hmARC-1 and hmARC-2 proteins. B, the N-reductive activity determined after initial cyanide treatment of hmARC-1 and hmARC-2 and subsequent sulfide/dithionite treatment. The N-reductive activities were measured in the presence of cyt b5 and cyt b5R using the model substrate benzamidoxime as substrate. C, recombinant AAO1 from A. thaliana was subjected to identical treatments and served as a control for cyanide inactivation and sulfide/dithionite reactivation of XO family enzymes.
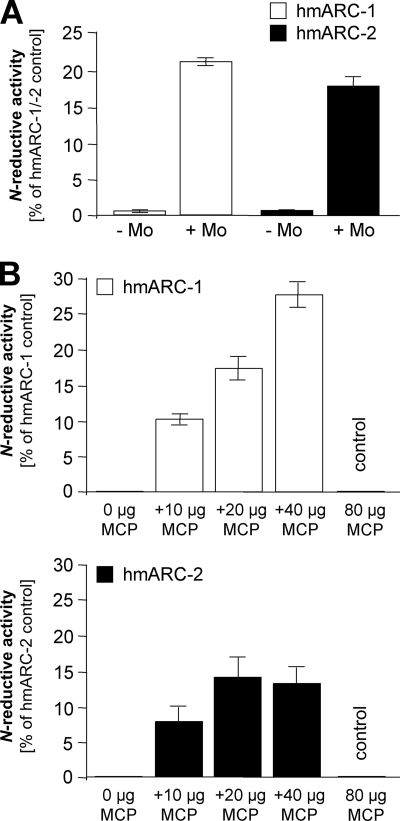
FIGURE 6.
Reconstitution of the N-reductive activity of Moco-free hmARC-1 and hmARC-2 proteins. A, reconstitution of the N-reductive activity of Moco-free but MPT-loaded hmARC-1 and hmARC-2 as expressed in E. coli RK5206 by molybdate treatment. B, reconstitution of the N-reductive activity of MPT/Moco-free hmARC-1 and hmARC-2 as expressed in E. coli RK5204 by coincubation with different amounts of Moco-loaded Moco carrier protein MCP. The N-reductive activities were measured in the presence of cyt b5 and cyt b5R using the model substrate benzamidoxime as substrate.
TABLE 3.
EPR parameters of signals seen with hmARC-1 and hmARC-2
Also shown are the parameters for the low pH signals seen with chicken sulfite oxidase, the C207S mutant of human sulfite oxidase, and spinach nitrate reductase.
| Enzyme | g1,2,3 | gav | A |
|---|---|---|---|
| MHz | |||
| hmARC-1 | 1.9983, 1.9691, 1.9585 | 1.9753 | Aav = 23.9 |
| C246S hmARC-1 | 1.9990, 1.9693, 1.9587 | 1.9757 | Aav = 23.9 |
| hmARC-2 | 1.9994, 1.9658, 1.9616 | 1.9756 | A1,2,3 = 24.6, 39.0, 31.0 |
| Low pH chicken sulfite oxidasea | 2.0037, 1.9720, 1.9658 | 1.9805 | A1,2,3 = 39.5, 37.7, 46.4 |
| C207S human sulfite oxidaseb | 1.9789, 1.9654, 1.9545 | 1.9663 | A1,2,3 = 6, 12, 6 |
| Spinach nitrate reductasec | 1.9957, 1.9692, 1.9652 | 1.9767 | A1,2,3 = 55.9, 59.0, 68.6 |
Quantification of Moco-bound Cyanolyzable Sulfur
The sulfur ligand of Moco as present in enzymes of the xanthine oxidase family can be liberated by cyanide treatment, which results in formation of thiocyanate (SCN−). To ensure that the mARC proteins are completely free from other sulfur-containing molecules that may be bound to cysteinyl residues and give unspecific background, the proteins were preincubated with 20 mm of the reducing agent Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) overnight at 4 °C. TCEP was removed by gel filtration on a Sephadex G-50 NICKTM column (Amersham Biosciences) equilibrated in 0.1 m Tris acetate, pH 8.6. Subsequently, 450 μl of the eluted protein fraction were incubated with 50 μl of 0.5 m potassium cyanide overnight at 22 °C. Formed SCN− was separated from the protein with a 5-kDa molecular mass cutoff Vivaspin concentrator (Sartorius, Göttingen, Germany), and 450 μl of ferric nitrate reagent (100 g of Fe(NO3)3 × 9H2O and 200 ml of 65% HNO3/150 ml) were added to 450 μl of the flow-through. The formed complex was quantified at 460 nm using a KSCN standard curve.
Conversion of mARC-bound MPT to Moco by Molybdate Treatment
The mARC proteins as purified after expression in the MPT-accumulating E. coli strain RK5206 were incubated in the presence of 10 mm sodium molybdate for 20 min in a final volume of 400 μl of nit-1 buffer. Excess molybdate was removed by dialysis against 5 liters of nit-1 buffer for 3 h. The conversion of MPT to active Moco was tested by subjecting the molybdate-treated proteins to the nit-1 NADH nitrate reductase reconstitution assay and by determination of the N-reductive activity with benzamidoxime as substrate.
Moco Transfer from MCP to Cofactor-free mARC Protein
20 μg of MPT/Moco-free mARC proteins as purified after expression in the E. coli strain RK5204 were coincubated with different amounts (10, 20, 40, and 80 μg) or equimolar amounts (10 μg), respectively, of Moco-loaded MCP from C. reinhardtii. Transfer of Moco from MCP to mARC proteins was analyzed by measuring the N-reductive activity of the respective mARC protein with benzamidoxime as substrate.
Isolation of Mitochondria from Mouse Tissues
Mitochondria from mouse liver and kidney were isolated by use of the MitoIso1 Kit (Sigma) according to the manufacturer's instructions.
Immunoblot Analysis of hmARC-1 and hmARC-2 in Different Mouse Tissues
For immunoblot analysis, the proteins were electrophoresed on 12% polyacrylamide gels in the presence of sodium dodecylsulfate. After blotting of proteins onto nitrocellulose membrane, the respective primary antibody was used (anti-MOSC-1, diluted 1:1000; anti-MOSC-2, diluted 1:2000). As secondary antibody, horseradish peroxidase-conjugated anti-rabbit IgG (Sigma) was used in a 1:10,000 dilution, and chemiluminescence was detected using the enhanced chemoluminiscence system (Amersham Biosciences).
Immunofluorescence Localization of hmARC-2 in Mouse Embryonic Fibroblasts
Mouse embryonic fibroblasts were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. For fluorescence microscopy, the cells were grown on coverslips, fixed 24–48 h after seeding by treatment with 4% formaldehyde, and permeabilized with 0.2% Triton X-100. For indirect immunofluorescence, the cells were rinsed three times in PBS; incubated with anti-MOSC-1 antibodies against hmARC-1 or anti-MOSC-2 antibodies against hmARC-2 proteins (Sigma), respectively, for 1 h; and washed three times in PBS. Subsequently, the cells were incubated with an anti-rabbit IgG Cy2-antibody conjugated with fluorescein as secondary antibody. Mitochondria were stained by MitoTrackerTM (Molecular Probes, Göttingen, Germany). After three washes, the cells were mounted in Mowiol (Hoechst, Frankfurt, Germany). Fluorescence images were taken with a conventional fluorescence microscope (Axiophot, Zeiss, Oberkochen, Germany) equipped with a digital CCD camera (MicroMax, Princeton Instruments, Trenton, NJ) and processed by MetaMorph Imaging software (Visitron Systems, Puchheim, Germany).
EPR Spectroscopy
EPR samples were prepared by incubating 1 μm cyt b5, 1 μm cyt b5R, and 100 μm hmARC in 50 mm NaPO4, pH 7.0, containing 300 mm NaCl and 10% glycerol for >1 h at 4 °C to remove resting signals. The samples were made anaerobic at 4 °C and subsequently transferred to argon-flushed, septum-sealed EPR tubes. An anaerobic solution of 1 mm NADH was added to 1 e− equivalent (more than six additions with ∼10 s mixing in between) and then frozen in an acetone/liquid nitrogen bath. The intermolecular electron transfer rates for cyt b5/hmARC were found to be quite slow (data not shown); thus incremental additions of NADH were required for greatest accumulation of the MoV species. Attempts to partially reduce hmARC via titration with solutions of sodium dithionite failed because the protein precipitated on contact. EPR spectra were recorded using a Brüker ER 300 spectrometer equipped with an ER 035 m gaussmeter and a HP 5352B microwave frequency counter and fitted with a Brüker ST 4102 X-band cavity. Temperature was controlled using a Brüker ER 4111 VT variable temperature unit and liquid nitrogen cryostat. The spectra were collected at 150 K with the following instrument settings: microwave power, 4.0 milliwatt; modulation amplitude, 0.3 millitesla. The spectra were recorded as the averages of 100 separate scans. Simulations were performed using the EasySpin 3.1.0 software package (31). No attempt was made to simulate the 95Mo/97Mo contributions to the spectra.
RESULTS
Cloning of hmARC-1 and hmARC-2 cDNAs and Their Tandem Arrangement in the Genome
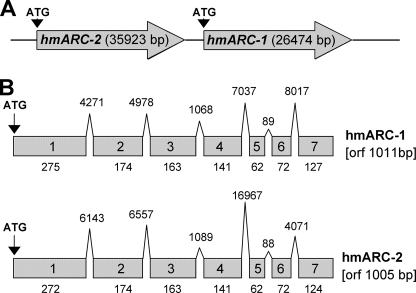
The human genome harbors two genes that encode for proteins with striking sequence similarities to the recently identified mARC protein from pig liver mitochondria (13), which is a member of the MOSC protein family (14). According to their homologies to mARC from pig, these genes were named hmARC-1 (human mARC-1) and hmARC-2 (human mARC-2), with the numbering following the preliminary database entries of the MOSC-1 (GeneID 64757) and MOSC-2 (GeneID 54996) genes. Both genes are arranged in a tandem orientation on chromosome 1 with their reading frames pointing in the same direction (Fig. 1A) and with an interspace of only 5044 base pairs. The predicted genes span regions of 26,474 (hmARC-1) and 35,923 base pairs (hmARC-2), respectively, and their predicted open reading frames share similar sizes of 1011 base pairs (NCBI reference sequence NM_022746) and 1005 base pairs (NCBI reference sequence NM_017898) and an overall sequence identity/similarity of 66/80%. Although both genes are interrupted by six introns of varying sizes, the structure and size of exons are highly conserved between both mARC genes. With the exception of the human Moco sulfurase HMCS, which is required for activating the Mo-enzymes AO and XOR (9) and which is the name-giving member of the MOSC family, no other homolog of hmARC-1 and hmARC-2 is found in the human genome. Cloning of hmARC-1 and hmARC-2 cDNAs derived from HepG2 cells (16, 17) basically confirmed the predicted sequences of the protein-encoding open reading frames as deposited in the database, with the exception of two reproducible polymorphisms that were identified in the hmARC-1 cDNA causing substitutions of methionine 187 by lysine (protein entry NP_073583 versus AAH10619) and alanine 165 by threonine (protein entry ACB21046 versus NP_073583).
FIGURE 1.
Genomic and cDNA structure of hmARC-1 and hmARC-2. A, tandem orientation of hmARC-1 and hmARC-2 genes on chromosome 1. B, exon-intron structure of hmARC-1 and hmARC-2 genes. The overall sizes of the respective open reading frames (orf) are indicated, and the relative sizes of exons (boxes) and introns (peaks) are shown; their lengths are indicated as numbers of base pairs.
Expression and Purification of Recombinant hmARC-1, hmARC-2, Cytochrome b5, and Cytochrome b5 Reductase
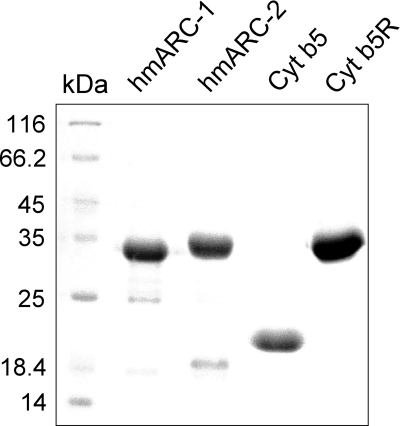
Preliminary studies have shown that the full-length proteins of hmARC-1 and hmARC-2 were characterized by poor solubility, weak saturation with Moco (∼0.02 mol/mol protein), and weak activity when expressed in E. coli.3 Because mitochondrial presequences are well known to impair expression of recombinant proteins in E. coli, the predicted mitochondrial targeting signals were removed from the N-terminal ends of both human mARC proteins in new expression constructs. In fact, in their truncated forms both human mARC proteins showed improved solubility, and thus, a simple purification by affinity chromatography was sufficient to obtain recombinant hmARC-1 and hmARC-2 proteins in concentrations of 10–15 mg/liter expression culture and in satisfying purity (Fig. 2). After purification, both proteins displayed major bands of ∼33 and ∼35 kDa in Coomassie-stained SDS-polyacrylamide gels, thereby corresponding well to the calculated molecular masses of 32.94 and 33.43 kDa for the deduced His6-tagged hmARC-1 and hmARC-2 monomers, respectively.
FIGURE 2.
Purity of recombinant hmARC-1, hmARC-2, cyt b5, and cyt b5R. 20 μg of recombinantly expressed hmARC-1, hmARC-2, cyt b5, and cyt b5R were separated on a 15% SDS-PA gel and stained with Coomassie Brilliant Blue.
cyt b5 is an amphiphatic protein consisting of two domains: an N-terminal water-soluble heme-binding domain and a C-terminal hydrophobic membrane-anchoring domain (32). Mammalian genomes encode two such proteins, one being anchored to the membrane of the endoplasmic reticulum and the other being anchored to the outer membrane of mitochondria (33). To improve the solubility and thus the expression and purity of the mitochondrial cyt b5 protein used in this study, the protein was partially truncated at the C terminus to remove the hydrophobic membrane-anchoring domain. Moreover, the heme precursor aminolevolinic acid was added to the expression culture to support the biosynthesis of heme in E. coli DL41 during expression of cyt b5. Thereby, recombinant cyt b5 expressed in these cells and purified by affinity chromatography was obtained in a soluble form of ∼15 kDa characterized by high purity (Fig. 2) and a dark red color, which indicated the binding of heme.
Human NADH-cyt b5R is presumed to exist in two different forms, a full-length protein of 301 amino acids, which contains an N-terminal membrane-binding domain, and a truncated version of 278 amino acids (reference sequence NP_015565) lacking the membrane-binding domain because of alternative splicing of the cyt b5R-3 gene (GeneID 1727). For the present study, the cyt b5R isoform 2 was expressed in E. coli DL41 and purified by affinity chromatography, whereby a soluble protein of ∼33 kDa was obtained (Fig. 2), which was characterized by a light yellow color indicative of bound FAD.
Biochemical Characterization of Recombinant hmARC-1, hmARC-2, Cytochrome b5, and Cytochrome b5 Reductase
Upon being subjected to size exclusion chromatography, all recombinant proteins of the three-component enzyme system eluted at nearly identical retention times, which indicates that all of the proteins used in this study, hmARC-1, hmARC-2, cyt b5, and cyt b5R, are characterized by similar molecular masses under nondenaturing conditions. This was confirmed by proper calculation of the respective molecular mass based on a calibration curve with standard proteins; a molecular mass of ∼33 kDa was determined for both hmARC-1 and hmARC-2, which corresponds well to the molecular mass of the monomers of hmARC-1 and hmARC-2 and indicates that these proteins do not form oligomers under the used conditions. Similar to the mARC proteins cyt b5R also appeared to be monomeric under native conditions, because the molecular mass of the protein as determined by size exclusion chromatography was ∼33 kDa, which is in agreement to the molecular mass of 32 kDa as deduced from the cyt b5R amino acid sequence. In contrast, cyt b5 has been found to form homodimeric complexes because the vast majority of the 15-kDa cyt b5 monomers eluted in the 33-kDa fraction.
Upon characterization of the recombinantly generated proteins, hmARC-1 and hmARC-2 have been subjected to FormA-dephospho analysis, which allows the common quantification of bound Moco and its metal-free precursor MPT. In fact, for both mARC proteins, high saturation with MPT/Moco has been found, with hmARC-1 and hmARC-1 showing average saturations of 84 and 59%, respectively (Table 1). However, FormA-dephospho analysis does not allow discrimination between molybdenum-free MPT and active Moco, and thus, the Moco/MPT ratio was estimated by the nit-1 NR reconstitution assay. This assay is based on the reconstitution of the apo-NR protein in extracts of the Moco-deficient N. crassa nit-1 mutant by the transfer of Moco and/or MPT derived from an exogenous source in the absence or presence of molybdate (24). Theoretically, in the absence of molybdate, only active Moco is able to reconstitute apo-NR, whereas the addition of molybdate to the reconstitution approach will convert also MPT to active Moco, which leads to an increase in nit-1 reconstitution activity. In fact, both hmARC-1 and hmARC-2 showed nit-1 reconstitution activity in the absence as well as in the presence of molybdate, thereby suggesting that not only Moco but also MPT is bound to these proteins (data not shown). However, the nit-1 assay is not suited to determine a precise ratio between MPT and Moco, and therefore, the amount of active Moco was determined by measuring the amount of protein-bound molybdenum. Based on inductively plasma-coupled mass spectrometry analysis, the average molybdenum content for both proteins turned out to be 0.4 mol of molybdenum/mol protein (Table 1), which indicates that ∼50% of the cofactor bound to hmARC-1 (84% FormA dephospho) and more than 65% of the cofactor bound to hmARC-2 (59% FormA dephospho) is present in its active molybdenum-loaded form. Determination of cyt b5-bound heme revealed that in average preparations 5% of the proteins were saturated with heme as calculated from the specific extinction coefficient (27) and the sodium dithionite-reduced minus the oxidized spectrum (30) (Table 1). However, single preparations yielded up to 70% heme saturation, indicating that heme incorporation is highly variable from preparation to preparation. The ratio of FAD to cyt b5R was likewise constituted by use of the specific molar extinction coefficient (28), which showed that recombinant cyt b5R is characterized by an average FAD content of ∼29% (Table 1).
TABLE 1.
Cofactor content of recombinant hmARC-1, hmARC-2, cyt b5, and cyt b5R
| Protein | Cofactor content | Metal contenta | Saturation with active cofactor |
|---|---|---|---|
| mol/mol protein | mol/mol protein | ||
| hmARC-1 | 0.84 ± 0.13 FormAb | 0.45 ± 0.04 Mo | 45% Moco |
| hmARC-2 | 0.59 ± 0.08 FormAb | 0.37 ± 0.04 Mo | 37% Moco |
| cyt b5 | 0.05 ± 0.01 hemec | 5% heme | |
| cyt b5R | 0.29 ± 0.09 FADd | 29% FAD |
a Determined via inductively plasma-coupled mass spectrometry (n = 4).
b Representing Moco and its metal-free precursor MPT (n = 12).
c Determined via differential spectra and extinction coefficient (n = 4).
d Determined via extinction coefficient (n = 7).
Spectroscopic Properties of Recombinant hmARC-1, hmARC-2, cyt b5, and cyt b5R
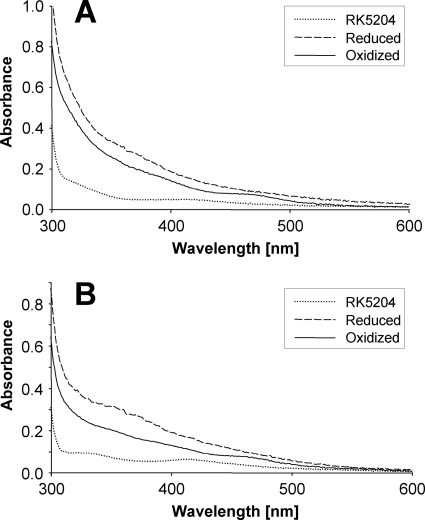
UV-visible absorption spectra of Moco-loaded hmARC-1 and hmARC-2 proteins as expressed in the Moco-accumulating E. coli strain TP1000 (18) showed a broad absorption shoulder at ∼350 nm in the TCEP-reduced form. When the reduced proteins were oxidized by exposure to air, the shoulder shifted to a region between 370 and 400 nm with an appearance of an additional shoulder at ∼465 nm (Fig. 3), indicating that both proteins are redox active. Surprisingly, also Moco-free hmARC-1 and hmARC-2 proteins as expressed in the Moco-deficient E. coli strain RK5204 (19) revealed absorption. However, absorption at ∼330 and 420 nm of the RK5204 derived proteins were different from the absorption of the TP1000-derived proteins and less pronounced. Moreover, FormA-dephospho analysis clearly demonstrated that they do not derive from Moco or MPT (data not shown), suggesting that some other as yet unknown molecule is bound to recombinant hmARC-1 and hmARC-2.
FIGURE 3.
UV-visible absorption spectra of hmARC-1 and hmARC-2. All spectra of hmARC-1 (A) and hmARC-2 (B) were recorded with 4 mg/ml protein in elution buffer (50 mm sodium phosphate, pH 8.0, containing 300 mm sodium chloride, 250 mm imidazole, 10% glycerol). Proteins obtained after expression in E. coli TP1000 were reduced with 10 mm Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and re-oxidized by exposure to air for 20 h. Spectra of proteins obtained after expression in E. coli RK5204 were recorded with the respective proteins as purified.
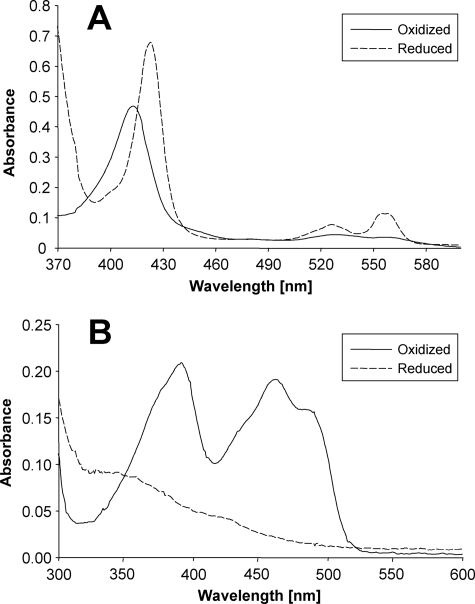
Typical absorption spectra were obtained for the recombinant human cyt b5 protein with a pronounced high intensity Soret peak at 413 nm and faint absorbance between 520 and 560 nm in the oxidized state (Fig. 4A). Upon reduction with dithionite, the Soret peak shifted to 423 nm, and two minor peaks occurred at 527 and 557 nm, which has been shown to be a general feature of cyt b5 proteins (27). When recombinant human cyt b5R was subjected to UV-visible absorption spectroscopy, two distinct absorption maxima at 391 and 462 nm accompanied by a shoulder at ∼480 nm were identified for the purified protein in the oxidized state (Fig. 4B). After reduction by dithionite, however, all of these absorption bands were eliminated and replaced by weak but broad absorption between 320 and 500 nm. The spectra of cyt b5 and cyt b5R indicate that these heterologuously expressed proteins meet the demands of electron carrier proteins as required for establishing a defined N-reductive system.
FIGURE 4.
UV-visible absorption spectra of recombinant human cyt b5 and cyt b5R. A, oxidized cyt b5 (4 μm) was measured using the purified protein; for reduction the protein was treated with dithionite. B, the spectrum of oxidized cyt b5R (18 μm) was recorded using the purified protein; for reduction the protein was treated with 2 mm dithionite. All of the spectra were recorded in elution buffer (50 mm sodium phosphate, pH 8.0, containing 300 mm sodium chloride, 250 mm imidazole, 10% glycerol).
Substrate Specificity of hmARC-1 and hmARC-2
Because the specific substrate preferences of the two human mARC proteins have not been reported as yet, a set of N-hydroxylated substrates was analyzed for their potential in serving as substrates for recombinant hmARC-1 and hmARC-2, respectively, in concert with cyt b5 and cyt b5R. All of the tested substrates were converted by both hmARC-1 and hmARC-2, although some differences in the efficiency of catalysis (Vmax/Km) were observed. Although hmARC-1 and hmARC-2 catalyzed the reduction of benzamidoxime with similar efficiency, N-hydroxy-cytosine was clearly more efficiently reduced by hmARC-1 (Table 2). By contrast, N4-hydroxy-l-arginine was more efficiently converted by hmARC-2 than by hmARC-1 (17). Varying efficiencies have likewise been observed for various N-hydroxylated sulfonamides (16), which demonstrates that although hmARC-1 and hmARC-2 act on the same substrates, each isoform has its own set of preferred substrates.
TABLE 2.
Kinetic parameters of the reduction of benzamidoxime and N-hydroxy-cytosine by hmARC-1 and hmARC-2
The specific activities of hmARC-1 and hmARC-2 were determined in the presence of cyt b5 and cyt b5 R as electron carriers and NADH as cosubstrate. Substrate concentrations between 0.1 and 8.6 mm and between 0.1 and 5.0 mm were used for N-hydroxy-cytosine and benzamidoxime, respectively.
| Substrate | Parameters | hmARC-1 | hmARC-2 |
|---|---|---|---|
| Benzamidoxime | Kma | 0.11 ± 0.04 | 0.27 ± 0.08 |
| Vmaxb | 424.3 ± 26.9 | 419.7 ± 27.4 | |
| Efficiencyc | 3860 | 1550 | |
| N-Hydroxy-cytosine | Kma | 0.52 ± 0.14 | 1.52 ± 0.28 |
| Vmaxb | 535.4 ± 38.9 | 219.8 ± 14.8 | |
| Efficiencyc | 1030 | 140 |
a Unit of measure is mm.
b Unit of measure is nmol min−1 mg mARC−1.
c Efficiency of catalysis (Vmax/Km).
Distinction of hmARC-1 and hmARC-2 from Typical Members of the Xanthine Oxidase Family of Molybdenum Enzymes
Mo-enzymes belonging to the XO family are characterized by a typical active site molybdenum-sulfur group (4). The sulfido group can be removed by cyanide treatment, which releases the terminal sulfur in the form of thiocyanate and results in inactivation of the enzyme (34). The sulfido group can be reintroduced by sulfide/dithionite treatment of cyanide-inactivated or otherwise desulfurated XO family enzymes, restoring activity (35). It is therefore reasonable to conclude that if mARC proteins are members of the XO family, they will be sensitive to cyanide. In contrast to a control experiment with the AAO1 protein, in which activity was lost on treatment of the enzyme with cyanide, however, no decrease in the N-reductive activity was observed with either of the two human mARC proteins when similarly treated (Fig. 5, A and C), indicating that the mARC proteins do not require a terminal sulfur ligand. The absence of sulfur in the active molybdenum-site is further supported by the observation that no thiocyanate formation could be detected upon cyanide treatment (data not shown). Moreover, sulfide/dithionite treatment did not result in enhanced activity of the purified mARC proteins (Fig. 5A). Nearly identical results were obtained when hmARC-1 and hmARC-2 were treated with sulfide/dithionite subsequent to treatment with cyanide (Fig. 5B). These results indicate that the molybdenum center of human mARC proteins differs from the molybdenum center of typical XO family enzymes such as AO and XOR in that they lack a terminal sulfido ligand.
Reconstitution of the N-Reductive Activity of Moco-free hmARC-1 and hmARC-2 Proteins
After expression in the E. coli strain TP1000, which accumulates eukaryotic Moco, hmARC-1 and hmARC-2 were capable of reducing N-hydroxylated substrates (Table 2). However, this activity was absent when the proteins were expressed in the Moco-free/MPT-accumulating E. coli strain RK5206 (Fig. 6A), thereby demonstrating the dependence of mARC proteins on Moco. Incubation of MPT-loaded hmARC-1 and hmARC-2 purified from E. coli RK5206 with molybdate and subsequent dialysis partially restored the N-reductive activity (Fig. 6A), which can be ascribed to a nonenzymatic conversion of MPT to Moco by molybdate. In support of this, hmARC-1 and hmARC-2 expressed in E. coli RK5204 were free from Moco and MPT and thus did not exhibit any N-reductive activity (Fig. 6B). After coincubation with the recombinant Moco carrier protein MCP from C. reinhardtii (21), however, the N-reductive activity with benzamidoxime as substrate was likewise restored (Fig. 6B). Because MCP itself did not exhibit N-reductive activity on its own, the Moco of MCP must have been transferred to the mARC proteins, with recovery of activity. In these experiments, the addition of molybdate to MPT is likely to generate a “trioxo” Moco, with the molybdenum metal initially being ligated to the two dithiolene sulfurs of the pterin, two molybdenum-oxygen groups, and a Mo-OH group. The same type of Moco is bound by MCP as identified by extended x-ray absorption fine structure analysis.4 However, it remains unclear whether one of the oxygen ligands is subsequently replaced by a cysteine residue after incorporation of the molybdate into MPT or after transfer of Moco from MCP to mARC, such as occurs with enzymes of the SO family. Even coordination of the molybdenum metal by other yet unknown amino acid ligands cannot be excluded.
We note that neither approach to reconstitution shown in Fig. 6 was supplemented with an inorganic sulfur source, and thus, spontaneous sulfuration of the reconstituted Moco can be excluded. These experiments therefore confirm the findings from the cyanide and sulfide/dithionite treatments (Fig. 5) and indicate that hmARC-1 and hmARC-2 are unlikely to represent members of the XO family.
EPR Studies of Human mARC-1 and mARC-2
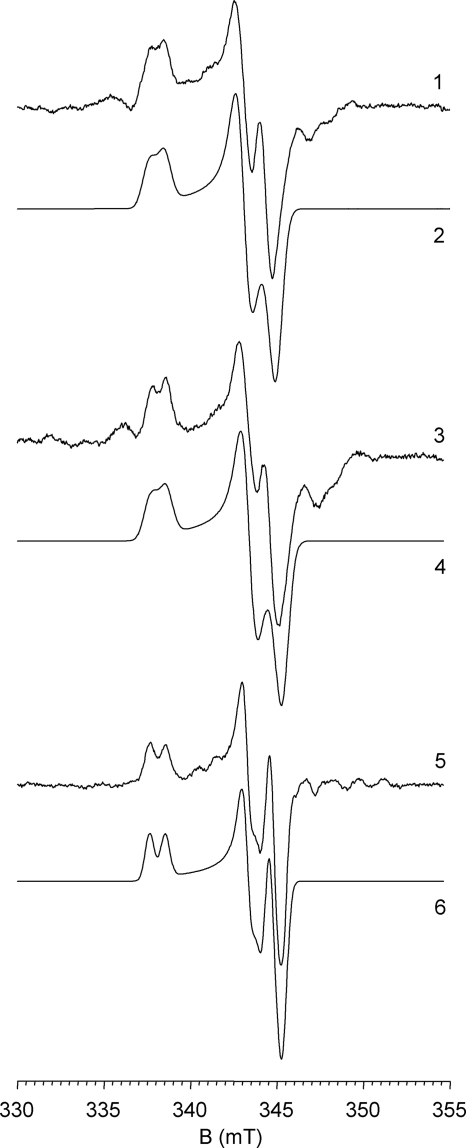
Neither hmARC-1 nor hmARC-2 was EPR-active as isolated (data not shown). Upon partial reduction with NADH/cyt b5R/cyt b5 (see “Experimental Procedures”), however, both proteins developed EPR signals characteristic of the Mo(V) (d1) state, as shown in Fig. 7. The signals were very similar (although not identical; Table 3), with rhombic symmetry, gav ∼1.975, and approximately isotropic superhyperfine coupling to a single exchangeable proton (Aav ∼24 MHz for hmARC-1, Aav ∼32 MHz for hmARC-2). The spectra closely resemble the so-called “low pH” EPR signal seen with SO and, in particular, NR (see spectral parameters given in Table 3).
FIGURE 7.
Molybdenum (V) EPR spectra exhibited by hmARC-1 and hmARC-2. Shown are the experimental spectra (spectra 1, 3, and 5) and simulations (spectra 2, 4, and 6) for wild-type hmARC-1 (spectra 1 and 2), the C246S variant of hmARC-1 (spectra 3 and 4), and wild-type hmARC-2 (spectra 5 and 6). The spectroscopic parameters used in each simulation, obtained as described under “Experimental Procedures,” are given in Table 3. The spectra were aligned to a microwave frequency of 9.4556 GHz.
Given the similarity in EPR signals seen with the hmARC proteins and members of the SO family of Mo-enzymes, an attempt was made to identify the putative cysteine residue that coordinates the molybdenum. Substitution of this cysteine with serine in human SO significantly perturbs the EPR signal, with g1,2,3 = 2.0037, 1.9720, 1.9658 seen with the wild-type protein shifting to g1,2,3 = 1.9789, 1.9654, 1.9545 and with much weaker proton splitting in the mutant (36). This being the case, we undertook to substitute each of the nine cysteine residues in hmARC-1 and an additional nonconserved residue in hmARC-2 to determine which might be coordinated to the molybdenum in the protein. Surprisingly, none of the substitutions perturbed the EPR signal to any significant degree. Fig. 7 (center) shows the spectrum of the hmARC-1/C246S variant as an example, which may be compared with the spectrum of the wild-type protein (Fig. 7, top). The relatively low gav of 1.9753 to 1.9757 seen for the hmARC proteins notwithstanding, it appears that they lack a third sulfur ligand to the molybdenum as seen in members of either the XO or SO families.
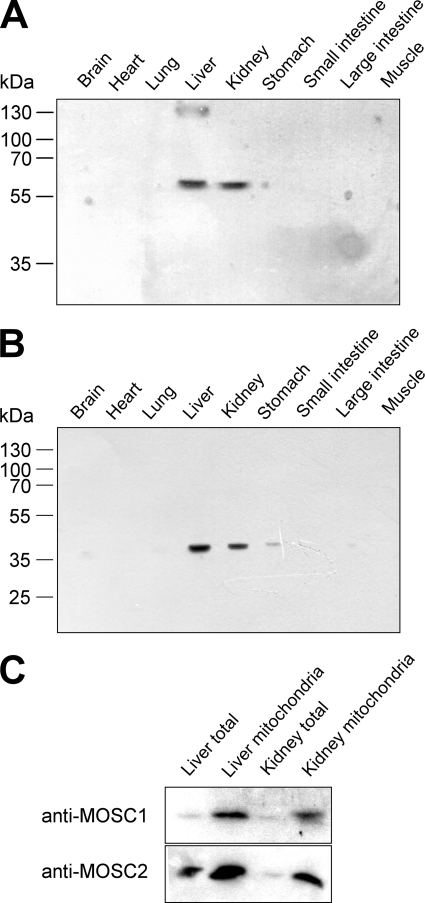
Localization of Native Murine and Human mARC-2
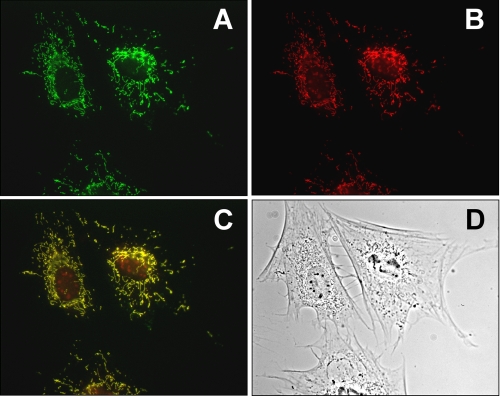
It was one aim of this work to investigate whether mARC proteins exclusively localize to mitochondria, as suggested by previous reports (13, 37), or whether they also reside in other compartments, and another aim was to show the expression levels of native mammalian mARC proteins in different tissues. By use of antibodies raised against specific peptide sequences of human mARC-1 and mARC-2 (anti-MOSC-1 and anti-MOSC-2), cross-reacting proteins were identified in murine total cell extracts from liver and kidney (Fig. 8, A and B) but also from stomach in the case of hmARC-2 (Fig. 8B). Although the molecular mass of the proteins identified by anti-hmARC-2 antibodies corresponded well with the expected molecular mass of the native murine mARC-2 protein of ∼35 kDa (Fig. 8B), the anti-hmARC-1 antibodies exclusively detected proteins of 60–65 kDa (Fig. 8A). The same cross-reacting proteins were found to be enriched in mitochondria isolated from liver and kidney (Fig. 8C), indicating that the detected proteins indeed localize to mitochondria rather than to the cytosol. This observation was supported for mARC-2 by immunofluorescence analysis because the fluorescence of the anti-MOSC-2/Cy2-coupled antibodies in mouse embryonic fibroblasts overlapped exclusively with the mitochondria-specific fluorescence of the Mitotracker dye and did not occur in other compartments of the cell (Fig. 9). Although this specific fluorescence pattern was detected in all anti-MOSC-2/Cy2-treated cells, only a minority of the analyzed anti-MOSC-1-treated cells showed Cy2-coupled fluorescence in the mitochondria and nonassignable fluorescence elsewhere in the cell (data not shown). However, the intensity of the fluorescence was not significant, and thus, a reliable determination of the subcellular localization of murine mARC-1 was impossible.
FIGURE 8.
Tissue-specific localization of mouse mARC-1 and mARC-2. Total protein extracts from different tissues of mouse (100 μg each) were subjected to SDS-PAGE and subsequent immunoblot analysis using anti-MOSC-1 (A) and anti-MOSC-2 (B) antibodies for detection of mouse mARC-1 and mARC-2, respectively. In isolated mitochondria prepared from liver and kidney, anti-MOSC-1 and anti-MOSC-2 cross-reacting proteins were enriched relative to the respective total extracts (C), indicating a mitochondrial localization of mouse mARC-1 and mARC-2 proteins (lanes were loaded with 30 μg of mitochondrial protein and 30 μg of total protein, respectively).
FIGURE 9.
Immunolocalization of mARC-2 in mouse embryonic fibroblast. Localization of endogenous mARC-2 in mouse embryonic fibroblasts was visualized by use of anti-MOSC-2 antibodies in combination with IgG Cy2-coupled antibodies. A, fluorescence of the Cy2-coupled antibody. B, counterstaining of the mitochondria by MitoTrackerTM. C, merging of A and B reveals colocalization of mouse mARC-2 exclusively with the mitochondria. D, phase contrast image of the analyzed fibroblasts.
DISCUSSION
Proteins of the MOSC family of Mo-enzymes share a common domain that contains a strictly conserved cysteine residue that has been predicted to function as sulfur carrier and to be required for assembly of diverse metal-sulfur clusters (14). In humans, members of the MOSC family include the Moco sulfurase protein HMCS and two highly homologous MOSC domain proteins, MOSC-1 and MOSC-2, which have been renamed hmARC-1 and hmARC-2 on the basis of their reactivity with amidoxime substrates (Refs. 15–17 and this work). Although the Moco sulfurase consists of an N-terminal pyridoxal phosphate-binding domain responsible for abstracting sulfur from l-cysteine and a C-terminal Moco-binding domain onto which the sulfur is transferred (11, 12), both hmARC proteins are stand-alone proteins lacking the Moco sulfurase-typical N-terminal domain. Interestingly, all eukaryotic organisms, with the exception of certain specialized Moco-independent yeast species (38), appear to harbor the same set of MOSC family proteins, suggesting a highly conserved function for each of these proteins. However, the physiological function is known only for the Moco sulfurase, which is required for the post-translational sulfuration and activation of the Moco of AO and XOR proteins. In E. coli, the MOSC domain proteins YcbX and YiiM have been shown to be required for detoxification of N-hydroxylated base analogs given that mutations in the YcbX and YiiM genes lead to hypersensitivity to 6-N-hydroxylaminopurine (39, 40). Based on the finding that hmARC-1 and hmARC-2 likewise catalyze the reduction of N-hydroxylated base analogs with high efficiency (N-hydroxy-cytosine; Table 2), it appears likely that one of the physiological functions of human mARC proteins could be to prevent accumulation of such mutagenic substances in the cell. In addition, hmARC-1 and hmARC-2 have recently been suggested to act as regulators for the l-arginine-dependent biosynthesis of NO by catalyzing the controlled elimination of the NO precursor N4-hydroxy-l-arginine (17). However, because immunolocalization of mARC proteins in mouse cells has shown that at least mARC-2 localizes exclusively to the mitochondria (Fig. 9), the putative reduction of N4-hydroxy-l-arginine is likely to be limited to this organelle. Consistent with this, in addition to endothelial and neuronal NO synthases, a mitochondrial NO synthase has been identified (41) that might be regulated by mARC proteins. A role for mammalian mARC proteins in the detoxification of N-hydroxylated substrates appears likely, given the relatively high abundance of these proteins in typical detoxification organs such as liver and kidney (Fig. 8).
Both hmARC-1 and hmARC-2 elute as monomers in native size exclusion chromatography, which contrasts with all other known eukaryotic Mo-enzymes, which are dimers (summarized in Ref. 3). Because the recombinant human mARC proteins were truncated at their N-terminal ends to improve expression in E. coli, an influence on the oligomerization cannot be excluded. Still, the N-terminal truncations were limited to the predicted mitochondrial targeting sequences, which are highly unlikely to serve as a dimerization domain. Thus, hmARC-1 and hmARC-2 presently must be considered the smallest eukaryotic Mo-enzymes regarding the number of monomers per active enzyme complex as well as the molecular mass of each hmARC monomer (∼33 kDa, in contrast to the monomers of plant SO, ∼45 kDa; animal SO, ∼52 kDa; NR, ∼110 kDa; and AO and XOR, ∼145 kDa).
A comparative analysis of different substrates used in this and previous studies showed that recombinant hmARC-1 and hmARC-2 act on the same N-hydroxylated compounds, albeit with different efficiencies. With regard to the efficiency of catalysis (Vmax/Km), both proteins showed nearly identical reduction rates for benzamidoxime (Table 2), whereas hmARC-1 reacted more efficiently with N-hydroxylated sulfonamides (N-hydroxy-valdecoxib and N-hydroxybenzene sulfon-amide) (16) and N-hydroxy-cytosine (Table 2), and hmARC-2 reacted more efficiently with N-hydroxy-l-arginine (17). Obviously, in vitro both proteins have remarkable overlapping substrate preferences with differences only in the specific activity for some of the substrates. However, the physiological substrates for hmARC-1 and hmARC-2 are as yet unknown, and it therefore remains to be shown whether or not both proteins also in vivo act on the same or on different substrates. In this respect, it should be kept in mind that all hitherto analyzed mammalian genomes harbor two mARC genes, which suggests the evolutionary need for the function of each protein. One could speculate that the two mARC proteins either have identical tissue and/or subcellular localizations and act on different substrates or are localized differently and act on identical substrates. Alternatively, they could be differentially regulated. In fact, mouse mARC-2 is exclusively localized in the mitochondrion (Fig. 9), consistent with the identification of both murine mARC proteins in the inner mitochondrial membrane (37) and porcine mARC-2 in the outer mitochondrial membrane (13). Although all of these findings suggest a localization of mammalian mARC proteins at one of the mitochondrial membranes, they leave open the issue of in which mitochondrial subcompartment(s) mARC proteins are localized and whether or not both mARC proteins localize to the same subcompartment(s). Moreover, the fact that the anti-MOSC-1 antibodies used in this study did not detect a mouse mARC-1 homolog with the expected molecular mass of ∼33 kDa in any of the analyzed tissues but instead a protein of ∼65 kDa (Fig. 8) may indicate some other difference between mARC-1 and mARC-2. Either mARC-1 is subject to post-translational modification or participates in highly stable protein-protein interactions, increasing the molecular mass observed in our experiments. Alternatively, the immunostained band obtained by use of the anti-MOSC-1 antibody may not be mARC-1 but an unrelated protein that presents mARC-1-like epitopes. Still, because recombinant hmARC-1 is detected by this specific antibody (supplemental Fig. S1), the possibility must be considered that mARC-1 is simply not expressed in the analyzed tissues or that expression of mARC-1 requires a specific induction.
Surprisingly, our results suggest that hmARC-1 and hmARC-2 do not belong to any of the two known eukaryotic Mo-enzyme families. Neither protein possesses the terminal sulfido ligand that is typical for members of the XO family, because cyanide treatment neither released sulfur in the form of thiocyanate nor significantly affected the activities of hmARC-1 and hmARC-2 (Fig. 5). Furthermore, reconstitution of the N-reductive activity of MPT-loaded mARC proteins by molybdate treatment and of cofactor-free mARC proteins by coincubation with Moco-loaded MCP from C. reinhardtii in the absence of a sulfur source (Fig. 6) confirms that the human mARC proteins do not require a sulfurated Moco for activity. On the other hand, a cysteine ligand provided by the polypeptide chain can be excluded on the basis of the mutational work described here with hmARC-1 and hmARC-2. These cysteine-to-serine substitutions did not alter the EPR spectra of hmARC-1 and hmARC-2 (Fig. 7) and did not affect their N-reductive activity. Thus, notwithstanding the similarity of the EPR signals seen with hmARC-1 and hmARC-2 to the low pH Mo(V) signals exhibited by SO and NR, they cannot be regarded as members of the SO family. There are two likely possibilities for the molybdenum coordination sphere of the hmARC proteins. The first is a trioxo species LMoVIO3 (with L representing the bidentate enedithiolate ligand) that protonates upon reduction to LMoO2(OH). The second is an LMoO2X species, with X being either an as yet unknown inorganic or alternatively a protein-derived ligand other than cysteine. In either case, the coordination of the molybdenum in the Moco of hmARC-1 and hmARC-2 differs fundamentally from that seen in enzymes of the SO and XO families, and thus, mARC/MOSC proteins appear to represent a new family of Mo-enzymes in eukaryotes.
Acknowledgments
We give our thanks to Marion Kirchner, Victoria Michael (Technische Universität Braunschweig, Plant Biology), Adelina Calean (Technische Universität Braunschweig, Environmental Geology), and Petra Köster (Christian-Albrechts-Universität Kiel) for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
B. Wahl, R. R. Mendel, and F. Bittner, unpublished data.
G. Schwarz, K. Fischer, and G. George, personal communication.
- Moco
- molybdenum cofactor
- AO
- aldehyde oxidase
- cyt b5
- cytochrome b5
- cyt b5R
- cyt b5 reductase
- MCP
- Moco carrier protein
- MPT
- molybdopterin
- NR
- nitrate reductase
- SO
- sulfite oxidase
- TCEP
- Tris(2-carboxyethyl) phosphine hydrochloride
- XOR
- xanthine oxidoreductase
- XO
- xanthine oxidase
- Mo-enzyme
- Moco-containing enzyme.
REFERENCES
- 1.Schwarz G., Mendel R. R., Ribbe M. W. (2009) Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 2.Kisker C., Schindelin H., Rees D. C. (1997) Annu. Rev. Biochem. 66, 233–267 [DOI] [PubMed] [Google Scholar]
- 3.Mendel R. R., Bittner F. (2006) Biochim. Biophys. Acta 1763, 621–635 [DOI] [PubMed] [Google Scholar]
- 4.Hille R. (2005) Arch. Biochem. Biophys. 433, 107–116 [DOI] [PubMed] [Google Scholar]
- 5.Terao M., Kurosaki M., Marini M., Vanoni M. A., Saltini G., Bonetto V., Bastone A., Federico C., Saccone S., Fanelli R., Salmona M., Garattini E. (2001) J. Biol. Chem. 276, 46347–46363 [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez-Trelles F., Tarrío R., Ayala F. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13413–13417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hille R. (1996) Chem. Rev. 96, 2757–2816 [DOI] [PubMed] [Google Scholar]
- 8.Watanabe T., Ihara N., Itoh T., Fujita T., Sugimoto Y. (2000) J. Biol. Chem. 275, 21789–21792 [DOI] [PubMed] [Google Scholar]
- 9.Ichida K., Matsumura T., Sakuma R., Hosoya T., Nishino T. (2001) Biochem. Biophys. Res. Commun. 282, 1194–1200 [DOI] [PubMed] [Google Scholar]
- 10.Bittner F., Oreb M., Mendel R. R. (2001) J. Biol. Chem. 276, 40381–40384 [DOI] [PubMed] [Google Scholar]
- 11.Heidenreich T., Wollers S., Mendel R. R., Bittner F. (2005) J. Biol. Chem. 280, 4213–4218 [DOI] [PubMed] [Google Scholar]
- 12.Wollers S., Heidenreich T., Zarepour M., Zachmann D., Kraft C., Zhao Y., Mendel R. R., Bittner F. (2008) J. Biol. Chem. 283, 9642–9650 [DOI] [PubMed] [Google Scholar]
- 13.Havemeyer A., Bittner F., Wollers S., Mendel R., Kunze T., Clement B. (2006) J. Biol. Chem. 281, 34796–34802 [DOI] [PubMed] [Google Scholar]
- 14.Anantharaman V., Aravind L. (2002) FEMS Microbiol. Lett. 207, 55–61 [DOI] [PubMed] [Google Scholar]
- 15.Gruenewald S., Wahl B., Bittner F., Hungeling H., Kanzow S., Kotthaus J., Schwering U., Mendel R. R., Clement B. (2008) J. Med. Chem. 51, 8173–8177 [DOI] [PubMed] [Google Scholar]
- 16.Havemeyer A., Gruenewald S., Wahl B., Bittner F., Mendel R. R., Erdélyi P., Fischer J., Clement B. (2010) Drug Met. Disp. 38, 1917–1921 [DOI] [PubMed] [Google Scholar]
- 17.Kotthaus J., Wahl B., Havemeyer A., Kotthaus J., Schade D., Bittner F., Mendel R. R., Clement B.Biochem. J., in press [DOI] [PubMed] [Google Scholar]
- 18.Palmer T., Santini C. L., Iobbi-Nivol C., Eaves D. J., Boxer D. H., Giordano G. (1996) Mol. Microbiol. 20, 875–884 [DOI] [PubMed] [Google Scholar]
- 19.Stewart V., MacGregor C. H. (1982) J. Bacteriol. 151, 788–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koiwai H., Akaba S., Seo M., Komano T., Koshiba T. (2000) J. Biochem. 127, 659–664 [DOI] [PubMed] [Google Scholar]
- 21.Fischer K., Llamas A., Tejada-Jimenez M., Schrader N., Kuper J., Ataya F. S., Galvan A., Mendel R. R., Fernandez E., Schwarz G. (2006) J. Biol. Chem. 281, 30186–30194 [DOI] [PubMed] [Google Scholar]
- 22.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 24.Nason A., Lee K. Y., Pan S. S., Ketchum P. A., Lamberti A., DeVries J. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 3242–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson J. L., Rajagopalan K. V. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 6856–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz G., Boxer D. H., Mendel R. R. (1997) J. Biol. Chem. 272, 26811–26814 [DOI] [PubMed] [Google Scholar]
- 27.Strittmatter P., Velick S. F. (1956) J. Biol. Chem. 221, 253–264 [PubMed] [Google Scholar]
- 28.Whitby L. G. (1953) Biochem. J. 54, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihara K., Sato R. (1978) Methods Enzymol. 52, 102–108 [DOI] [PubMed] [Google Scholar]
- 30.Estabrook R. W., Werringloer J. (1978) Methods Enzymol. 52, 212–220 [DOI] [PubMed] [Google Scholar]
- 31.Stoll S., Schweiger A. (2006) J. Magn. Reson. 178, 42–55 [DOI] [PubMed] [Google Scholar]
- 32.Vergères G., Waskell L. (1992) J. Biol. Chem. 267, 12583–12591 [PubMed] [Google Scholar]
- 33.Altuve A., Silchenko S., Lee K. H., Kuczera K., Terzyan S., Zhang X., Benson D. R., Rivera M. (2001) Biochemistry 40, 9469–9483 [DOI] [PubMed] [Google Scholar]
- 34.Massey V., Edmondson D. (1970) J. Biol. Chem. 245, 6595–6598 [PubMed] [Google Scholar]
- 35.Wahl R. C., Rajagopalan K. V. (1982) J. Biol. Chem. 257, 1354–1359 [PubMed] [Google Scholar]
- 36.George G. N., Garrett R. M., Prince R. C., Rajagopalan K. V. (2004) Inorg. Chem. 43, 8456–8460 [DOI] [PubMed] [Google Scholar]
- 37.Da Cruz S., Xenarios I., Langridge J., Vilbois F., Parone P. A., Martinou J. C. (2003) J. Biol. Chem. 278, 41566–41571 [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Gladyshev V. N. (2008) J. Mol. Biol. 379, 881–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozmin S. G., Leroy P., Pavlov Y. I., Schaaper R. M. (2008) Mol. Microbiol. 68, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozmin S. G., Schaaper R. M. (2007) Mutat. Res. 619, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bates T. E., Loesch A., Burnstock G., Clark J. B. (1995) Biochem. Biophys. Res. Commun. 213, 896–900 [DOI] [PubMed] [Google Scholar]
- 42.Lamy M. T., Gutteridge S., Bary R. C. (1980) Biochem. J. 185, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutteridge S., Bray R. C., Notton B. A., Fido R. J., Hewitt E. J. (1983) Biochem. J. 213, 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]